Ovarian Cancer: A Landscape of Mitochondria with Emphasis on Mitochondrial Dynamics
Abstract
1. Introduction
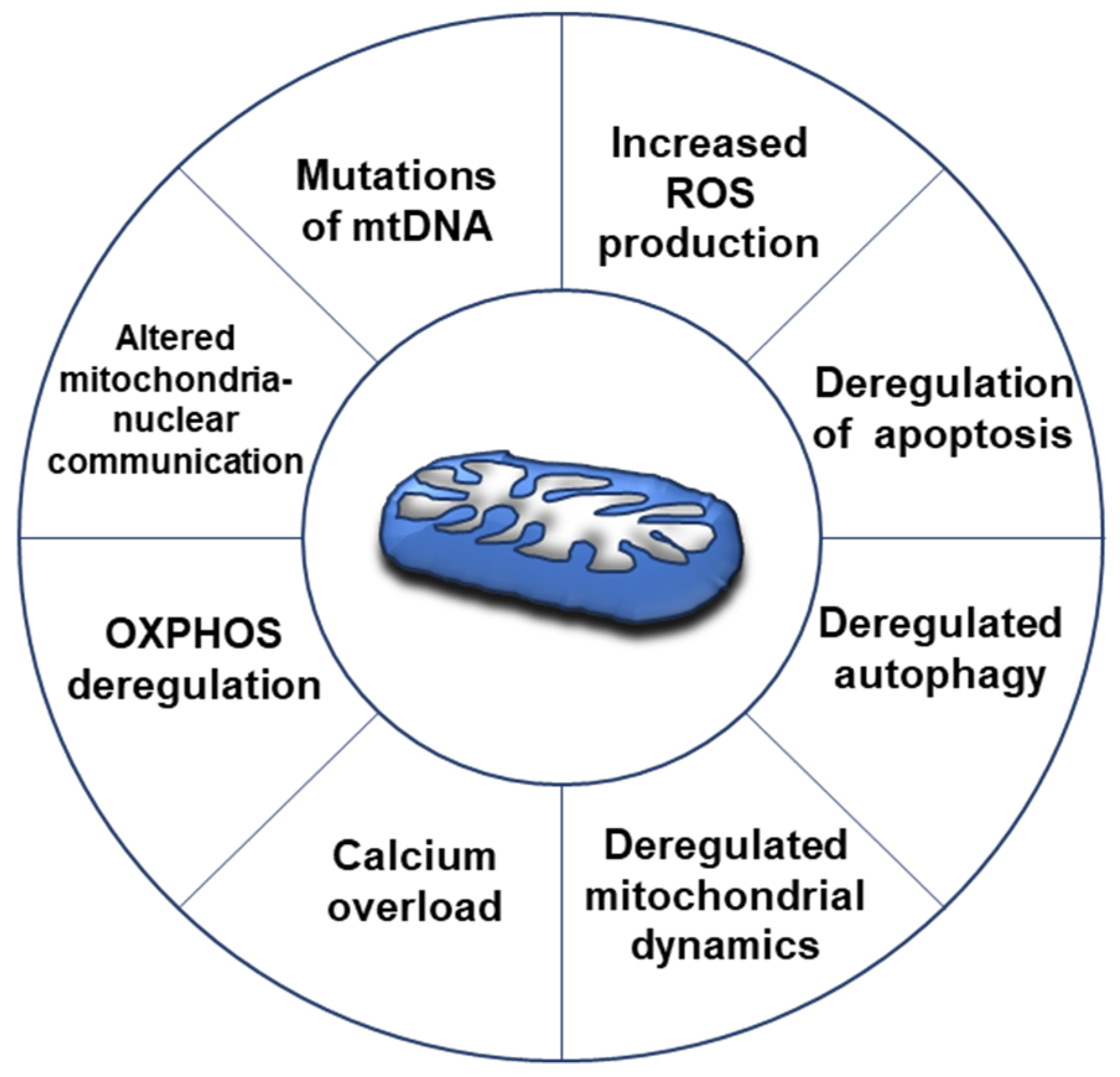
2. Mitochondrial Overview in Cancer
3. Mitochondrial Alterations in Ovarian Cancer
4. Mitochondrial Dynamics in Ovarian Cancer
4.1. Proteins Involved in Mitochondrial Dynamic Machinery
4.2. Alteration of Mitochondrial Dynamics in Ovarian Cancer
4.3. Mitochondrial Morphology and Chemoresistance in Ovarian Cancer
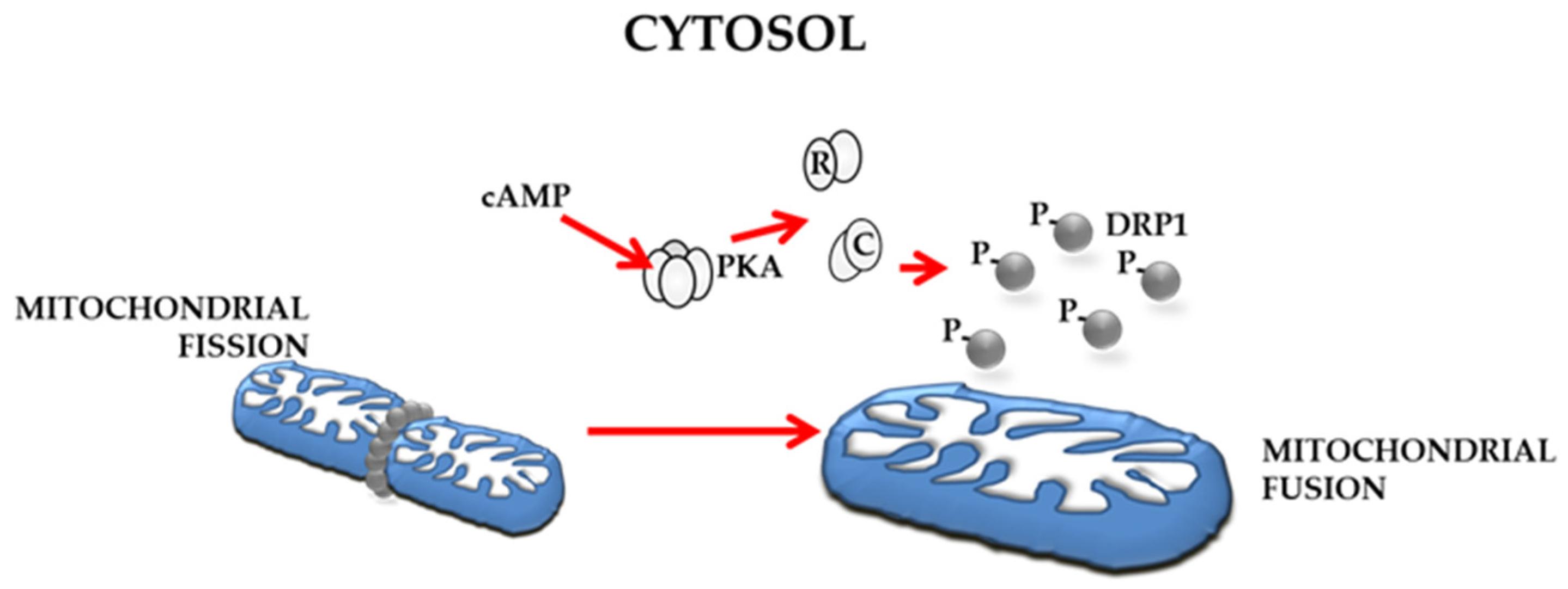
4.4. Underscoring the Possible Importance of cAMP/PKA Signalling in Regulation of Mitochondrial Dynamics in OC
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Testa, U.; Petrucci, E.; Pasquini, L.; Castelli, G.; Pelosi, E. Ovarian Cancers: Genetic Abnormalities, Tumor Heterogeneity and Progression, Clonal Evolution and Cancer Stem Cells. Medicines 2018, 5, 16. [Google Scholar] [CrossRef] [PubMed]
- Kamal, R.; Hamed, S.; Mansour, S.; Mounir, Y.; Abdel Sallam, S. Ovarian Cancer Screening-Ultrasound; Impact on Ovarian Cancer Mortality. Br. J. Radiol. 2018, 91, 20170571. [Google Scholar] [CrossRef] [PubMed]
- De Leo, A.; Santini, D.; Ceccarelli, C.; Santandrea, G.; Palicelli, A.; Acquaviva, G.; Chiarucci, F.; Rosini, F.; Ravegnini, G.; Pession, A.; et al. What Is New on Ovarian Carcinoma: Integrated Morphologic and Molecular Analysis Following the New 2020 World Health Organization Classification of Female Genital Tumors. Diagnostics 2021, 11, 697. [Google Scholar] [CrossRef] [PubMed]
- Vang, R.; Shih, I.-M.; Kurman, R.J. Ovarian Low-Grade and High-Grade Serous Carcinoma: Pathogenesis, Clinicopathologic and Molecular Biologic Features, and Diagnostic Problems. Adv. Anat. Pathol. 2009, 16, 267–282. [Google Scholar] [CrossRef]
- Goundiam, O.; Gestraud, P.; Popova, T.; De la Motte Rouge, T.; Fourchotte, V.; Gentien, D.; Hupé, P.; Becette, V.; Houdayer, C.; Roman-Roman, S.; et al. Histo-Genomic Stratification Reveals the Frequent Amplification/Overexpression of CCNE1 and BRD4 Genes in Non-BRCAness High Grade Ovarian Carcinoma. Int. J. Cancer 2015, 137, 1890–1900. [Google Scholar] [CrossRef]
- Bernards, S.S.; Pennington, K.P.; Harrell, M.I.; Agnew, K.J.; Garcia, R.L.; Norquist, B.M.; Swisher, E.M. Clinical Characteristics and Outcomes of Patients with BRCA1 or RAD51C Methylated versus Mutated Ovarian Carcinoma. Gynecol. Oncol. 2018, 148, 281–285. [Google Scholar] [CrossRef]
- Ebata, T.; Yamashita, S.; Takeshima, H.; Yoshida, H.; Kawata, Y.; Kino, N.; Yasugi, T.; Terao, Y.; Yonemori, K.; Kato, T.; et al. DNA Methylation of the Immediate Upstream Region of BRCA1 Major Transcription Start Sites Is an Independent Favorable Prognostic Factor in Patients with High-Grade Serous Ovarian Cancer. Gynecol. Oncol. 2022, 167, 513–518. [Google Scholar] [CrossRef]
- Kim, S.I.; Lee, M.; Kim, H.S.; Chung, H.H.; Kim, J.-W.; Park, N.H.; Song, Y.-S. Effect of BRCA Mutational Status on Survival Outcome in Advanced-Stage High-Grade Serous Ovarian Cancer. J. Ovarian Res. 2019, 12, 40. [Google Scholar] [CrossRef]
- Konecny, G.E.; Winterhoff, B.; Wang, C. Gene-Expression Signatures in Ovarian Cancer: Promise and Challenges for Patient Stratification. Gynecol. Oncol. 2016, 141, 379–385. [Google Scholar] [CrossRef]
- Rattanapan, Y.; Korkiatsakul, V.; Kongruang, A.; Siriboonpiputtana, T.; Rerkamnuaychoke, B.; Chareonsirisuthigul, T. MicroRNA Expression Profiling of Epithelial Ovarian Cancer Identifies New Markers of Tumor Subtype. Microrna 2020, 9, 289–294. [Google Scholar] [CrossRef]
- Berkel, C.; Cacan, E. Transcriptomic Analysis Reveals Tumor Stage- or Grade-Dependent Expression of MiRNAs in Serous Ovarian Cancer. Hum. Cell 2021, 34, 862–877. [Google Scholar] [CrossRef] [PubMed]
- Vaupel, P.; Schmidberger, H.; Mayer, A. The Warburg Effect: Essential Part of Metabolic Reprogramming and Central Contributor to Cancer Progression. Int. J. Radiat. Biol. 2019, 95, 912–919. [Google Scholar] [CrossRef] [PubMed]
- Zong, W.-X.; Rabinowitz, J.D.; White, E. Mitochondria and Cancer. Mol. Cell 2016, 61, 667–676. [Google Scholar] [CrossRef] [PubMed]
- Burke, P.J. Mitochondria, Bioenergetics and Apoptosis in Cancer. Trends Cancer 2017, 3, 857–870. [Google Scholar] [CrossRef]
- Jelic, M.D.; Mandic, A.D.; Maricic, S.M.; Srdjenovic, B.U. Oxidative Stress and Its Role in Cancer. J. Cancer Res. Ther. 2021, 17, 22–28. [Google Scholar] [CrossRef]
- DeBerardinis, R.J.; Chandel, N.S. Fundamentals of Cancer Metabolism. Sci. Adv. 2016, 2, e1600200. [Google Scholar] [CrossRef]
- Strobbe, D.; Sharma, S.; Campanella, M. Links between Mitochondrial Retrograde Response and Mitophagy in Pathogenic Cell Signalling. Cell. Mol. Life Sci. 2021, 78, 3767–3775. [Google Scholar] [CrossRef]
- Li, N.; Li, H.; Wang, Y.; Cao, L.; Zhan, X. Quantitative Proteomics Revealed Energy Metabolism Pathway Alterations in Human Epithelial Ovarian Carcinoma and Their Regulation by the Antiparasite Drug Ivermectin: Data Interpretation in the Context of 3P Medicine. EPMA J. 2020, 11, 661–694. [Google Scholar] [CrossRef]
- Li, N.; Zhan, X. Signaling Pathway Network Alterations in Human Ovarian Cancers Identified with Quantitative Mitochondrial Proteomics. EPMA J. 2019, 10, 153–172. [Google Scholar] [CrossRef]
- Shukla, P.; Singh, K.K. The Mitochondrial Landscape of Ovarian Cancer: Emerging Insights. Carcinogenesis 2021, 42, 663–671. [Google Scholar] [CrossRef]
- Signorile, A.; De Rasmo, D.; Cormio, A.; Musicco, C.; Rossi, R.; Fortarezza, F.; Palese, L.L.; Loizzi, V.; Resta, L.; Scillitani, G.; et al. Human Ovarian Cancer Tissue Exhibits Increase of Mitochondrial Biogenesis and Cristae Remodeling. Cancers 2019, 11, 1350. [Google Scholar] [CrossRef]
- Hecker, D. Enzyme histochemical and electron microscopic studies on the problem of infiltrating (invasive) tumor growth. 2. Electron microscopic studies. Gegenbaurs Morphol. Jahrb. 1977, 123, 51–64. [Google Scholar]
- Dier, U.; Shin, D.-H.; Hemachandra, L.P.M.P.; Uusitalo, L.M.; Hempel, N. Bioenergetic Analysis of Ovarian Cancer Cell Lines: Profiling of Histological Subtypes and Identification of a Mitochondria-Defective Cell Line. PLoS ONE 2014, 9, e98479. [Google Scholar] [CrossRef]
- Kobayashi, H. Recent Advances in Understanding the Metabolic Plasticity of Ovarian Cancer: A Systematic Review. Heliyon 2022, 8, e11487. [Google Scholar] [CrossRef]
- Shen, L.; Zhan, X. Mitochondrial Dysfunction Pathway Alterations Offer Potential Biomarkers and Therapeutic Targets for Ovarian Cancer. Oxidative Med. Cell. Longev. 2022, 2022, 5634724. [Google Scholar] [CrossRef]
- Pernas, L.; Scorrano, L. Mito-Morphosis: Mitochondrial Fusion, Fission, and Cristae Remodeling as Key Mediators of Cellular Function. Annu. Rev. Physiol. 2016, 78, 505–531. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, T.; Ferraz, L.S. Therapeutic Potential of Targeting Mitochondrial Dynamics in Cancer. Biochem. Pharmacol. 2020, 182, 114282. [Google Scholar] [CrossRef]
- Kong, B.; Tsuyoshi, H.; Orisaka, M.; Shieh, D.-B.; Yoshida, Y.; Tsang, B.K. Mitochondrial Dynamics Regulating Chemoresistance in Gynecological Cancers. Ann. N. Y. Acad. Sci. 2015, 1350, 1–16. [Google Scholar] [CrossRef]
- Ul Fatima, N.; Ananthanarayanan, V. Mitochondrial Movers and Shapers: Recent Insights into Regulators of Fission, Fusion and Transport. Curr. Opin. Cell Biol. 2022, 80, 102150. [Google Scholar] [CrossRef] [PubMed]
- Papa, S.; Martino, P.L.; Capitanio, G.; Gaballo, A.; De Rasmo, D.; Signorile, A.; Petruzzella, V. The Oxidative Phosphorylation System in Mammalian Mitochondria. Adv. Exp. Med. Biol. 2012, 942, 3–37. [Google Scholar] [CrossRef] [PubMed]
- Raimondi, V.; Ciccarese, F.; Ciminale, V. Oncogenic Pathways and the Electron Transport Chain: A DangeROS Liaison. Br. J. Cancer 2020, 122, 168–181. [Google Scholar] [CrossRef] [PubMed]
- Brand, M.D. Mitochondrial Generation of Superoxide and Hydrogen Peroxide as the Source of Mitochondrial Redox Signaling. Free Radic. Biol. Med. 2016, 100, 14–31. [Google Scholar] [CrossRef] [PubMed]
- Jackson, M.J.; Papa, S.; Bolaños, J.; Bruckdorfer, R.; Carlsen, H.; Elliott, R.M.; Flier, J.; Griffiths, H.R.; Heales, S.; Holst, B.; et al. Antioxidants, Reactive Oxygen and Nitrogen Species, Gene Induction and Mitochondrial Function. Mol. Aspects Med. 2002, 23, 209–285. [Google Scholar] [CrossRef] [PubMed]
- Rekatsina, M.; Paladini, A.; Piroli, A.; Zis, P.; Pergolizzi, J.V.; Varrassi, G. Correction to: Pathophysiologic Approach to Pain Therapy for Complex Pain Entities: A Narrative Review. Pain Ther. 2020, 9, 23. [Google Scholar] [CrossRef]
- Peoples, J.N.; Saraf, A.; Ghazal, N.; Pham, T.T.; Kwong, J.Q. Mitochondrial Dysfunction and Oxidative Stress in Heart Disease. Exp. Mol. Med. 2019, 51, 1–13. [Google Scholar] [CrossRef]
- Tuli, H.S.; Kaur, J.; Vashishth, K.; Sak, K.; Sharma, U.; Choudhary, R.; Behl, T.; Singh, T.; Sharma, S.; Saini, A.K.; et al. Molecular Mechanisms behind ROS Regulation in Cancer: A Balancing Act between Augmented Tumorigenesis and Cell Apoptosis. Arch. Toxicol. 2022, 97, 103–120. [Google Scholar] [CrossRef]
- Patel, P.S.; Castelow, C.; Patel, D.S.; Bhattacharya, S.K.; Kuscu, C.; Kuscu, C.; Makowski, L.; Eason, J.D.; Bajwa, A. Mitochondrial Role in Oncogenesis and Potential Chemotherapeutic Strategy of Mitochondrial Infusion in Breast Cancer. Int. J. Mol. Sci. 2022, 23, 12993. [Google Scholar] [CrossRef]
- Xia, M.; Zhang, Y.; Jin, K.; Lu, Z.; Zeng, Z.; Xiong, W. Communication between Mitochondria and Other Organelles: A Brand-New Perspective on Mitochondria in Cancer. Cell Biosci. 2019, 9, 27. [Google Scholar] [CrossRef]
- Ryu, J.; Thomas, S.N. Quantitative Mass Spectrometry-Based Proteomics for Biomarker Development in Ovarian Cancer. Molecules 2021, 26, 2674. [Google Scholar] [CrossRef]
- Yang, D.; Kim, J. Mitochondrial Retrograde Signalling and Metabolic Alterations in the Tumour Microenvironment. Cells 2019, 8, 275. [Google Scholar] [CrossRef]
- Ma, Y.; Wang, L.; Jia, R. The Role of Mitochondrial Dynamics in Human Cancers. Am. J. Cancer Res. 2020, 10, 1278–1293. [Google Scholar]
- Kumar, S.; Ashraf, R.; Aparna, C.K. Mitochondrial Dynamics Regulators: Implications for Therapeutic Intervention in Cancer. Cell Biol. Toxicol. 2022, 38, 377–406. [Google Scholar] [CrossRef]
- Jia, D.; Park, J.H.; Jung, K.H.; Levine, H.; Kaipparettu, B.A. Elucidating the Metabolic Plasticity of Cancer: Mitochondrial Reprogramming and Hybrid Metabolic States. Cells 2018, 7, 21. [Google Scholar] [CrossRef] [PubMed]
- Kuo, C.-L.; Ponneri Babuharisankar, A.; Lin, Y.-C.; Lien, H.-W.; Lo, Y.K.; Chou, H.-Y.; Tangeda, V.; Cheng, L.-C.; Cheng, A.N.; Lee, A.Y.-L. Mitochondrial Oxidative Stress in the Tumor Microenvironment and Cancer Immunoescape: Foe or Friend? J. Biomed. Sci. 2022, 29, 74. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, L.; Supuran, C.T.; Alfarouk, K.O. The Warburg Effect and the Hallmarks of Cancer. Anticancer Agents Med. Chem. 2017, 17, 164–170. [Google Scholar] [CrossRef]
- Di Gregorio, J.; Petricca, S.; Iorio, R.; Toniato, E.; Flati, V. Mitochondrial and Metabolic Alterations in Cancer Cells. Eur. J. Cell Biol. 2022, 101, 151225. [Google Scholar] [CrossRef] [PubMed]
- Gentric, G.; Mieulet, V.; Mechta-Grigoriou, F. Heterogeneity in Cancer Metabolism: New Concepts in an Old Field. Antioxid. Redox Signal. 2017, 26, 462–485. [Google Scholar] [CrossRef] [PubMed]
- Green, D.R. The Mitochondrial Pathway of Apoptosis Part II: The BCL-2 Protein Family. Cold Spring Harb. Perspect. Biol. 2022, 14, a041046. [Google Scholar] [CrossRef]
- Romani, A.M.P. Cisplatin in Cancer Treatment. Biochem. Pharmacol. 2022, 206, 115323. [Google Scholar] [CrossRef]
- Kaloni, D.; Diepstraten, S.T.; Strasser, A.; Kelly, G.L. BCL-2 Protein Family: Attractive Targets for Cancer Therapy. Apoptosis 2022. [Google Scholar] [CrossRef]
- Favaloro, B.; Allocati, N.; Graziano, V.; Di Ilio, C.; De Laurenzi, V. Role of Apoptosis in Disease. Aging 2012, 4, 330–349. [Google Scholar] [CrossRef]
- Li, N.; Zhan, X. Mass spectrometry-based mitochondrial proteomics in human ovarian cancers. Mass. Spectrom. Rev. 2020, 39, 471–498. [Google Scholar] [CrossRef] [PubMed]
- Liu, V.W.; Shi, H.H.; Cheung, A.N.; Chiu, P.M.; Leung, T.W.; Nagley, P.; Wong, L.C.; Ngan, H.Y. High Incidence of Somatic Mitochondrial DNA Mutations in Human Ovarian Carcinomas. Cancer Res. 2001, 61, 5998–6001. [Google Scholar]
- Wu, Y.; Zhang, X.; Wang, Z.; Zheng, W.; Cao, H.; Shen, W. Targeting Oxidative Phosphorylation as an Approach for the Treatment of Ovarian Cancer. Front. Oncol. 2022, 12, 971479. [Google Scholar] [CrossRef] [PubMed]
- Ni, J.; Wang, Y.; Cheng, X.; Teng, F.; Wang, C.; Han, S.; Chen, X.; Guo, W. Pathogenic Heteroplasmic Somatic Mitochondrial DNA Mutation Confers Platinum-Resistance and Recurrence of High-Grade Serous Ovarian Cancer. Cancer Manag. Res. 2020, 12, 11085–11093. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, V.W.S.; Xue, W.C.; Cheung, A.N.Y.; Ngan, H.Y.S. Association of Decreased Mitochondrial DNA Content with Ovarian Cancer Progression. Br. J. Cancer 2006, 95, 1087–1091. [Google Scholar] [CrossRef]
- Nayak, A.P.; Kapur, A.; Barroilhet, L.; Patankar, M.S. Oxidative Phosphorylation: A Target for Novel Therapeutic Strategies Against Ovarian Cancer. Cancers 2018, 10, 337. [Google Scholar] [CrossRef]
- Tondo-Steele, K.; McLean, K. The “Sweet Spot” of Targeting Tumor Metabolism in Ovarian Cancers. Cancers 2022, 14, 4696. [Google Scholar] [CrossRef]
- De Luise, M.; Sollazzo, M.; Lama, E.; Coadă, C.A.; Bressi, L.; Iorio, M.; Cavina, B.; D’Angelo, L.; Milioni, S.; Marchio, L.; et al. Inducing Respiratory Complex I Impairment Elicits an Increase in PGC1α in Ovarian Cancer. Sci. Rep. 2022, 12, 8020. [Google Scholar] [CrossRef] [PubMed]
- Gentric, G.; Kieffer, Y.; Mieulet, V.; Goundiam, O.; Bonneau, C.; Nemati, F.; Hurbain, I.; Raposo, G.; Popova, T.; Stern, M.-H.; et al. PML-Regulated Mitochondrial Metabolism Enhances Chemosensitivity in Human Ovarian Cancers. Cell Metab. 2019, 29, 156–173.e10. [Google Scholar] [CrossRef]
- Matassa, D.S.; Criscuolo, D.; Avolio, R.; Agliarulo, I.; Sarnataro, D.; Pacelli, C.; Scrima, R.; Colamatteo, A.; Matarese, G.; Capitanio, N.; et al. Regulation of Mitochondrial Complex III Activity and Assembly by TRAP1 in Cancer Cells. Cancer Cell Int. 2022, 22, 402. [Google Scholar] [CrossRef] [PubMed]
- Kang, I.; Chu, C.T.; Kaufman, B.A. The Mitochondrial Transcription Factor TFAM in Neurodegeneration: Emerging Evidence and Mechanisms. FEBS Lett. 2018, 592, 793–811. [Google Scholar] [CrossRef] [PubMed]
- Signorile, A.; Sgaramella, G.; Bellomo, F.; De Rasmo, D. Prohibitins: A Critical Role in Mitochondrial Functions and Implication in Diseases. Cells 2019, 8, 71. [Google Scholar] [CrossRef] [PubMed]
- Ande, S.R.; Nguyen, K.H.; Padilla-Meier, G.P.; Wahida, W.; Nyomba, B.L.G.; Mishra, S. Prohibitin Overexpression in Adipocytes Induces Mitochondrial Biogenesis, Leads to Obesity Development, and Affects Glucose Homeostasis in a Sex-Specific Manner. Diabetes 2014, 63, 3734–3741. [Google Scholar] [CrossRef] [PubMed]
- Oyang, L.; Li, J.; Jiang, X.; Lin, J.; Xia, L.; Yang, L.; Tan, S.; Wu, N.; Han, Y.; Yang, Y.; et al. The Function of Prohibitins in Mitochondria and the Clinical Potentials. Cancer Cell Int. 2022, 22, 343. [Google Scholar] [CrossRef]
- Papa, S.; Scacco, S.; De Rasmo, D.; Signorile, A.; Papa, F.; Panelli, D.; Nicastro, A.; Scaringi, R.; Santeramo, A.; Roca, E.; et al. CAMP-Dependent Protein Kinase Regulates Post-Translational Processing and Expression of Complex I Subunits in Mammalian Cells. Biochim. Biophys. Acta 2010, 1797, 649–658. [Google Scholar] [CrossRef]
- Kingnate, C.; Charoenkwan, K.; Kumfu, S.; Chattipakorn, N.; Chattipakorn, S.C. Possible Roles of Mitochondrial Dynamics and the Effects of Pharmacological Interventions in Chemoresistant Ovarian Cancer. EBioMedicine 2018, 34, 256–266. [Google Scholar] [CrossRef]
- Frattaruolo, L.; Brindisi, M.; Curcio, R.; Marra, F.; Dolce, V.; Cappello, A.R. Targeting the Mitochondrial Metabolic Network: A Promising Strategy in Cancer Treatment. Int. J. Mol. Sci. 2020, 21, 6014. [Google Scholar] [CrossRef]
- Signorile, A.; Santeramo, A.; Tamma, G.; Pellegrino, T.; D’Oria, S.; Lattanzio, P.; De Rasmo, D. Mitochondrial CAMP Prevents Apoptosis Modulating Sirt3 Protein Level and OPA1 Processing in Cardiac Myoblast Cells. Biochim. Biophys. Acta BBA Mol. Cell Res. 2017, 1864, 355–366. [Google Scholar] [CrossRef]
- Giacomello, M.; Pyakurel, A.; Glytsou, C.; Scorrano, L. The Cell Biology of Mitochondrial Membrane Dynamics. Nat. Rev. Mol. Cell Biol. 2020, 21, 204–224. [Google Scholar] [CrossRef]
- Zhao, S.; Heng, N.; Wang, H.; Wang, H.; Zhang, H.; Gong, J.; Hu, Z.; Zhu, H. Mitofusins: From Mitochondria to Fertility. Cell. Mol. Life. Sci. 2022, 79, 370. [Google Scholar] [CrossRef]
- Detmer, S.A.; Chan, D.C. Complementation between Mouse Mfn1 and Mfn2 Protects Mitochondrial Fusion Defects Caused by CMT2A Disease Mutations. J. Cell Biol. 2007, 176, 405–414. [Google Scholar] [CrossRef] [PubMed]
- Guillery, O.; Malka, F.; Landes, T.; Guillou, E.; Blackstone, C.; Lombès, A.; Belenguer, P.; Arnoult, D.; Rojo, M. Metalloprotease-Mediated OPA1 Processing Is Modulated by the Mitochondrial Membrane Potential. Biol. Cell 2008, 100, 315–325. [Google Scholar] [CrossRef] [PubMed]
- Olichon, A.; Baricault, L.; Gas, N.; Guillou, E.; Valette, A.; Belenguer, P.; Lenaers, G. Loss of OPA1 Perturbates the Mitochondrial Inner Membrane Structure and Integrity, Leading to Cytochrome c Release and Apoptosis. J. Biol. Chem. 2003, 278, 7743–7746. [Google Scholar] [CrossRef] [PubMed]
- Del Dotto, V.; Fogazza, M.; Carelli, V.; Rugolo, M.; Zanna, C. Eight Human OPA1 Isoforms, Long and Short: What Are They For? Biochim. Biophys. Acta BBA Bioenerg. 2018, 1859, 263–269. [Google Scholar] [CrossRef]
- Quintana-Cabrera, R.; Manjarrés-Raza, I.; Vicente-Gutiérrez, C.; Corrado, M.; Bolaños, J.P.; Scorrano, L. Opa1 Relies on Cristae Preservation and ATP Synthase to Curtail Reactive Oxygen Species Accumulation in Mitochondria. Redox Biol. 2021, 41, 101944. [Google Scholar] [CrossRef]
- Olichon, A.; Elachouri, G.; Baricault, L.; Delettre, C.; Belenguer, P.; Lenaers, G. OPA1 Alternate Splicing Uncouples an Evolutionary Conserved Function in Mitochondrial Fusion from a Vertebrate Restricted Function in Apoptosis. Cell Death Differ. 2007, 14, 682–692. [Google Scholar] [CrossRef]
- Ishihara, N.; Fujita, Y.; Oka, T.; Mihara, K. Regulation of Mitochondrial Morphology through Proteolytic Cleavage of OPA1. EMBO J. 2006, 25, 2966–2977. [Google Scholar] [CrossRef]
- Cogliati, S.; Frezza, C.; Soriano, M.E.; Varanita, T.; Quintana-Cabrera, R.; Corrado, M.; Cipolat, S.; Costa, V.; Casarin, A.; Gomes, L.C.; et al. Mitochondrial Cristae Shape Determines Respiratory Chain Supercomplexes Assembly and Respiratory Efficiency. Cell 2013, 155, 160–171. [Google Scholar] [CrossRef]
- Anand, R.; Wai, T.; Baker, M.J.; Kladt, N.; Schauss, A.C.; Rugarli, E.; Langer, T. The I-AAA Protease YME1L and OMA1 Cleave OPA1 to Balance Mitochondrial Fusion and Fission. J. Cell Biol. 2014, 204, 919–929. [Google Scholar] [CrossRef]
- Cipolat, S.; Rudka, T.; Hartmann, D.; Costa, V.; Serneels, L.; Craessaerts, K.; Metzger, K.; Frezza, C.; Annaert, W.; D’Adamio, L.; et al. Mitochondrial Rhomboid PARL Regulates Cytochrome c Release during Apoptosis via OPA1-Dependent Cristae Remodeling. Cell 2006, 126, 163–175. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Shangguan, X.; Zhou, W.; Cao, Y.; Zheng, Q.; Tu, J.; Hu, G.; Liang, Z.; Jiang, C.; Deng, L.; et al. Glucose Limitation Activates AMPK Coupled SENP1-Sirt3 Signalling in Mitochondria for T Cell Memory Development. Nat. Commun. 2021, 12, 4371. [Google Scholar] [CrossRef]
- MacVicar, T.; Langer, T. OPA1 Processing in Cell Death and Disease—The Long and Short of It. J. Cell Sci. 2016, 129, 2297–2306. [Google Scholar] [CrossRef]
- Navaratnarajah, T.; Anand, R.; Reichert, A.S.; Distelmaier, F. The Relevance of Mitochondrial Morphology for Human Disease. Int. J. Biochem. Cell Biol. 2021, 134, 105951. [Google Scholar] [CrossRef] [PubMed]
- Viana, M.P.; Levytskyy, R.M.; Anand, R.; Reichert, A.S.; Khalimonchuk, O. Protease OMA1 Modulates Mitochondrial Bioenergetics and Ultrastructure through Dynamic Association with MICOS Complex. iScience 2021, 24, 102119. [Google Scholar] [CrossRef] [PubMed]
- Pellegrini, L.; Scorrano, L. A Cut Short to Death: Parl and Opa1 in the Regulation of Mitochondrial Morphology and Apoptosis. Cell Death Differ. 2007, 14, 1275–1284. [Google Scholar] [CrossRef] [PubMed]
- Wai, T.; Saita, S.; Nolte, H.; Müller, S.; König, T.; Richter-Dennerlein, R.; Sprenger, H.-G.; Madrenas, J.; Mühlmeister, M.; Brandt, U.; et al. The Membrane Scaffold SLP2 Anchors a Proteolytic Hub in Mitochondria Containing PARL and the I-AAA Protease YME1L. EMBO Rep. 2016, 17, 1844–1856. [Google Scholar] [CrossRef]
- Mishra, P.; Chan, D.C. Mitochondrial Dynamics and Inheritance during Cell Division, Development and Disease. Nat. Rev. Mol. Cell Biol. 2014, 15, 634–646. [Google Scholar] [CrossRef]
- Ren, L.; Chen, X.; Chen, X.; Li, J.; Cheng, B.; Xia, J. Mitochondrial Dynamics: Fission and Fusion in Fate Determination of Mesenchymal Stem Cells. Front. Cell. Dev. Biol. 2020, 8, 580070. [Google Scholar] [CrossRef]
- Cribbs, J.T.; Strack, S. Reversible Phosphorylation of Drp1 by Cyclic AMP-Dependent Protein Kinase and Calcineurin Regulates Mitochondrial Fission and Cell Death. EMBO Rep. 2007, 8, 939–944. [Google Scholar] [CrossRef]
- Cereghetti, G.M.; Stangherlin, A.; Martins de Brito, O.; Chang, C.R.; Blackstone, C.; Bernardi, P.; Scorrano, L. Dephosphorylation by Calcineurin Regulates Translocation of Drp1 to Mitochondria. Proc. Natl. Acad. Sci. USA 2008, 105, 15803–15808. [Google Scholar] [CrossRef] [PubMed]
- Otera, H.; Wang, C.; Cleland, M.M.; Setoguchi, K.; Yokota, S.; Youle, R.J.; Mihara, K. Mff Is an Essential Factor for Mitochondrial Recruitment of Drp1 during Mitochondrial Fission in Mammalian Cells. J. Cell Biol. 2010, 191, 1141–1158. [Google Scholar] [CrossRef] [PubMed]
- Tait, S.W.G.; Green, D.R. Mitochondrial Regulation of Cell Death. Cold Spring Harb. Perspect. Biol. 2013, 5, a008706. [Google Scholar] [CrossRef] [PubMed]
- Picard, M.; McManus, M.J.; Csordás, G.; Várnai, P.; Dorn, G.W.; Williams, D.; Hajnóczky, G.; Wallace, D.C. Trans-Mitochondrial Coordination of Cristae at Regulated Membrane Junctions. Nat. Commun. 2015, 6, 6259. [Google Scholar] [CrossRef]
- Bonora, M.; Giorgi, C.; Pinton, P. Molecular Mechanisms and Consequences of Mitochondrial Permeability Transition. Nat. Rev. Mol. Cell Biol. 2022, 23, 266–285. [Google Scholar] [CrossRef]
- Youle, R.J. Morphology of Mitochondria during Apoptosis: Worms-to-Beetles in Worms. Dev. Cell 2005, 8, 298–299. [Google Scholar] [CrossRef]
- Cleland, M.M.; Norris, K.L.; Karbowski, M.; Wang, C.; Suen, D.-F.; Jiao, S.; George, N.M.; Luo, X.; Li, Z.; Youle, R.J. Bcl-2 Family Interaction with the Mitochondrial Morphogenesis Machinery. Cell Death Differ. 2011, 18, 235–247. [Google Scholar] [CrossRef]
- Brooks, C.; Cho, S.-G.; Wang, C.-Y.; Yang, T.; Dong, Z. Fragmented Mitochondria Are Sensitized to Bax Insertion and Activation during Apoptosis. Am. J. Physiol. Cell Physiol. 2011, 300, C447–C455. [Google Scholar] [CrossRef]
- Frank, S.; Gaume, B.; Bergmann-Leitner, E.S.; Leitner, W.W.; Robert, E.G.; Catez, F.; Smith, C.L.; Youle, R.J. The Role of Dynamin-Related Protein 1, a Mediator of Mitochondrial Fission, in Apoptosis. Dev. Cell 2001, 1, 515–525. [Google Scholar] [CrossRef]
- Sheridan, C.; Delivani, P.; Cullen, S.P.; Martin, S.J. Bax- or Bak-Induced Mitochondrial Fission Can Be Uncoupled from Cytochrome C Release. Mol. Cell 2008, 31, 570–585. [Google Scholar] [CrossRef]
- Lee, Y.; Jeong, S.-Y.; Karbowski, M.; Smith, C.L.; Youle, R.J. Roles of the Mammalian Mitochondrial Fission and Fusion Mediators Fis1, Drp1, and Opa1 in Apoptosis. Mol. Biol. Cell 2004, 15, 5001–5011. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, Y.; Jiang, X.; Gu, Y.; Zheng, H.; Wang, X.; Zhang, H.; Wu, J.; Cheng, Y. OPA1 supports mitochondrial dynamics and immune evasion to CD8+ T cell in lung adenocarcinoma. PeerJ 2022, 10, e14543. [Google Scholar] [CrossRef] [PubMed]
- Mannella, C.A.; Lederer, W.J.; Jafri, M.S. The Connection between Inner Membrane Topology and Mitochondrial Function. J. Mol. Cell. Cardiol. 2013, 62, 51–57. [Google Scholar] [CrossRef]
- Varanita, T.; Soriano, M.E.; Romanello, V.; Zaglia, T.; Quintana-Cabrera, R.; Semenzato, M.; Menabò, R.; Costa, V.; Civiletto, G.; Pesce, P.; et al. The OPA1-Dependent Mitochondrial Cristae Remodeling Pathway Controls Atrophic, Apoptotic, and Ischemic Tissue Damage. Cell Metab. 2015, 21, 834–844. [Google Scholar] [CrossRef] [PubMed]
- Frezza, C.; Cipolat, S.; Martins de Brito, O.; Micaroni, M.; Beznoussenko, G.V.; Rudka, T.; Bartoli, D.; Polishuck, R.S.; Danial, N.N.; De Strooper, B.; et al. OPA1 Controls Apoptotic Cristae Remodeling Independently from Mitochondrial Fusion. Cell 2006, 126, 177–189. [Google Scholar] [CrossRef]
- Scorrano, L.; Ashiya, M.; Buttle, K.; Weiler, S.; Oakes, S.A.; Mannella, C.A.; Korsmeyer, S.J. A Distinct Pathway Remodels Mitochondrial Cristae and Mobilizes Cytochrome c during Apoptosis. Dev. Cell 2002, 2, 55–67. [Google Scholar] [CrossRef] [PubMed]
- Snigirevskaya, E.S.; Komissarchik, Y.Y. Ultrastructural Traits of Apoptosis. Cell Biol. Int. 2019, 43, 728–738. [Google Scholar] [CrossRef]
- Deshwal, S.; Fiedler, K.U.; Langer, T. Mitochondrial Proteases: Multifaceted Regulators of Mitochondrial Plasticity. Annu. Rev. Biochem. 2020, 89, 501–528. [Google Scholar] [CrossRef]
- Guo, X.-Y.; Guo, H.-F.; Guo, H.-M. Clinical Significance of SLP-2 in Epithelial Ovarian Cancer and Its Regulatory Effect on the Notch Signaling Pathway. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 1666–1671. [Google Scholar] [CrossRef]
- Cheng, M.; Yu, H.; Kong, Q.; Wang, B.; Shen, L.; Dong, D.; Sun, L. The Mitochondrial PHB2/OMA1/DELE1 Pathway Cooperates with Endoplasmic Reticulum Stress to Facilitate the Response to Chemotherapeutics in Ovarian Cancer. Int. J. Mol. Sci. 2022, 23, 1320. [Google Scholar] [CrossRef]
- Tsai, H.-W.; Li, C.-J.; Lin, L.-T.; Chiang, A.-J.; Chen, S.-N.; Wen, Z.-H.; Tsui, K.-H. Expression Status and Prognostic Significance of Mitochondrial Dynamics OPA3 in Human Ovarian Cancer. Aging 2022, 14, 3874–3886. [Google Scholar] [CrossRef]
- Ryu, S.-W.; Jeong, H.J.; Choi, M.; Karbowski, M.; Choi, C. Optic Atrophy 3 as a Protein of the Mitochondrial Outer Membrane Induces Mitochondrial Fragmentation. Cell. Mol. Life Sci. 2010, 67, 2839–2850. [Google Scholar] [CrossRef] [PubMed]
- Gegg, M.E.; Cooper, J.M.; Chau, K.-Y.; Rojo, M.; Schapira, A.H.V.; Taanman, J.-W. Mitofusin 1 and Mitofusin 2 Are Ubiquitinated in a PINK1/Parkin-Dependent Manner upon Induction of Mitophagy. Hum. Mol. Genet. 2010, 19, 4861–4870. [Google Scholar] [CrossRef] [PubMed]
- Yu, R.; Liu, T.; Jin, S.-B.; Ankarcrona, M.; Lendahl, U.; Nistér, M.; Zhao, J. MIEF1/2 Orchestrate Mitochondrial Dynamics through Direct Engagement with Both the Fission and Fusion Machineries. BMC Biol. 2021, 19, 229. [Google Scholar] [CrossRef]
- Chakraborty, P.K.; Murphy, B.; Mustafi, S.B.; Dey, A.; Xiong, X.; Rao, G.; Naz, S.; Zhang, M.; Yang, D.; Dhanasekaran, D.N.; et al. Cystathionine β-Synthase Regulates Mitochondrial Morphogenesis in Ovarian Cancer. FASEB J. 2018, 32, 4145–4157. [Google Scholar] [CrossRef] [PubMed]
- Ashraf, R.; Kumar, S. Mfn2-Mediated Mitochondrial Fusion Promotes Autophagy and Suppresses Ovarian Cancer Progression by Reducing ROS through AMPK/MTOR/ERK Signaling. Cell. Mol. Life Sci. 2022, 79, 573. [Google Scholar] [CrossRef]
- Tanwar, D.K.; Parker, D.J.; Gupta, P.; Spurlock, B.; Alvarez, R.D.; Basu, M.K.; Mitra, K. Crosstalk between the Mitochondrial Fission Protein, Drp1, and the Cell Cycle Is Identified across Various Cancer Types and Can Impact Survival of Epithelial Ovarian Cancer Patients. Oncotarget 2016, 7, 60021–60037. [Google Scholar] [CrossRef]
- Bandopadhyay, S.; Prasad, P.; Ray, U.; Das Ghosh, D.; Roy, S.S. SIRT6 Promotes Mitochondrial Fission and Subsequent Cellular Invasion in Ovarian Cancer. FEBS Open Bio 2022, 12, 1657–1676. [Google Scholar] [CrossRef]
- Kong, B.; Wang, Q.; Fung, E.; Xue, K.; Tsang, B.K. P53 Is Required for Cisplatin-Induced Processing of the Mitochondrial Fusion Protein L-Opa1 That Is Mediated by the Mitochondrial Metallopeptidase Oma1 in Gynecologic Cancers. J. Biol. Chem. 2014, 289, 27134–27145. [Google Scholar] [CrossRef]
- Kong, B.; Han, C.Y.; Kim, S.I.; Patten, D.A.; Han, Y.; Carmona, E.; Shieh, D.-B.; Cheung, A.C.; Mes-Masson, A.-M.; Harper, M.-E.; et al. Prohibitin 1 Interacts with P53 in the Regulation of Mitochondrial Dynamics and Chemoresistance in Gynecologic Cancers. J. Ovarian Res. 2022, 15, 70. [Google Scholar] [CrossRef]
- Alavi, M.V. Targeted OMA1 Therapies for Cancer. Int. J. Cancer 2019, 145, 2330–2341. [Google Scholar] [CrossRef]
- Zou, G.-P.; Yu, C.-X.; Shi, S.-L.; Li, Q.-G.; Wang, X.-H.; Qu, X.-H.; Yang, Z.-J.; Yao, W.-R.; Yan, D.-D.; Jiang, L.-P.; et al. Mitochondrial Dynamics Mediated by DRP1 and MFN2 Contributes to Cisplatin Chemoresistance in Human Ovarian Cancer SKOV3 Cells. J. Cancer 2021, 12, 7358–7373. [Google Scholar] [CrossRef] [PubMed]
- Tsuyoshi, H.; Wong, V.K.W.; Han, Y.; Orisaka, M.; Yoshida, Y.; Tsang, B.K. Saikosaponin-d, a Calcium Mobilizing Agent, Sensitizes Chemoresistant Ovarian Cancer Cells to Cisplatin-Induced Apoptosis by Facilitating Mitochondrial Fission and G2/M Arrest. Oncotarget 2017, 8, 99825–99840. [Google Scholar] [CrossRef] [PubMed]
- Farrand, L.; Kim, J.Y.; Im-Aram, A.; Suh, J.-Y.; Lee, H.J.; Tsang, B.K. An Improved Quantitative Approach for the Assessment of Mitochondrial Fragmentation in Chemoresistant Ovarian Cancer Cells. PLoS ONE 2013, 8, e74008. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, S.R.; Ray, U.; Chatterjee, B.P.; Roy, S.S. Targeted Apoptosis in Ovarian Cancer Cells through Mitochondrial Dysfunction in Response to Sambucus Nigra Agglutinin. Cell Death Dis. 2017, 8, e2762. [Google Scholar] [CrossRef] [PubMed]
- Gong, L.-H.; Chen, X.-X.; Wang, H.; Jiang, Q.-W.; Pan, S.-S.; Qiu, J.-G.; Mei, X.-L.; Xue, Y.-Q.; Qin, W.-M.; Zheng, F.-Y.; et al. Piperlongumine Induces Apoptosis and Synergizes with Cisplatin or Paclitaxel in Human Ovarian Cancer Cells. Oxidative Med. Cell. Longev. 2014, 2014, 906804. [Google Scholar] [CrossRef] [PubMed]
- Gupta, N.; Huang, T.-T.; Horibata, S.; Lee, J.-M. Cell Cycle Checkpoints and beyond: Exploiting the ATR/CHK1/WEE1 Pathway for the Treatment of PARP Inhibitor-Resistant Cancer. Pharmacol. Res. 2022, 178, 106162. [Google Scholar] [CrossRef]
- Lao, M.; Zhang, X.; Yang, H.; Bai, X.; Liang, T. RCAN1-Mediated Calcineurin Inhibition as a Target for Cancer Therapy. Mol. Med. 2022, 28, 69. [Google Scholar] [CrossRef]
- Ventura, J.-J.; Nebreda, A.R. Protein Kinases and Phosphatases as Therapeutic Targets in Cancer. Clin. Transl. Oncol. 2006, 8, 153–160. [Google Scholar] [CrossRef]
- Turdo, A.; D’Accardo, C.; Glaviano, A.; Porcelli, G.; Colarossi, C.; Colarossi, L.; Mare, M.; Faldetta, N.; Modica, C.; Pistone, G.; et al. Targeting Phosphatases and Kinases: How to Checkmate Cancer. Front. Cell Dev. Biol. 2021, 9, 690306. [Google Scholar] [CrossRef]
- Ould Amer, Y.; Hebert-Chatelain, E. Mitochondrial CAMP-PKA Signaling: What Do We Really Know? Biochim. Biophys. Acta BBA Bioenerg. 2018, 1859, 868–877. [Google Scholar] [CrossRef] [PubMed]
- Di Benedetto, G.; Lefkimmiatis, K.; Pozzan, T. The Basics of Mitochondrial CAMP Signalling: Where, When, Why. Cell Calcium 2021, 93, 102320. [Google Scholar] [CrossRef] [PubMed]
- Lefkimmiatis, K.; Zaccolo, M. CAMP Signaling in Subcellular Compartments. Pharmacol. Ther. 2014, 143, 295–304. [Google Scholar] [CrossRef]
- Zippin, J.H.; Chen, Y.; Nahirney, P.; Kamenetsky, M.; Wuttke, M.S.; Fischman, D.A.; Levin, L.R.; Buck, J. Compartmentalization of Bicarbonate-Sensitive Adenylyl Cyclase in Distinct Signaling Microdomains. FASEB J. 2003, 17, 82–84. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, M.B.; Alghamdi, A.A.A.; Islam, S.U.; Lee, J.-S.; Lee, Y.-S. CAMP Signaling in Cancer: A PKA-CREB and EPAC-Centric Approach. Cells 2022, 11, 2020. [Google Scholar] [CrossRef] [PubMed]
- De Rasmo, D.; Gattoni, G.; Papa, F.; Santeramo, A.; Pacelli, C.; Cocco, T.; Micelli, L.; Sardaro, N.; Larizza, M.; Scivetti, M.; et al. The β-Adrenoceptor Agonist Isoproterenol Promotes the Activity of Respiratory Chain Complex I and Lowers Cellular Reactive Oxygen Species in Fibroblasts and Heart Myoblasts. Eur. J. Pharmacol. 2011, 652, 15–22. [Google Scholar] [CrossRef]
- Bellomo, F.; Piccoli, C.; Cocco, T.; Scacco, S.; Papa, F.; Gaballo, A.; Boffoli, D.; Signorile, A.; D’Aprile, A.; Scrima, R.; et al. Regulation by the CAMP Cascade of Oxygen Free Radical Balance in Mammalian Cells. Antioxid. Redox Signal. 2006, 8, 495–502. [Google Scholar] [CrossRef] [PubMed]
- Signorile, A.; Pacelli, C.; Palese, L.L.; Santeramo, A.; Roca, E.; Cocco, T.; De Rasmo, D. CAMP/PKA Signaling Modulates Mitochondrial Supercomplex Organization. Int. J. Mol. Sci. 2022, 23, 9655. [Google Scholar] [CrossRef] [PubMed]
- Iwai-Kanai, E.; Hasegawa, K.; Araki, M.; Kakita, T.; Morimoto, T.; Sasayama, S. Alpha- and Beta-Adrenergic Pathways Differentially Regulate Cell Type-Specific Apoptosis in Rat Cardiac Myocytes. Circulation 1999, 100, 305–311. [Google Scholar] [CrossRef]
- Zhang, F.; Zhang, L.; Qi, Y.; Xu, H. Mitochondrial CAMP Signaling. Cell. Mol. Life Sci. 2016, 73, 4577–4590. [Google Scholar] [CrossRef]
- Simko, V.; Iuliano, F.; Sevcikova, A.; Labudova, M.; Barathova, M.; Radvak, P.; Pastorekova, S.; Pastorek, J.; Csaderova, L. Hypoxia Induces Cancer-Associated CAMP/PKA Signalling through HIF-Mediated Transcriptional Control of Adenylyl Cyclases VI and VII. Sci. Rep. 2017, 7, 10121. [Google Scholar] [CrossRef]
- Palorini, R.; De Rasmo, D.; Gaviraghi, M.; Sala Danna, L.; Signorile, A.; Cirulli, C.; Chiaradonna, F.; Alberghina, L.; Papa, S. Oncogenic K-Ras Expression Is Associated with Derangement of the CAMP/PKA Pathway and Forskolin-Reversible Alterations of Mitochondrial Dynamics and Respiration. Oncogene 2013, 32, 352–362. [Google Scholar] [CrossRef] [PubMed]
- McDaid, H.M.; Cairns, M.T.; Atkinson, R.J.; McAleer, S.; Harkin, D.P.; Gilmore, P.; Johnston, P.G. Increased Expression of the RIα Subunit of the CAMP-Dependent Protein Kinase A Is Associated with Advanced Stage Ovarian Cancer. Br. J. Cancer 1999, 79, 933–939. [Google Scholar] [CrossRef]
- Bai, F.; Feng, J.; Cheng, Y.; Shi, J.; Yang, R.; Cui, H. Analysis of Gene Expression Patterns of Ovarian Cancer Cell Lines with Different Metastatic Potentials. Int. J. Gynecol. Cancer 2006, 16, 202–209. [Google Scholar] [CrossRef]
- Alper, O.; Hacker, N.F.; Cho-Chung, Y.S. Protein Kinase A-Ialpha Subunit-Directed Antisense Inhibition of Ovarian Cancer Cell Growth: Crosstalk with Tyrosine Kinase Signaling Pathway. Oncogene 1999, 18, 4999–5004. [Google Scholar] [CrossRef] [PubMed]
- Cheadle, C.; Nesterova, M.; Watkins, T.; Barnes, K.C.; Hall, J.C.; Rosen, A.; Becker, K.G.; Cho-Chung, Y.S. Regulatory Subunits of PKA Define an Axis of Cellular Proliferation/Differentiation in Ovarian Cancer Cells. BMC Med. Genom. 2008, 1, 43. [Google Scholar] [CrossRef] [PubMed]
- McKenzie, A.J.; Campbell, S.L.; Howe, A.K. Protein Kinase A Activity and Anchoring Are Required for Ovarian Cancer Cell Migration and Invasion. PLoS ONE 2011, 6, e26552. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zhang, L.; Liang, Y.; Zhang, C.; Xu, Z.; Zhang, L.; Fuji, R.; Mu, W.; Li, L.; Jiang, J.; et al. Cyclic AMP Mimics the Anti-Ageing Effects of Calorie Restriction by Up-Regulating Sirtuin. Sci. Rep. 2015, 5, 12012. [Google Scholar] [CrossRef] [PubMed]
- Finley, L.W.S.; Carracedo, A.; Lee, J.; Souza, A.; Egia, A.; Zhang, J.; Teruya-Feldstein, J.; Moreira, P.I.; Cardoso, S.M.; Clish, C.B.; et al. SIRT3 Opposes Reprogramming of Cancer Cell Metabolism through HIF1α Destabilization. Cancer Cell 2011, 19, 416–428. [Google Scholar] [CrossRef]
- Zhao, Q.; Zhou, J.; Li, F.; Guo, S.; Zhang, L.; Li, J.; Qi, Q.; Shi, Y. The Role and Therapeutic Perspectives of Sirtuin 3 in Cancer Metabolism Reprogramming, Metastasis, and Chemoresistance. Front. Oncol. 2022, 12, 910963. [Google Scholar] [CrossRef]
- Dong, X.-C.; Jing, L.-M.; Wang, W.-X.; Gao, Y.-X. Down-Regulation of SIRT3 Promotes Ovarian Carcinoma Metastasis. Biochem. Biophys. Res. Commun. 2016, 475, 245–250. [Google Scholar] [CrossRef] [PubMed]
- Xiang, X.-Y.; Kang, J.-S.; Yang, X.-C.; Su, J.; Wu, Y.; Yan, X.-Y.; Xue, Y.-N.; Xu, Y.; Liu, Y.-H.; Yu, C.-Y.; et al. SIRT3 Participates in Glucose Metabolism Interruption and Apoptosis Induced by BH3 Mimetic S1 in Ovarian Cancer Cells. Int. J. Oncol. 2016, 49, 773–784. [Google Scholar] [CrossRef] [PubMed]
- Gong, S.; Chen, Y.; Meng, F.; Zhang, Y.; Wu, H.; Wu, F. Roflumilast Restores CAMP/PKA/CREB Signaling Axis for FtMt-Mediated Tumor Inhibition of Ovarian Cancer. Oncotarget 2017, 8, 112341–112353. [Google Scholar] [CrossRef] [PubMed]
- Lan, J.; Cadassou, O.; Corbet, C.; Riant, O.; Feron, O. Discovery of Mitochondrial Complex I Inhibitors as Anticancer and Radiosensitizer Drugs Based on Compensatory Stimulation of Lactate Release. Cancers 2022, 14, 5454. [Google Scholar] [CrossRef] [PubMed]
- Grieco, J.P.; Allen, M.E.; Perry, J.B.; Wang, Y.; Song, Y.; Rohani, A.; Compton, S.L.E.; Smyth, J.W.; Swami, N.S.; Brown, D.A.; et al. Progression-Mediated Changes in Mitochondrial Morphology Promotes Adaptation to Hypoxic Peritoneal Conditions in Serous Ovarian Cancer. Front. Oncol. 2020, 10, 600113. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
De Rasmo, D.; Cormio, A.; Cormio, G.; Signorile, A. Ovarian Cancer: A Landscape of Mitochondria with Emphasis on Mitochondrial Dynamics. Int. J. Mol. Sci. 2023, 24, 1224. https://doi.org/10.3390/ijms24021224
De Rasmo D, Cormio A, Cormio G, Signorile A. Ovarian Cancer: A Landscape of Mitochondria with Emphasis on Mitochondrial Dynamics. International Journal of Molecular Sciences. 2023; 24(2):1224. https://doi.org/10.3390/ijms24021224
Chicago/Turabian StyleDe Rasmo, Domenico, Antonella Cormio, Gennaro Cormio, and Anna Signorile. 2023. "Ovarian Cancer: A Landscape of Mitochondria with Emphasis on Mitochondrial Dynamics" International Journal of Molecular Sciences 24, no. 2: 1224. https://doi.org/10.3390/ijms24021224
APA StyleDe Rasmo, D., Cormio, A., Cormio, G., & Signorile, A. (2023). Ovarian Cancer: A Landscape of Mitochondria with Emphasis on Mitochondrial Dynamics. International Journal of Molecular Sciences, 24(2), 1224. https://doi.org/10.3390/ijms24021224




