Predicting Modifiers of Genotype-Phenotype Correlations in Craniofacial Development
Abstract
1. Introduction
2. Results
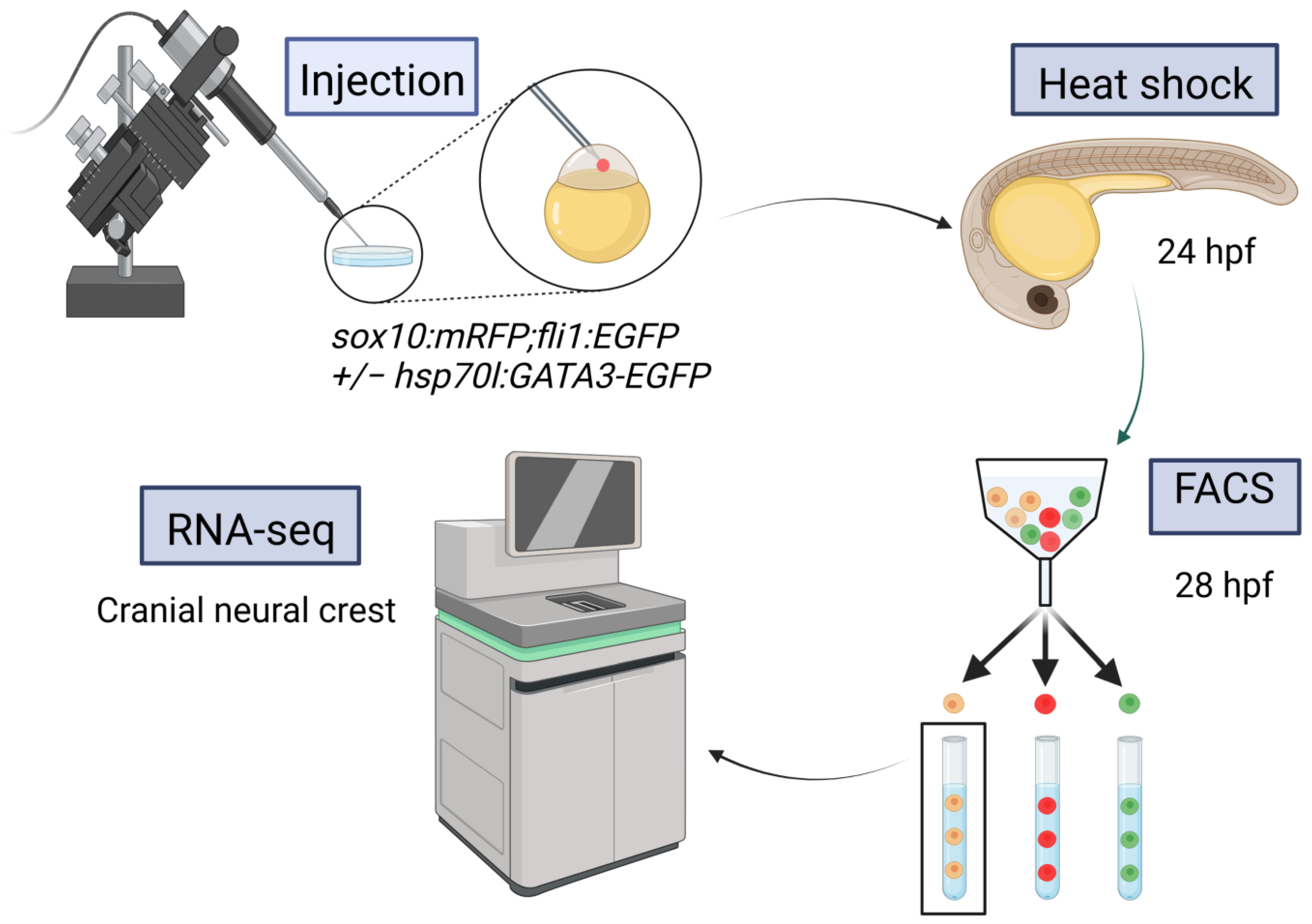
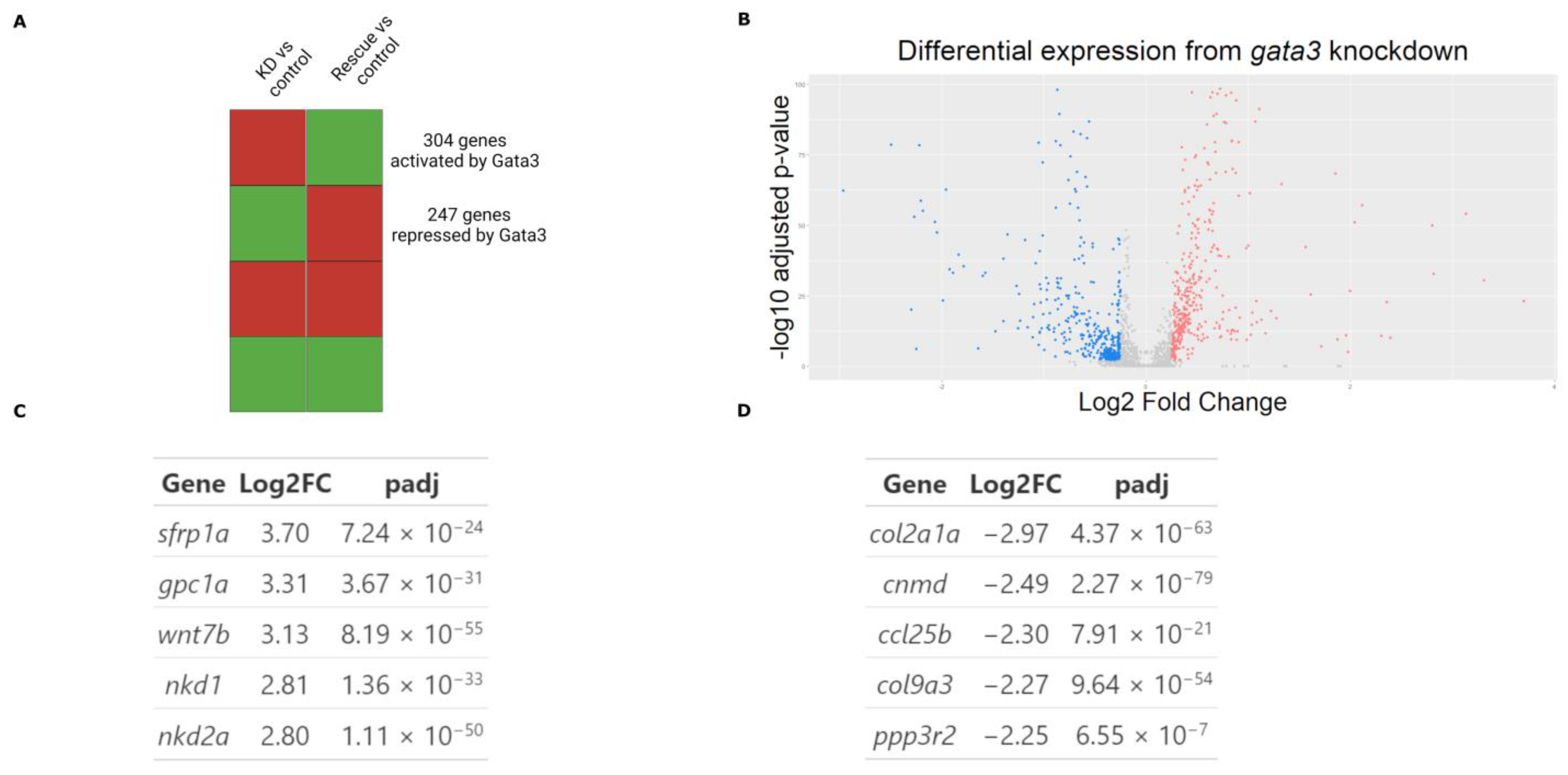
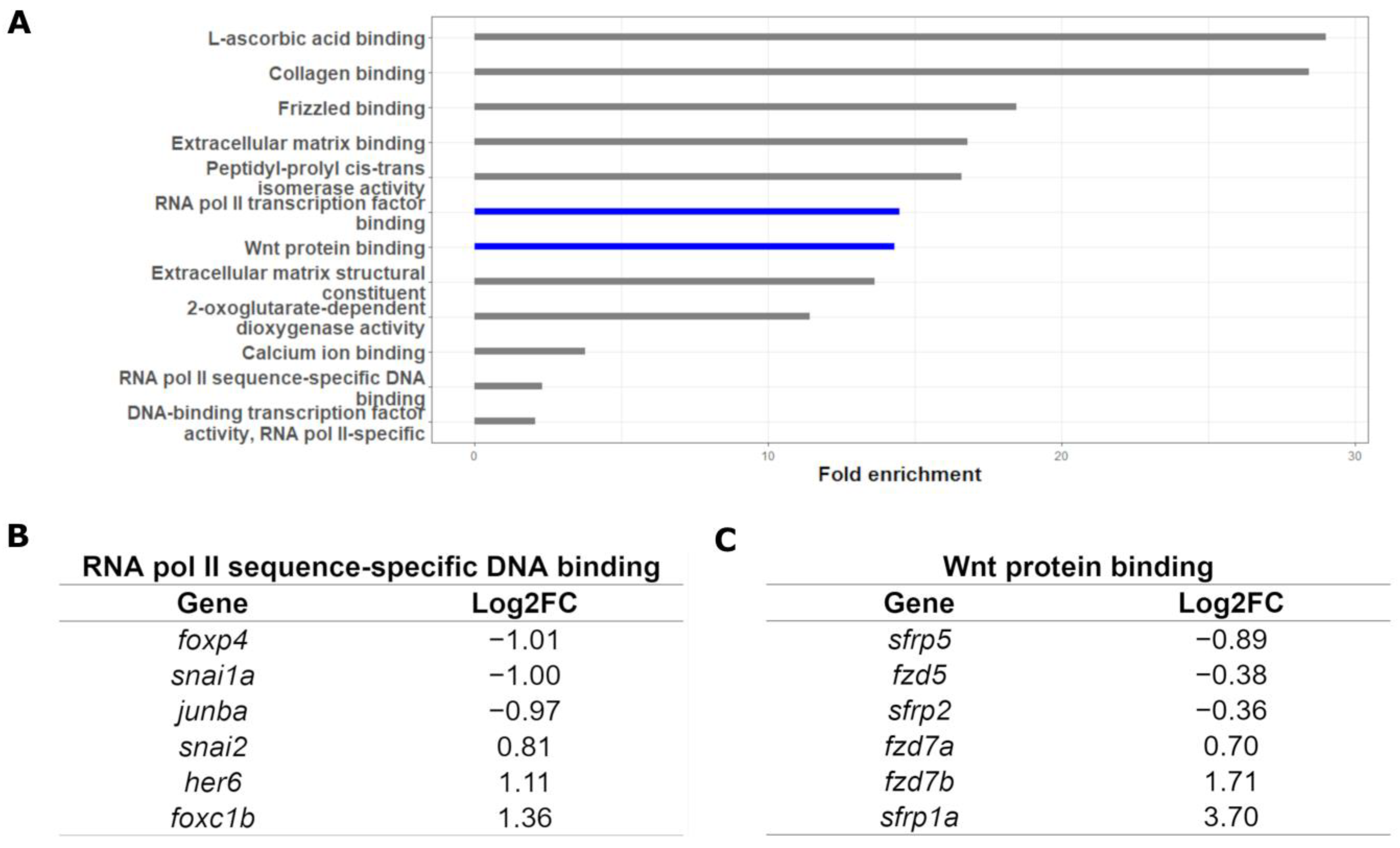
2.1. Targets of Gata3 in Cranial Neural Crest Cells
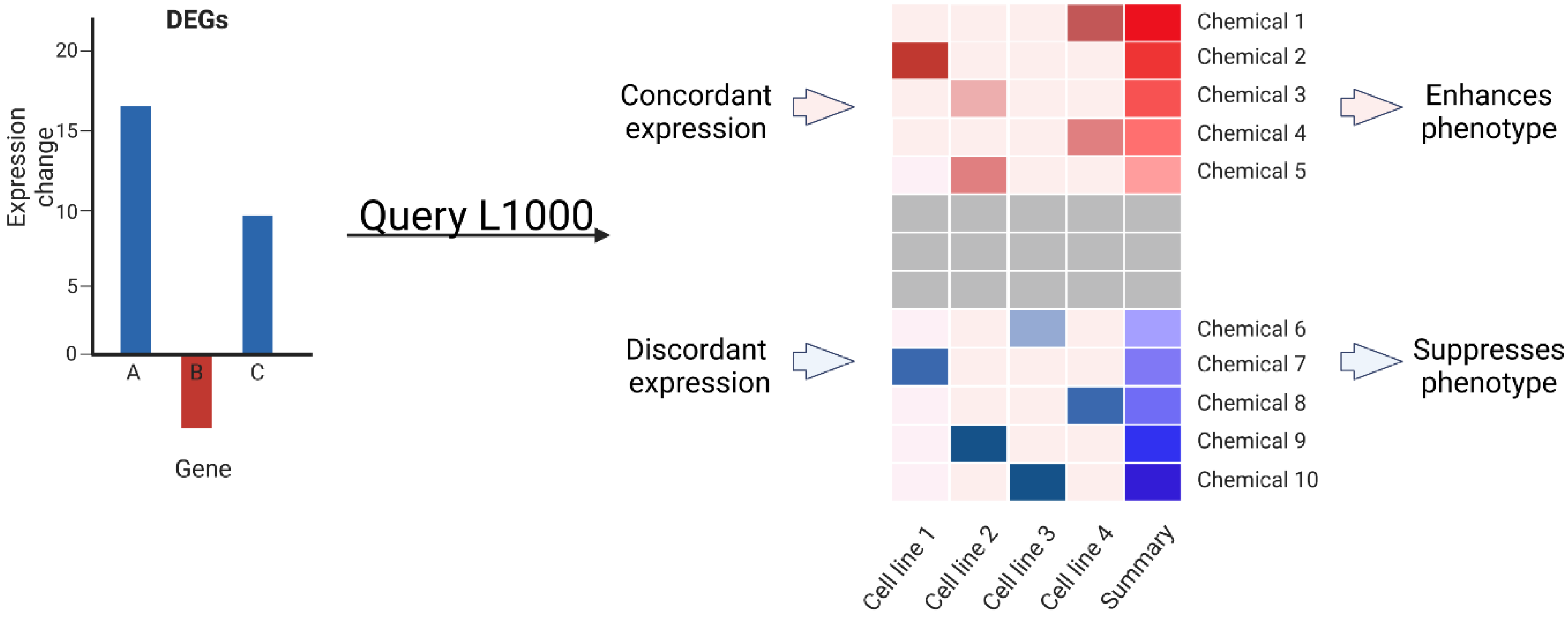
2.2. Transcriptomic Data Can Predict Chemical Modifiers of Genotype-Phenotype Correlations
3. Discussion
3.1. Prediction of Chemicals That Worsen Mutant Phenotypes
3.2. Identification of Chemicals That Are Protective against Deleterious Phenotypes
3.3. Identification of Functional Modules through RNA-seq
4. Materials and Methods
4.1. Animal Care and Use
4.2. Injections and Heat Shock
4.3. Sample Preparation and FACS
4.4. cDNA Library Preparation and RNA Sequencing
4.5. Differential Expression Analysis
4.6. Identification and Testing of Chemical Modifiers
4.7. Quantitative Reverse Transcription PCR (RT-qPCR)
4.8. Trabecular Measurements
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Watkins, S.E.; Meyer, R.E.; Strauss, R.P.; Aylsworth, A.S. Classification, epidemiology, and genetics of orofacial clefts. Clin. Plast. Surg. 2014, 41, 149–163. [Google Scholar] [CrossRef] [PubMed]
- Cordero, D.R.; Brugmann, S.; Chu, Y.; Bajpai, R.; Jame, M.; Helms, J.A. Cranial neural crest cells on the move: Their roles in craniofacial development. Am. J. Med. Genet. A 2011, 155A, 270–279. [Google Scholar] [CrossRef]
- Dixon, M.J.; Marazita, M.L.; Beaty, T.H.; Murray, J.C. Cleft lip and palate: Understanding genetic and environmental influences. Nat. Rev. Genet. 2011, 12, 167–178. [Google Scholar] [CrossRef] [PubMed]
- Eberhart, J.K.; Swartz, M.E.; Crump, J.G.; Kimmel, C.B. Early Hedgehog signaling from neural to oral epithelium organizes anterior craniofacial development. Development 2006, 133, 1069–1077. [Google Scholar] [CrossRef]
- Wada, N.; Javidan, Y.; Nelson, S.; Carney, T.J.; Kelsh, R.N.; Schilling, T.F. Hedgehog signaling is required for cranial neural crest morphogenesis and chondrogenesis at the midline in the zebrafish skull. Development 2005, 132, 3977–3988. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, N.; Wetherill, L.; Lovely, C.B.; Swartz, M.E.; Foroud, T.M.; Eberhart, J.K. Pdgfra protects against ethanol-induced craniofacial defects in a zebrafish model of FASD. Development 2013, 140, 3254–3265. [Google Scholar] [CrossRef] [PubMed]
- Dougherty, M.; Kamel, G.; Grimaldi, M.; Gfrerer, L.; Shubinets, V.; Ethier, R.; Hickey, G.; Cornell, R.A.; Liao, E.C. Distinct requirements for wnt9a and irf6 in extension and integration mechanisms during zebrafish palate morphogenesis. Development 2013, 140, 76–81. [Google Scholar] [CrossRef]
- Duncan, K.M.; Mukherjee, K.; Cornell, R.A.; Liao, E.C. Zebrafish models of orofacial clefts. Dev. Dyn. 2017, 246, 897–914. [Google Scholar] [CrossRef]
- Swartz, M.E.; Sheehan-Rooney, K.; Dixon, M.J.; Eberhart, J.K. Examination of a palatogenic gene program in zebrafish. Dev. Dyn. 2011, 240, 2204–2220. [Google Scholar] [CrossRef]
- Fletcher, G.; Jones, G.E.; Patient, R.; Snape, A. A role for GATA factors in Xenopus gastrulation movements. Mech. Dev. 2006, 123, 730–745. [Google Scholar] [CrossRef]
- Frelin, C.; Herrington, R.; Janmohamed, S.; Barbara, M.; Tran, G.; Paige, C.J.; Benveniste, P.; Zuniga-Pflucker, J.C.; Souabni, A.; Busslinger, M.; et al. GATA-3 regulates the self-renewal of long-term hematopoietic stem cells. Nat. Immunol. 2013, 14, 1037–1044. [Google Scholar] [CrossRef] [PubMed]
- Kuo, C.T.; Morrisey, E.E.; Anandappa, R.; Sigrist, K.; Lu, M.M.; Parmacek, M.S.; Soudais, C.; Leiden, J.M. GATA4 transcription factor is required for ventral morphogenesis and heart tube formation. Genes Dev. 1997, 11, 1048–1060. [Google Scholar] [CrossRef]
- Bernardini, L.; Sinibaldi, L.; Capalbo, A.; Bottillo, I.; Mancuso, B.; Torres, B.; Novelli, A.; Digilio, M.C.; Dallapiccola, B. HDR (Hypoparathyroidism, Deafness, Renal dysplasia) syndrome associated to GATA3 gene duplication. Clin. Genet. 2009, 76, 117–119. [Google Scholar] [CrossRef]
- Van Esch, H.; Groenen, P.; Nesbit, M.A.; Schuffenhauer, S.; Lichtner, P.; Vanderlinden, G.; Harding, B.; Beetz, R.; Bilous, R.W.; Holdaway, I.; et al. GATA3 haplo-insufficiency causes human HDR syndrome. Nature 2000, 406, 419–422. [Google Scholar] [CrossRef]
- Zhang, Y.B.; Hu, J.; Zhang, J.; Zhou, X.; Li, X.; Gu, C.; Liu, T.; Xie, Y.; Liu, J.; Gu, M.; et al. Genome-wide association study identifies multiple susceptibility loci for craniofacial microsomia. Nat. Commun. 2016, 7, 10605. [Google Scholar] [CrossRef] [PubMed]
- Lim, K.C.; Lakshmanan, G.; Crawford, S.E.; Gu, Y.; Grosveld, F.; Engel, J.D. Gata3 loss leads to embryonic lethality due to noradrenaline deficiency of the sympathetic nervous system. Nat. Genet. 2000, 25, 209–212. [Google Scholar] [CrossRef] [PubMed]
- Abe, M.; Cox, T.C.; Firulli, A.B.; Kanai, S.M.; Dahlka, J.; Lim, K.C.; Engel, J.D.; Clouthier, D.E. GATA3 is essential for separating patterning domains during facial morphogenesis. Development 2021, 148. [Google Scholar] [CrossRef]
- Sheehan-Rooney, K.; Swartz, M.E.; Zhao, F.; Liu, D.; Eberhart, J.K. Ahsa1 and Hsp90 activity confers more severe craniofacial phenotypes in a zebrafish model of hypoparathyroidism, sensorineural deafness and renal dysplasia (HDR). Dis. Model. Mech. 2013, 6, 1285–1291. [Google Scholar] [CrossRef]
- Hartsfield, J.K. Review of the etiologic heterogeneity of the oculo-auriculo-vertebral spectrum (Hemifacial Microsomia). Orthod. Craniofac. Res. 2007, 10, 121–128. [Google Scholar] [CrossRef]
- Swartz, M.E.; Lovely, C.B.; Eberhart, J.K. Variation in phenotypes from a Bmp-Gata3 genetic pathway is modulated by Shh signaling. PLoS Genet. 2021, 17, e1009579. [Google Scholar] [CrossRef]
- Barske, L.; Askary, A.; Zuniga, E.; Balczerski, B.; Bump, P.; Nichols, J.T.; Crump, J.G. Competition between Jagged-Notch and Endothelin1 Signaling Selectively Restricts Cartilage Formation in the Zebrafish Upper Face. PLoS Genet. 2016, 12, e1005967. [Google Scholar] [CrossRef] [PubMed]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Narayan, R.; Corsello, S.M.; Peck, D.D.; Natoli, T.E.; Lu, X.; Gould, J.; Davis, J.F.; Tubelli, A.A.; Asiedu, J.K.; et al. A Next Generation Connectivity Map: L1000 Platform and the First 1,000,000 Profiles. Cell 2017, 171, 1437–1452.e1417. [Google Scholar] [CrossRef] [PubMed]
- Fruscio, R.; de Haan, J.; Van Calsteren, K.; Verheecke, M.; Mhallem, M.; Amant, F. Ovarian cancer in pregnancy. Best Pract. Res. Clin. Obstet. Gynaecol. 2017, 41, 108–117. [Google Scholar] [CrossRef] [PubMed]
- Luci, S.; Giemsa, B.; Hause, G.; Kluge, H.; Eder, K. Clofibrate treatment in pigs: Effects on parameters critical with respect to peroxisome proliferator-induced hepatocarcinogenesis in rodents. BMC Pharmacol. 2007, 7, 6. [Google Scholar] [CrossRef] [PubMed]
- Ringseis, R.; Gutgesell, A.; Dathe, C.; Brandsch, C.; Eder, K. Feeding oxidized fat during pregnancy up-regulates expression of PPARalpha-responsive genes in the liver of rat fetuses. Lipids Health Dis. 2007, 6, 6. [Google Scholar] [CrossRef] [PubMed]
- Pandolfi, P.P.; Roth, M.E.; Karis, A.; Leonard, M.W.; Dzierzak, E.; Grosveld, F.G.; Engel, J.D.; Lindenbaum, M.H. Targeted disruption of the GATA3 gene causes severe abnormalities in the nervous system and in fetal liver haematopoiesis. Nat. Genet. 1995, 11, 40–44. [Google Scholar] [CrossRef]
- Ciani, L.; Krylova, O.; Smalley, M.J.; Dale, T.C.; Salinas, P.C. A divergent canonical WNT-signaling pathway regulates microtubule dynamics: Dishevelled signals locally to stabilize microtubules. J. Cell Biol. 2004, 164, 243–253. [Google Scholar] [CrossRef]
- Fukuda, Y.; Sano, O.; Kazetani, K.; Yamamoto, K.; Iwata, H.; Matsui, J. Tubulin is a molecular target of the Wnt-activating chemical probe. BMC Biochem. 2016, 17, 9. [Google Scholar] [CrossRef]
- Jordan, M.A.; Himes, R.H.; Wilson, L. Comparison of the effects of vinblastine, vincristine, vindesine, and vinepidine on microtubule dynamics and cell proliferation in vitro. Cancer Res. 1985, 45, 2741–2747. [Google Scholar]
- Hou, Z.Y.; Tong, X.P.; Peng, Y.B.; Zhang, B.K.; Yan, M. Broad targeting of triptolide to resistance and sensitization for cancer therapy. Biomed. Pharmacother. 2018, 104, 771–780. [Google Scholar] [CrossRef] [PubMed]
- Reno, T.A.; Kim, J.Y.; Raz, D.J. Triptolide Inhibits Lung Cancer Cell Migration, Invasion, and Metastasis. Ann. Thorac. Surg. 2015, 100, 1817–1824; discussion 1815–1824. [Google Scholar] [CrossRef]
- Ziaei, S.; Halaby, R. Immunosuppressive, anti-inflammatory and anti-cancer properties of triptolide: A mini review. Avicenna J. Phytomed. 2016, 6, 149–164. [Google Scholar]
- Vispe, S.; DeVries, L.; Creancier, L.; Besse, J.; Breand, S.; Hobson, D.J.; Svejstrup, J.Q.; Annereau, J.P.; Cussac, D.; Dumontet, C.; et al. Triptolide is an inhibitor of RNA polymerase I and II-dependent transcription leading predominantly to down-regulation of short-lived mRNA. Mol. Cancer Ther. 2009, 8, 2780–2790. [Google Scholar] [CrossRef] [PubMed]
- Okun, D.B.; Groncy, P.K.; Sieger, L.; Tanaka, K.R. Acute leukemia in pregnancy: Transient neonatal myelosuppression after combination chemotherapy in the mother. Med. Pediatr. Oncol. 1979, 7, 315–319. [Google Scholar] [CrossRef] [PubMed]
- Al-Aamri, H.M.; Ku, H.; Irving, H.R.; Tucci, J.; Meehan-Andrews, T.; Bradley, C. Time dependent response of daunorubicin on cytotoxicity, cell cycle and DNA repair in acute lymphoblastic leukaemia. BMC Cancer 2019, 19, 179. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Zhang, J.P.; Qian, J.Q.; Hu, C.Q. Cardiotoxicity evaluation of anthracyclines in zebrafish (Danio rerio). J. Appl. Toxicol. 2015, 35, 241–252. [Google Scholar] [CrossRef] [PubMed]
- Logan, K.; Ackerman, S. Effects of antibiotics on RNA polymerase III transcription. DNA 1988, 7, 483–491. [Google Scholar] [CrossRef]
- Pallis, M.; Burrows, F.; Whittall, A.; Boddy, N.; Seedhouse, C.; Russell, N. Efficacy of RNA polymerase II inhibitors in targeting dormant leukaemia cells. BMC Pharmacol. Toxicol. 2013, 14, 32. [Google Scholar] [CrossRef]
- Liang, X.; Xie, R.; Su, J.; Ye, B.; Wei, S.; Liang, Z.; Bai, R.; Chen, Z.; Li, Z.; Gao, X. Inhibition of RNA polymerase III transcription by Triptolide attenuates colorectal tumorigenesis. J. Exp. Clin. Cancer Res. 2019, 38, 217. [Google Scholar] [CrossRef]
- Dale, R.M.; Sisson, B.E.; Topczewski, J. The emerging role of Wnt/PCP signaling in organ formation. Zebrafish 2009, 6, 9–14. [Google Scholar] [CrossRef] [PubMed]
- Park, M.; Moon, R.T. The planar cell-polarity gene stbm regulates cell behaviour and cell fate in vertebrate embryos. Nat. Cell Biol. 2002, 4, 20–25. [Google Scholar] [CrossRef]
- Topczewski, J.; Dale, R.M.; Sisson, B.E. Planar cell polarity signaling in craniofacial development. Organogenesis 2011, 7, 255–259. [Google Scholar] [CrossRef] [PubMed]
- Kamel, G.; Hoyos, T.; Rochard, L.; Dougherty, M.; Kong, Y.; Tse, W.; Shubinets, V.; Grimaldi, M.; Liao, E.C. Requirement for frzb and fzd7a in cranial neural crest convergence and extension mechanisms during zebrafish palate and jaw morphogenesis. Dev. Biol. 2013, 381, 423–433. [Google Scholar] [CrossRef] [PubMed]
- Brinkmann, E.M.; Mattes, B.; Kumar, R.; Hagemann, A.I.; Gradl, D.; Scholpp, S.; Steinbeisser, H.; Kaufmann, L.T.; Ozbek, S. Secreted Frizzled-related Protein 2 (sFRP2) Redirects Non-canonical Wnt Signaling from Fz7 to Ror2 during Vertebrate Gastrulation. J. Biol. Chem. 2016, 291, 13730–13742. [Google Scholar] [CrossRef] [PubMed]
- Kimmel, C.B.; Ballard, W.W.; Kimmel, S.R.; Ullmann, B.; Schilling, T.F. Stages of embryonic development of the zebrafish. Dev. Dyn. 1995, 203, 253–310. [Google Scholar] [CrossRef]
- Lawson, N.D.; Weinstein, B.M. In vivo imaging of embryonic vascular development using transgenic zebrafish. Dev. Biol. 2002, 248, 307–318. [Google Scholar] [CrossRef] [PubMed]
- Kucenas, S.; Takada, N.; Park, H.C.; Woodruff, E.; Broadie, K.; Appel, B. CNS-derived glia ensheath peripheral nerves and mediate motor root development. Nat. Neurosci. 2008, 11, 143–151. [Google Scholar] [CrossRef]
- Yang, L.; Rastegar, S.; Strahle, U. Regulatory interactions specifying Kolmer-Agduhr interneurons. Development 2010, 137, 2713–2722. [Google Scholar] [CrossRef]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S.L. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013, 14, R36. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 3. [Google Scholar] [CrossRef]
- Zerbino, D.R.; Achuthan, P.; Akanni, W.; Amode, M.R.; Barrell, D.; Bhai, J.; Billis, K.; Cummins, C.; Gall, A.; Giron, C.G.; et al. Ensembl 2018. Nucleic Acids Res. 2018, 46, D754–D761. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 2012, 16, 284–287. [Google Scholar] [CrossRef] [PubMed]
- Walker, M.B.; Kimmel, C.B. A two-color acid-free cartilage and bone stain for zebrafish larvae. Biotech. Histochem. 2007, 82, 23–28. [Google Scholar] [CrossRef] [PubMed]

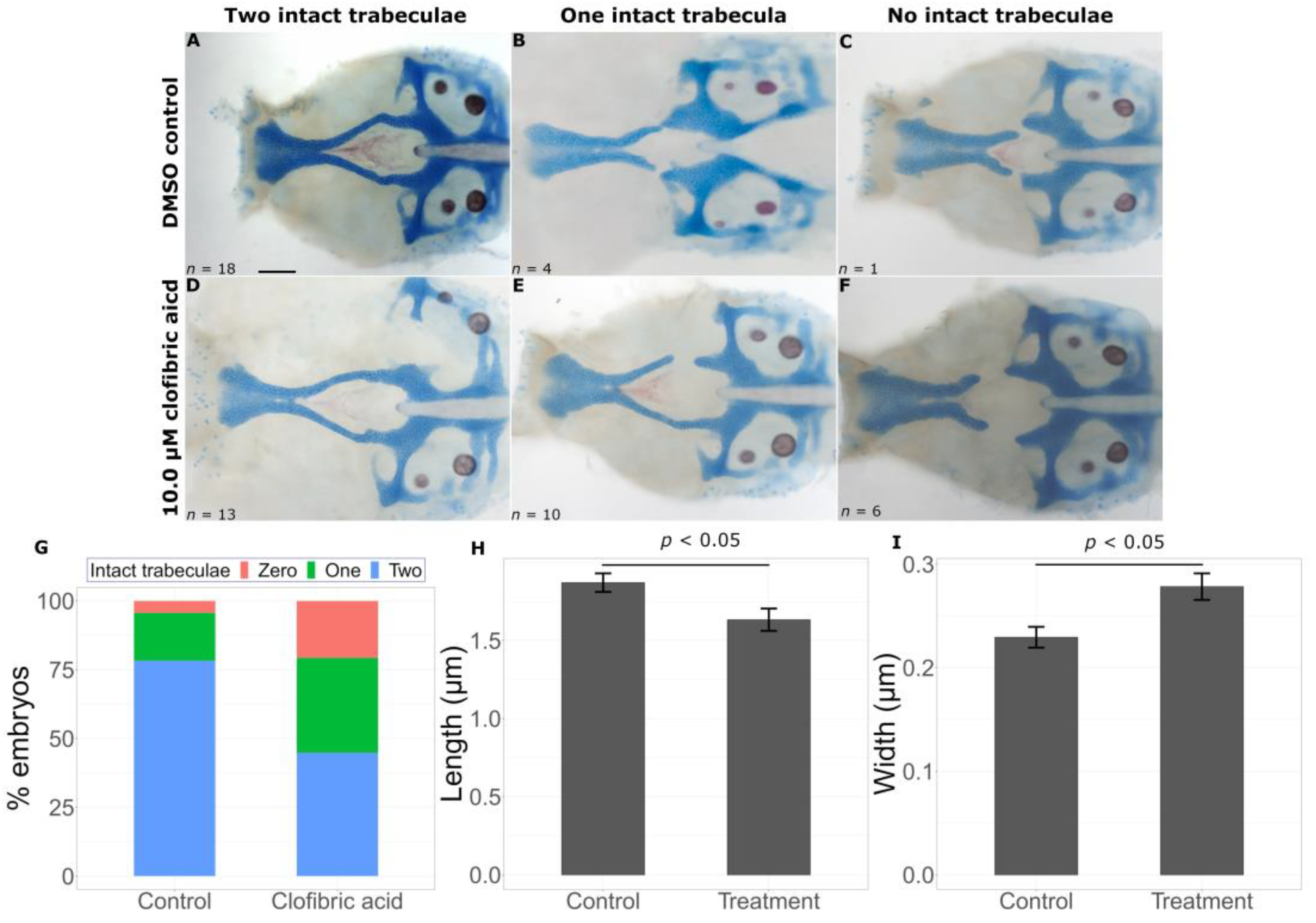
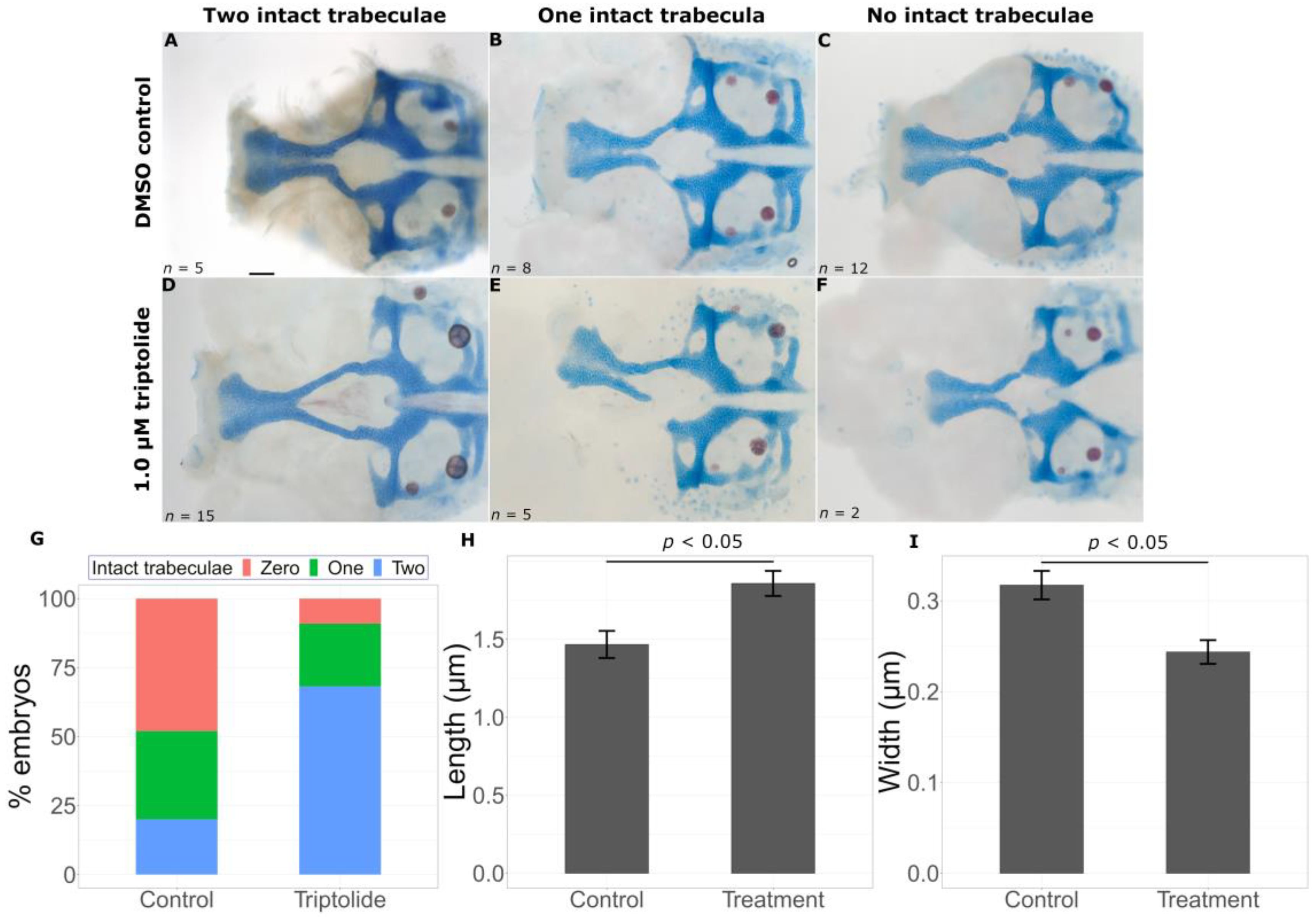
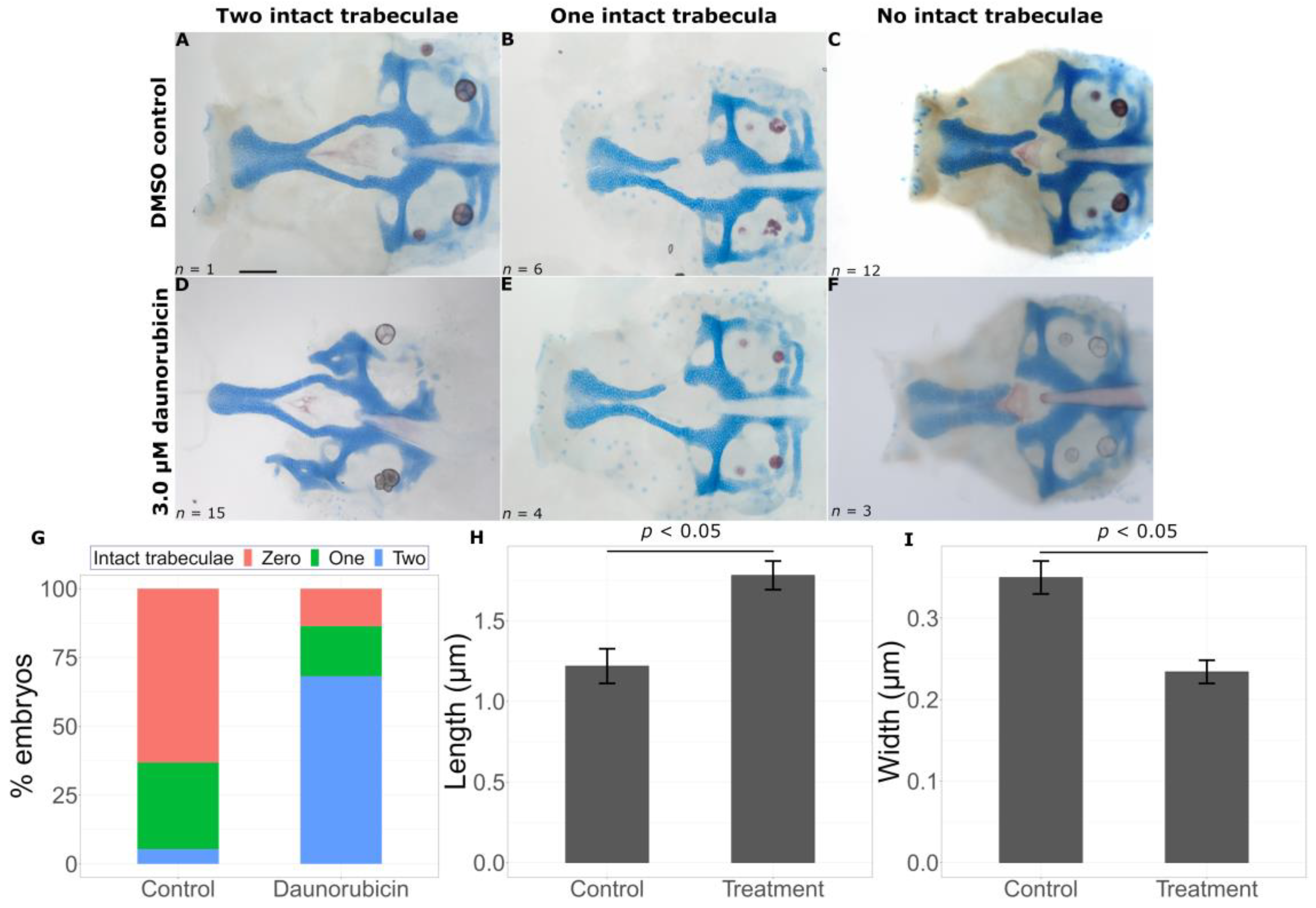
| Predicted Modification | Chemical | Function | CAS Registry Number |
|---|---|---|---|
| Phenotype enhancer | Flubendazole | Microtubule inhibitor, acetylcholinesterase inhibitor | 31430-15-6 |
| Rigosertib | Cell cycle inhibitor, PLK inhibitor | 1225497-78-8 | |
| Etacrynic acid | Na/K/Cl transporter | 58-54-8 | |
| Clofibric acid | PPAR receptor agonist | 882-09-7 | |
| Nocodazole | Tubulin inhibitor, tubulin polymerization inhibitor | 31430-18-9 | |
| Vinorelbine | Tubulin inhibitor, apoptosis stimulant | 71486-22-1 | |
| Vinblastine | Microtubule inhibitor, tubulin inhibitor | 865-21-4 | |
| Vincristine | Microtubule inhibitor, tubulin inhibitor | 57-22-7 | |
| Phenotype suppressor | PD-184352 | MEK inhibitor, MAP kinase | 212631-79-3 |
| Triptolide | RNA polymerase inhibitor | 38748-32-2 | |
| Belinostat | HDAC inhibitor, cell cycle inhibitor | 866323-14-0 | |
| BMS-191011 | Calcium activated potassium channel opener, potassium channel activator | 202821-81-6 | |
| ISOX | HDAC inhibitor, cell cycle inhibitor | 1045792-66-2 | |
| Daunorubicin | RNA synthesis inhibitor | 20830-81-3 | |
| Chromanol | Potassium channel blocker | 1481-93-2 | |
| Alvocidib | CDK inhibitor, apoptosis stimulant | 146426-40-6 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kar, R.D.; Eberhart, J.K. Predicting Modifiers of Genotype-Phenotype Correlations in Craniofacial Development. Int. J. Mol. Sci. 2023, 24, 1222. https://doi.org/10.3390/ijms24021222
Kar RD, Eberhart JK. Predicting Modifiers of Genotype-Phenotype Correlations in Craniofacial Development. International Journal of Molecular Sciences. 2023; 24(2):1222. https://doi.org/10.3390/ijms24021222
Chicago/Turabian StyleKar, Ranjeet D., and Johann K. Eberhart. 2023. "Predicting Modifiers of Genotype-Phenotype Correlations in Craniofacial Development" International Journal of Molecular Sciences 24, no. 2: 1222. https://doi.org/10.3390/ijms24021222
APA StyleKar, R. D., & Eberhart, J. K. (2023). Predicting Modifiers of Genotype-Phenotype Correlations in Craniofacial Development. International Journal of Molecular Sciences, 24(2), 1222. https://doi.org/10.3390/ijms24021222





