The Overlooked Microbiome—Considering Archaea and Eukaryotes Using Multiplex Nanopore-16S-/18S-rDNA-Sequencing: A Technical Report Focusing on Nasopharyngeal Microbiomes
Abstract
1. Introduction
2. Results and Discussion
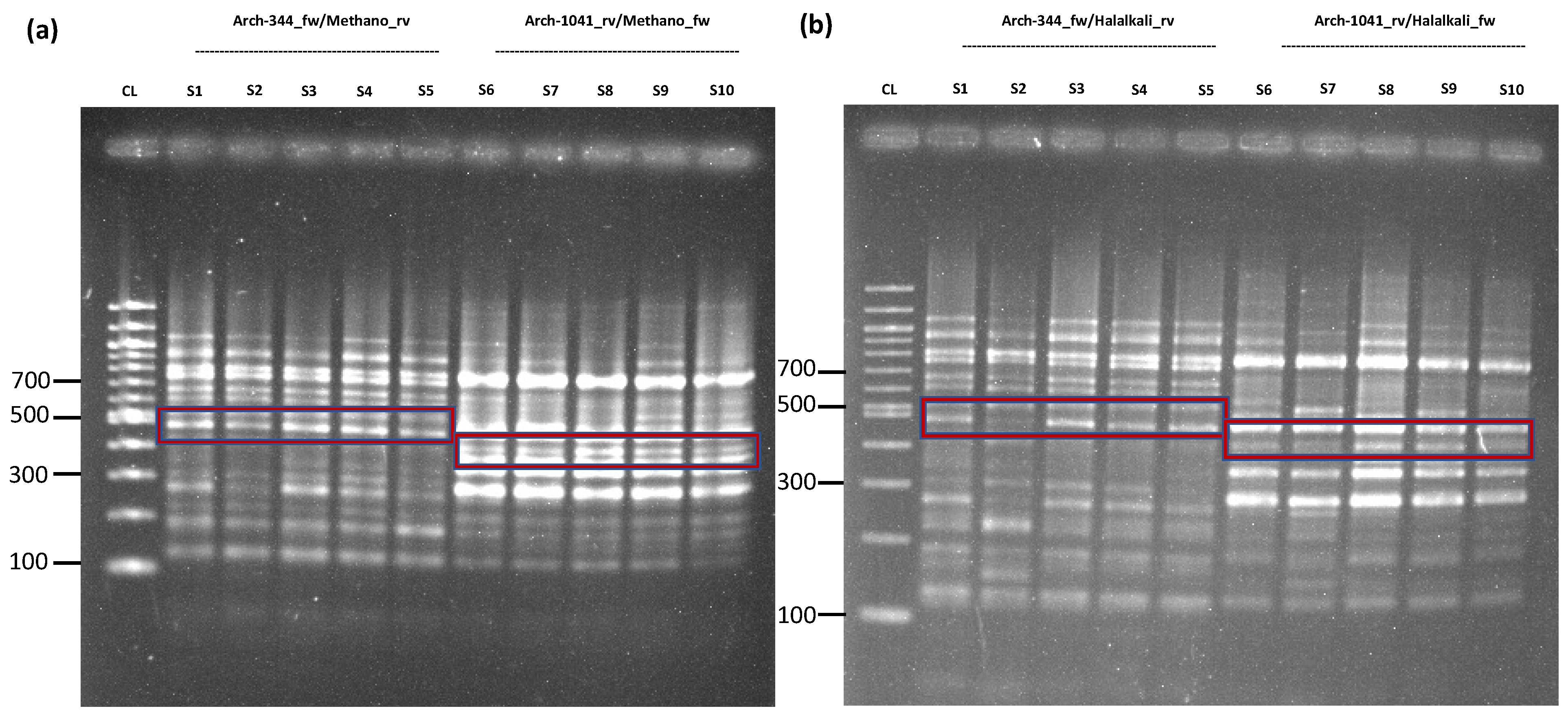
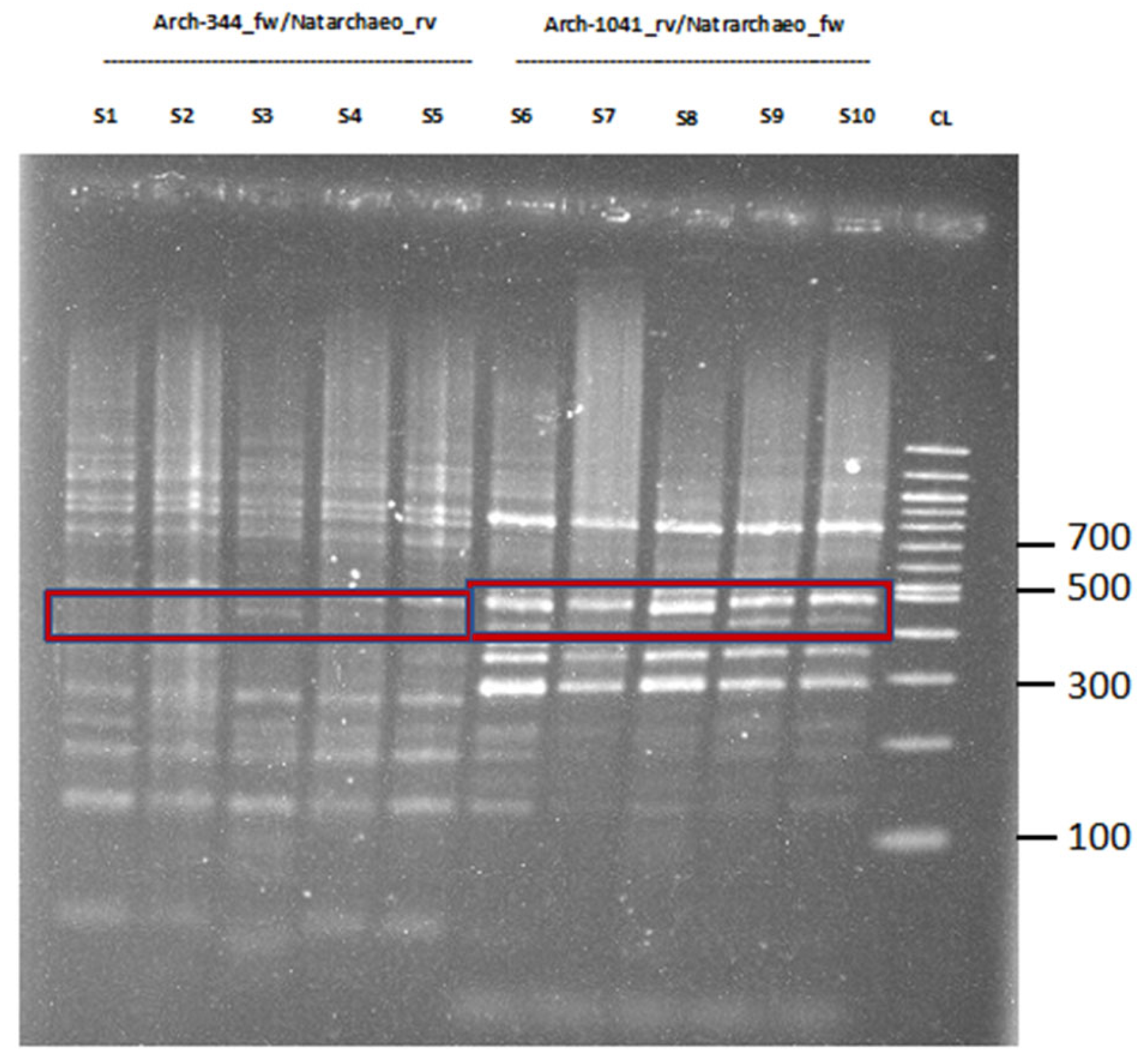
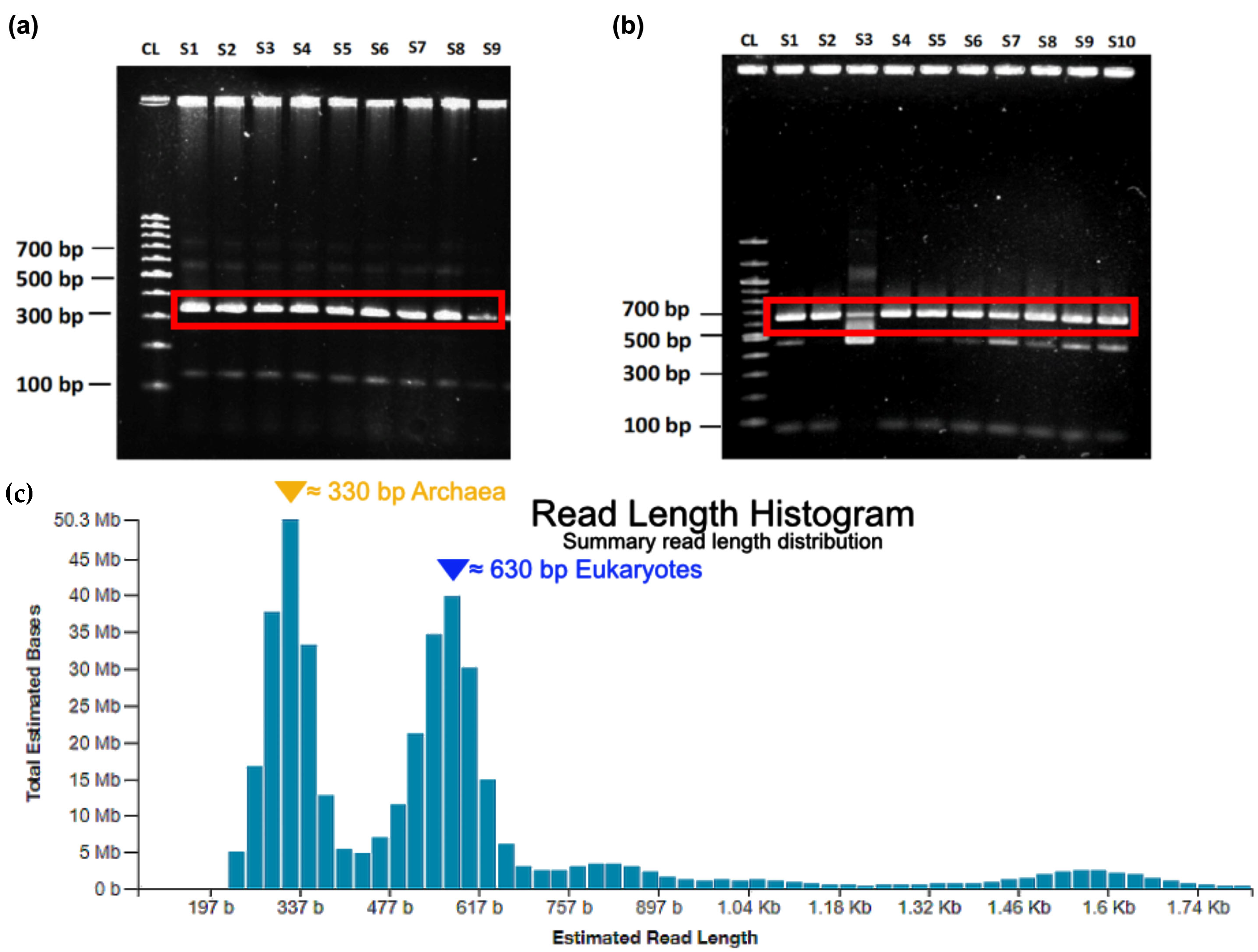
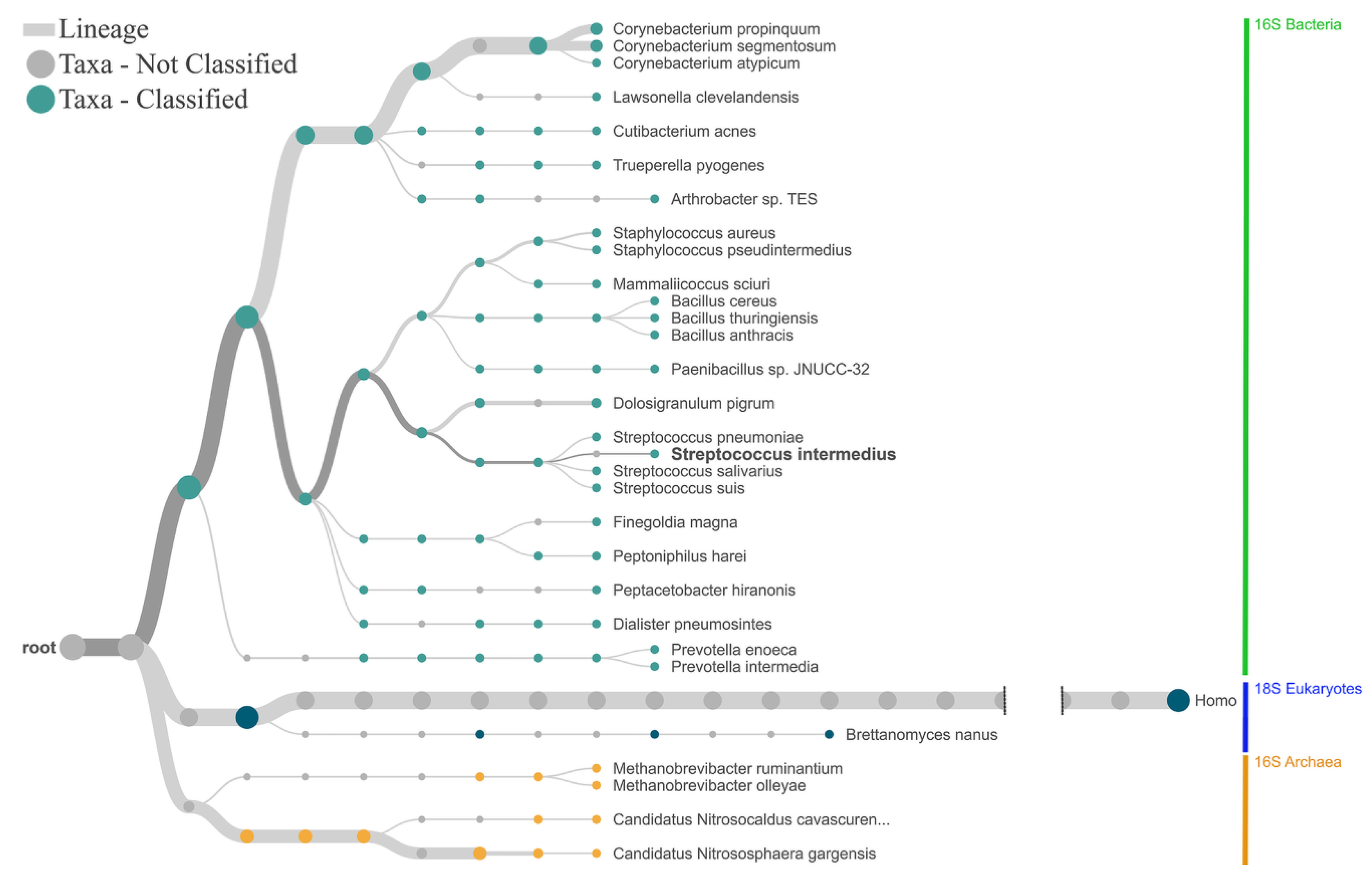
2.1. Proof of Concept and Validation
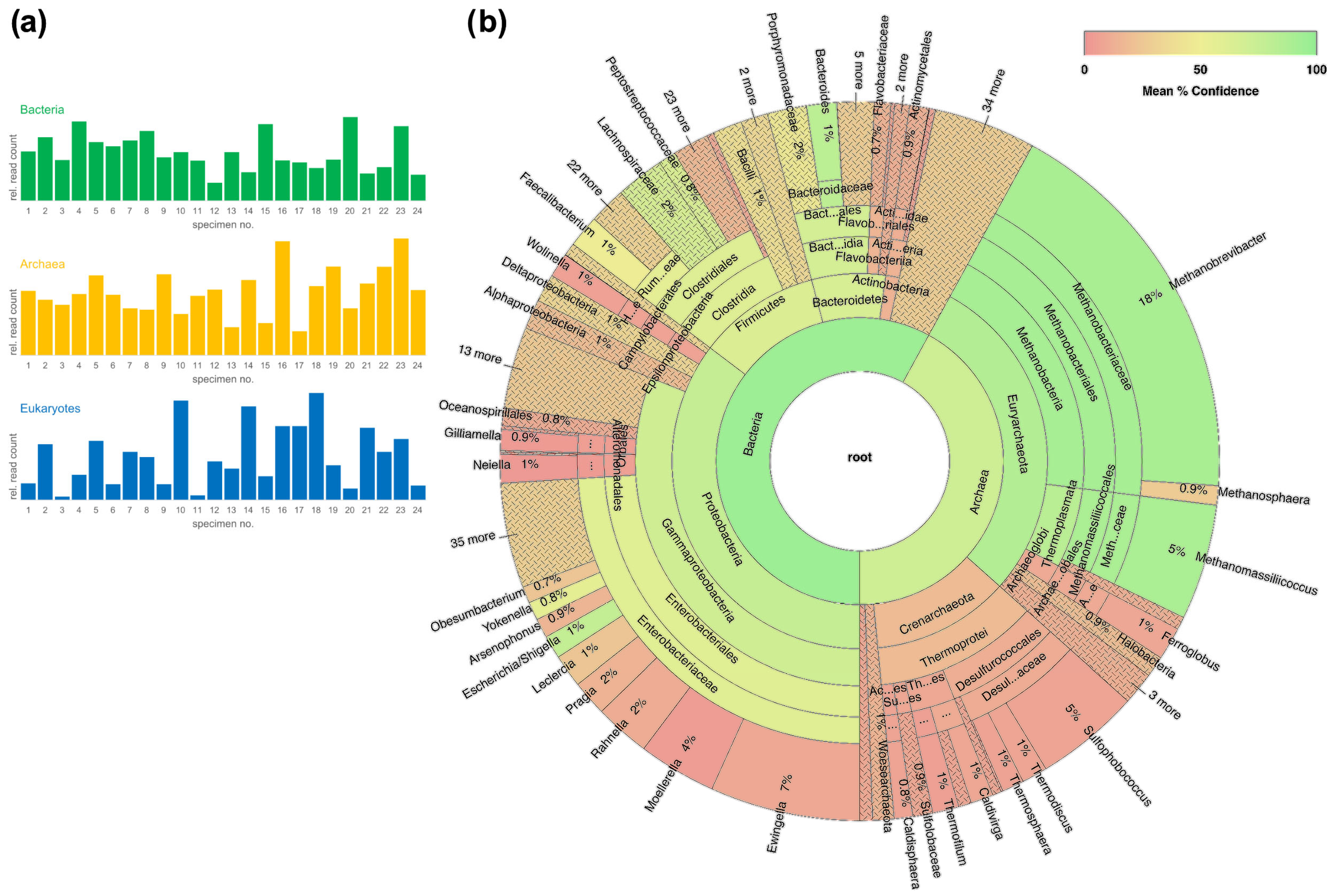
2.2. Expanded Testing of the Workflow on Fecal Specimens
2.3. Archaeal Cultures for Potential Translational Approaches
3. Material and Methods
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hu, H.; Natarajan, V.P.; Wang, F. Towards enriching and isolation of uncultivated archaea from marine sediments using a refned combination of conventional microbial cultivation methods. Mar. Life Sci. Technol. 2021, 3, 231–242. [Google Scholar] [CrossRef]
- Pausan, M.R.; Csorba, C.; Singer, G.; Till, H.; Schopf, V.; Santigli, E.; Klug, B.; Hogenauer, C.; Blohs, M.; Moissl-Eichinger, C. Exploring the Archaeome: Detection of Archaeal Signatures in the Human Body. Front Microbiol. 2019, 10, 2796. [Google Scholar] [CrossRef]
- Bang, C.; Schmitz, R.A. Archaea associated with human surfaces: Not to be underestimated. FEMS Microbiol. Rev. 2015, 39, 631–648. [Google Scholar] [CrossRef] [PubMed]
- Eckburg, P.B.; Lepp, P.W.; Relman, D.A. Archaea and their potential role in human disease. Infect. Immun. 2003, 71, 591–596. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Liu, Y.; Pan, J.; Wang, F.; Li, M. Perspectives on Cultivation Strategies of Archaea. Microb. Ecol. 2020, 79, 770–784. [Google Scholar] [CrossRef]
- Bruns, A.; Cypionka, H.; Overmann, J. Cyclic AMP and acyl homoserine lactones increase the cultivation efficiency of heterotrophic bacteria from the central Baltic Sea. Appl. Environ. Microbiol. 2002, 68, 3978–3987. [Google Scholar] [CrossRef]
- Huber, H.; Hohn, M.J.; Rachel, R.; Fuchs, T.; Wimmer, V.C.; Stetter, K.O. A new phylum of Archaea represented by a nanosized hyperthermophilic symbiont. Nature 2002, 417, 63–67. [Google Scholar] [CrossRef]
- Park, S.J.; Yoon, J.C.; Shin, K.S.; Kim, E.H.; Yim, S.; Cho, Y.J.; Sung, G.M.; Lee, D.G.; Kim, S.B.; Lee, D.U.; et al. Dominance of endospore-forming bacteria on a Rotating Activated Bacillus Contactor biofilm for advanced wastewater treatment. J. Microbiol. 2007, 45, 113–121. [Google Scholar]
- Nauhaus, K.; Albrecht, M.; Elvert, M.; Boetius, A.; Widdel, F. In vitro cell growth of marine archaeal-bacterial consortia during anaerobic oxidation of methane with sulfate. Environ. Microbiol. 2007, 9, 187–196. [Google Scholar] [CrossRef]
- Meulepas, R.J.; Jagersma, C.G.; Gieteling, J.; Buisman, C.J.; Stams, A.J.; Lens, P.N. Enrichment of anaerobic methanotrophs in sulfate-reducing membrane bioreactors. Biotechnol. Bioeng. 2009, 104, 458–470. [Google Scholar] [CrossRef]
- Bahram, M.; Anslan, S.; Hildebrand, F.; Bork, P.; Tedersoo, L. Newly designed 16S rRNA metabarcoding primers amplify diverse and novel archaeal taxa from the environment. Environ. Microbiol. Rep. 2019, 11, 487–494. [Google Scholar] [CrossRef]
- Cavicchioli, R.; Curmi, P.M.; Saunders, N.; Thomas, T. Pathogenic archaea: Do they exist? Bioessays 2003, 25, 1119–1128. [Google Scholar] [CrossRef]
- Wang, Y.; Tian, R.M.; Gao, Z.M.; Bougouffa, S.; Qian, P.Y. Optimal eukaryotic 18S and universal 16S/18S ribosomal RNA primers and their application in a study of symbiosis. PLoS ONE 2014, 9, e90053. [Google Scholar] [CrossRef]
- Cui, L.; Morris, A.; Ghedin, E. The human mycobiome in health and disease. Genome Med. 2013, 5, 63. [Google Scholar] [CrossRef]
- Hugerth, L.W.; Muller, E.E.; Hu, Y.O.; Lebrun, L.A.; Roume, H.; Lundin, D.; Wilmes, P.; Andersson, A.F. Systematic design of 18S rRNA gene primers for determining eukaryotic diversity in microbial consortia. PLoS ONE 2014, 9, e95567. [Google Scholar] [CrossRef]
- Baehren, C.; Buedding, E.; Bellm, A.; Schult, F.; Pembaur, A.; Wirth, S.; Ehrhardt, A.; Paulsen, F.; Postberg, J.; Aydin, M. The Relevance of the Bacterial Microbiome, Archaeome and Mycobiome in Pediatric Asthma and Respiratory Disorders. Cells 2022, 11, 1287. [Google Scholar] [CrossRef]
- Horz, H.P.; Conrads, G. Methanogenic Archaea and oral infections—Ways to unravel the black box. J. Oral. Microbiol. 2011, 3, 5940. [Google Scholar] [CrossRef]
- Huynh, H.T.; Pignoly, M.; Nkamga, V.D.; Drancourt, M.; Aboudharam, G. The repertoire of archaea cultivated from severe periodontitis. PLoS ONE 2015, 10, e0121565. [Google Scholar] [CrossRef]
- Moissl-Eichinger, C.; Pausan, M.; Taffner, J.; Berg, G.; Bang, C.; Schmitz, R.A. Archaea Are Interactive Components of Complex Microbiomes. Trends Microbiol. 2018, 26, 70–85. [Google Scholar] [CrossRef]
- Leigh, J.A.; Albers, S.V.; Atomi, H.; Allers, T. Model organisms for genetics in the domain Archaea: Methanogens, halophiles, Thermococcales and Sulfolobales. FEMS Microbiol. Rev. 2011, 35, 577–608. [Google Scholar] [CrossRef]
- Miller, T.L.; Wolin, M.J.; Conway de Macario, E.; Macario, A.J. Isolation of Methanobrevibacter smithii from human feces. Appl Environ. Microbiol. 1982, 43, 227–232. [Google Scholar] [CrossRef] [PubMed]
- Camara, A.; Konate, S.; Tidjani Alou, M.; Kodio, A.; Togo, A.H.; Cortaredona, S.; Henrissat, B.; Thera, M.A.; Doumbo, O.K.; Raoult, D.; et al. Clinical evidence of the role of Methanobrevibacter smithii in severe acute malnutrition. Sci. Rep. 2021, 11, 5426. [Google Scholar] [CrossRef] [PubMed]
- Hassani, Y.; Bregeon, F.; Aboudharam, G.; Drancourt, M.; Grine, G. Detection of Methanobrevobacter smithii and Methanobrevibacter oralis in Lower Respiratory Tract Microbiota. Microorganisms 2020, 8, 1866. [Google Scholar] [CrossRef] [PubMed]
- Drancourt, M.; Djemai, K.; Gouriet, F.; Grine, G.; Loukil, A.; Bedotto, M.; Levasseur, A.; Lepidi, H.; Bou-Khalil, J.; Khelaifia, S.; et al. Methanobrevibacter smithii Archaemia in Febrile Patients with Bacteremia, Including Those with Endocarditis. Clin. Infect. Dis. 2021, 73, e2571–e2579. [Google Scholar] [CrossRef] [PubMed]
- Miller, T.L.; Wolin, M.J. Methanosphaera stadtmaniae gen. nov., sp. nov.: A species that forms methane by reducing methanol with hydrogen. Arch. Microbiol. 1985, 141, 116–122. [Google Scholar] [CrossRef]
- Dridi, B.; Fardeau, M.L.; Ollivier, B.; Raoult, D.; Drancourt, M. Methanomassiliicoccus luminyensis gen. nov., sp. nov., a methanogenic archaeon isolated from human faeces. Int. J. Syst. Evol. Microbiol. 2012, 62, 1902–1907. [Google Scholar] [CrossRef]
- Annamaria Ferrari, T.B.; Rutili, A.; Canzi, E.; Biavati, B. Isolation and characterization of Methanobrevibacter oralis sp. nov. Curr. Microbiol. 1994, 29, 7–12. [Google Scholar] [CrossRef]
- Traore, S.I.; Khelaifia, S.; Armstrong, N.; Lagier, J.C.; Raoult, D. Isolation and culture of Methanobrevibacter smithii by co-culture with hydrogen-producing bacteria on agar plates. Clin. Microbiol. Infect. 2019, 25, e1561. [Google Scholar] [CrossRef]
- Khelaifia, S.; Raoult, D.; Drancourt, M. A versatile medium for cultivating methanogenic archaea. PLoS ONE 2013, 8, e61563. [Google Scholar] [CrossRef]
- Koskinen, K.; Pausan, M.R.; Perras, A.K.; Beck, M.; Bang, C.; Mora, M.; Schilhabel, A.; Schmitz, R.; Moissl-Eichinger, C. First Insights into the Diverse Human Archaeome: Specific Detection of Archaea in the Gastrointestinal Tract, Lung, and Nose and on Skin. mBio 2017, 8, e00824-17. [Google Scholar] [CrossRef]
- Huang, W.C.; Liu, Y.; Zhang, X.; Zhang, C.J.; Zou, D.; Zheng, S.; Xu, W.; Luo, Z.; Liu, F.; Li, M. Comparative genomic analysis reveals metabolic flexibility of Woesearchaeota. Nat. Commun. 2021, 12, 5281. [Google Scholar] [CrossRef]
- Dombrowski, N.; Lee, J.H.; Williams, T.A.; Offre, P.; Spang, A. Genomic diversity, lifestyles and evolutionary origins of DPANN archaea. FEMS Microbiol. Lett. 2019, 366, fnz008. [Google Scholar] [CrossRef]
- Hamm, J.N.; Erdmann, S.; Eloe-Fadrosh, E.A.; Angeloni, A.; Zhong, L.; Brownlee, C.; Williams, T.J.; Barton, K.; Carswell, S.; Smith, M.A.; et al. Unexpected host dependency of Antarctic Nanohaloarchaeota. Proc. Natl. Acad. Sci. USA 2019, 116, 14661–14670. [Google Scholar] [CrossRef]
- Jahn, U.; Gallenberger, M.; Paper, W.; Junglas, B.; Eisenreich, W.; Stetter, K.O.; Rachel, R.; Huber, H. Nanoarchaeum equitans and Ignicoccus hospitalis: New insights into a unique, intimate association of two archaea. J. Bacteriol. 2008, 190, 1743–1750. [Google Scholar] [CrossRef]
- Sakai, H.D.; Nur, N.; Kato, S.; Yuki, M.; Shimizu, M.; Itoh, T.; Ohkuma, M.; Suwanto, A.; Kurosawa, N. Insight into the symbiotic lifestyle of DPANN archaea revealed by cultivation and genome analyses. Proc. Natl. Acad. Sci. USA 2022, 119, e2115449119. [Google Scholar] [CrossRef]
- Lewis, W.H.; Tahon, G.; Geesink, P.; Sousa, D.Z.; Ettema, T.J.G. Innovations to culturing the uncultured microbial majority. Nat Rev. Microbiol. 2021, 19, 225–240. [Google Scholar] [CrossRef]
- Holmes, D.E.; Zhou, J.; Ueki, T.; Woodard, T.; Lovley, D.R. Mechanisms for Electron Uptake by Methanosarcina acetivorans during Direct Interspecies Electron Transfer. mBio 2021, 12, e0234421. [Google Scholar] [CrossRef]
- Shields, C.W.t.; Reyes, C.D.; Lopez, G.P. Microfluidic cell sorting: A review of the advances in the separation of cells from debulking to rare cell isolation. Lab Chip 2015, 15, 1230–1249. [Google Scholar] [CrossRef]
- Leung, K.; Zahn, H.; Leaver, T.; Konwar, K.M.; Hanson, N.W.; Page, A.P.; Lo, C.C.; Chain, P.S.; Hallam, S.J.; Hansen, C.L. A programmable droplet-based microfluidic device applied to multiparameter analysis of single microbes and microbial communities. Proc. Natl. Acad. Sci. USA 2012, 109, 7665–7670. [Google Scholar] [CrossRef]
- Aydin, M.; Weisser, C.; Rue, O.; Mariadassou, M.; Maass, S.; Behrendt, A.K.; Jaszczyszyn, Y.; Heilker, T.; Spaeth, M.; Vogel, S.; et al. The Rhinobiome of Exacerbated Wheezers and Asthmatics: Insights from a German Pediatric Exacerbation Network. Front. Allergy 2021, 2, 667562. [Google Scholar] [CrossRef]
| Targets | Archaea | |
|---|---|---|
| (Nested) PCR | PCR 1 | PCR 2 |
| Primer pair | Arch-344F/Arch-1041R | TOI-specific (Table S1) |
| 1st Denaturation | 95 °C 3 min | 95 °C 3 min |
| Number of cycles | 30 | 28 |
| Denaturation | 95 °C 30 s | 95 °C 3 s |
| Annealing | 55 °C 30 s | 55 °C 30 s |
| Elongation | 72 °C 30 s | 72 °C 30 s |
| Final Extension | 72 °C 5 min | 72 °C 5 min |
| Targets | Archaea | Eukaryotes | |
|---|---|---|---|
| (Nested) PCR | 1 | 2 | 1 |
| Primer pair | Arch-344F/Arch-1041R | Arch-519F/Arch-786Rtag | Euk-563F/Euk-1132Rtag |
| Amplicon size | 697 bp | 330 bp | 630 bp |
| 1st Denaturation | 95 °C 3 min | 95 °C 3 min | 95 °C 3 min |
| Number of cycles | 30 | 28 | 30 |
| Denaturation | 95 °C 30 s | 95 °C 3 s | 95 °C 30 s |
| Annealing | 55 °C 30 s | 55 °C 30 s | 55 °C 30 s |
| Elongation | 72 °C 30 s | 72 °C 30 s | 72 °C 30 s |
| Final Extension | 72 °C 5 min | 72 °C 5 min. | 72 °C 5 min. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Baehren, C.; Pembaur, A.; Weil, P.P.; Wewers, N.; Schult, F.; Wirth, S.; Postberg, J.; Aydin, M. The Overlooked Microbiome—Considering Archaea and Eukaryotes Using Multiplex Nanopore-16S-/18S-rDNA-Sequencing: A Technical Report Focusing on Nasopharyngeal Microbiomes. Int. J. Mol. Sci. 2023, 24, 1426. https://doi.org/10.3390/ijms24021426
Baehren C, Pembaur A, Weil PP, Wewers N, Schult F, Wirth S, Postberg J, Aydin M. The Overlooked Microbiome—Considering Archaea and Eukaryotes Using Multiplex Nanopore-16S-/18S-rDNA-Sequencing: A Technical Report Focusing on Nasopharyngeal Microbiomes. International Journal of Molecular Sciences. 2023; 24(2):1426. https://doi.org/10.3390/ijms24021426
Chicago/Turabian StyleBaehren, Carolin, Anton Pembaur, Patrick P. Weil, Nora Wewers, Frank Schult, Stefan Wirth, Jan Postberg, and Malik Aydin. 2023. "The Overlooked Microbiome—Considering Archaea and Eukaryotes Using Multiplex Nanopore-16S-/18S-rDNA-Sequencing: A Technical Report Focusing on Nasopharyngeal Microbiomes" International Journal of Molecular Sciences 24, no. 2: 1426. https://doi.org/10.3390/ijms24021426
APA StyleBaehren, C., Pembaur, A., Weil, P. P., Wewers, N., Schult, F., Wirth, S., Postberg, J., & Aydin, M. (2023). The Overlooked Microbiome—Considering Archaea and Eukaryotes Using Multiplex Nanopore-16S-/18S-rDNA-Sequencing: A Technical Report Focusing on Nasopharyngeal Microbiomes. International Journal of Molecular Sciences, 24(2), 1426. https://doi.org/10.3390/ijms24021426






