Comparative Transcriptome Analysis Reveals Complex Physiological Response and Gene Regulation in Peanut Roots and Leaves under Manganese Toxicity Stress
Abstract
1. Introduction
2. Results
2.1. Mn Toxicity Stress on Peanut Growth and Development
2.2. Influences of Mn Poisoning Stress on the Growth of Peanut Root
2.3. Influences of Mn Poisoning Stress on Peanut Physiological Response Indicators
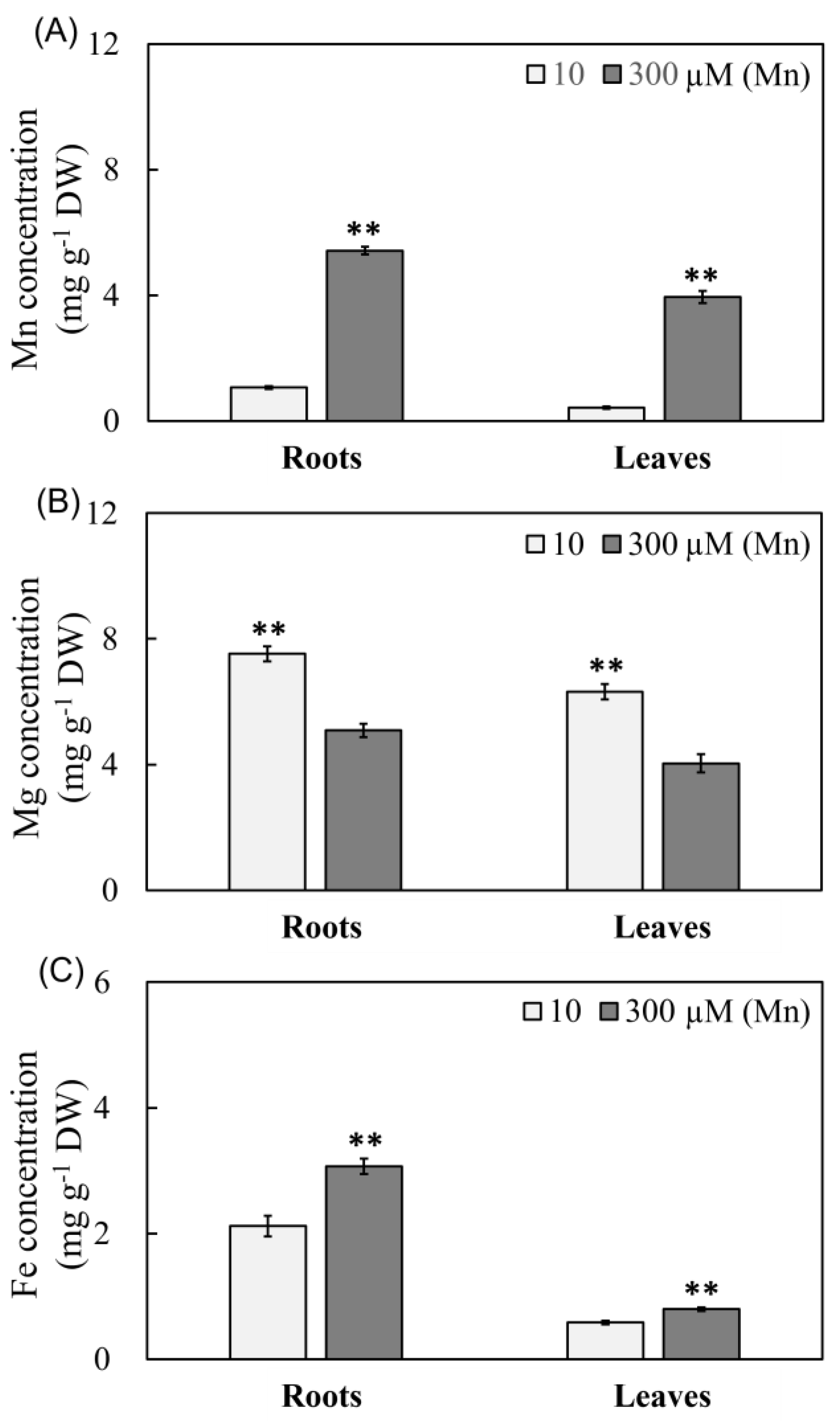
2.4. Results of Mn Effectiveness on Three Metal Ions Concentrations in Peanut Leaves and Roots
2.5. Peanut Root and Leaf Transcriptome Profiling in Response to Mn Poisoning
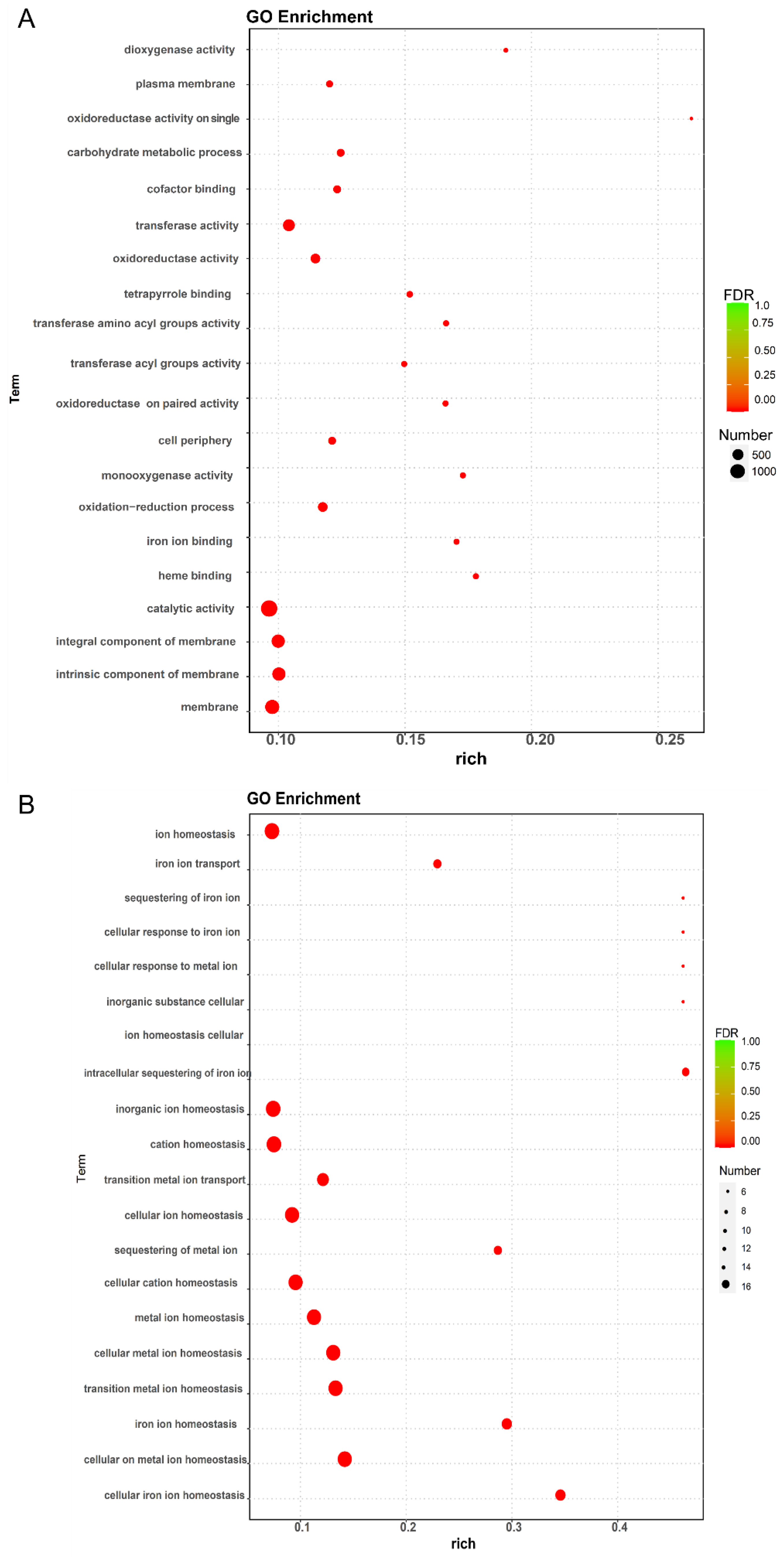
2.6. Analysis of Functional Enrichment of DEGs
2.7. Analysis of DEGs That Act as Conduits and Transporters
2.8. Distinguishing of DEGs That Act as Antioxidant Substances
2.9. Determination of DEGs That Serve as Transcription Factors
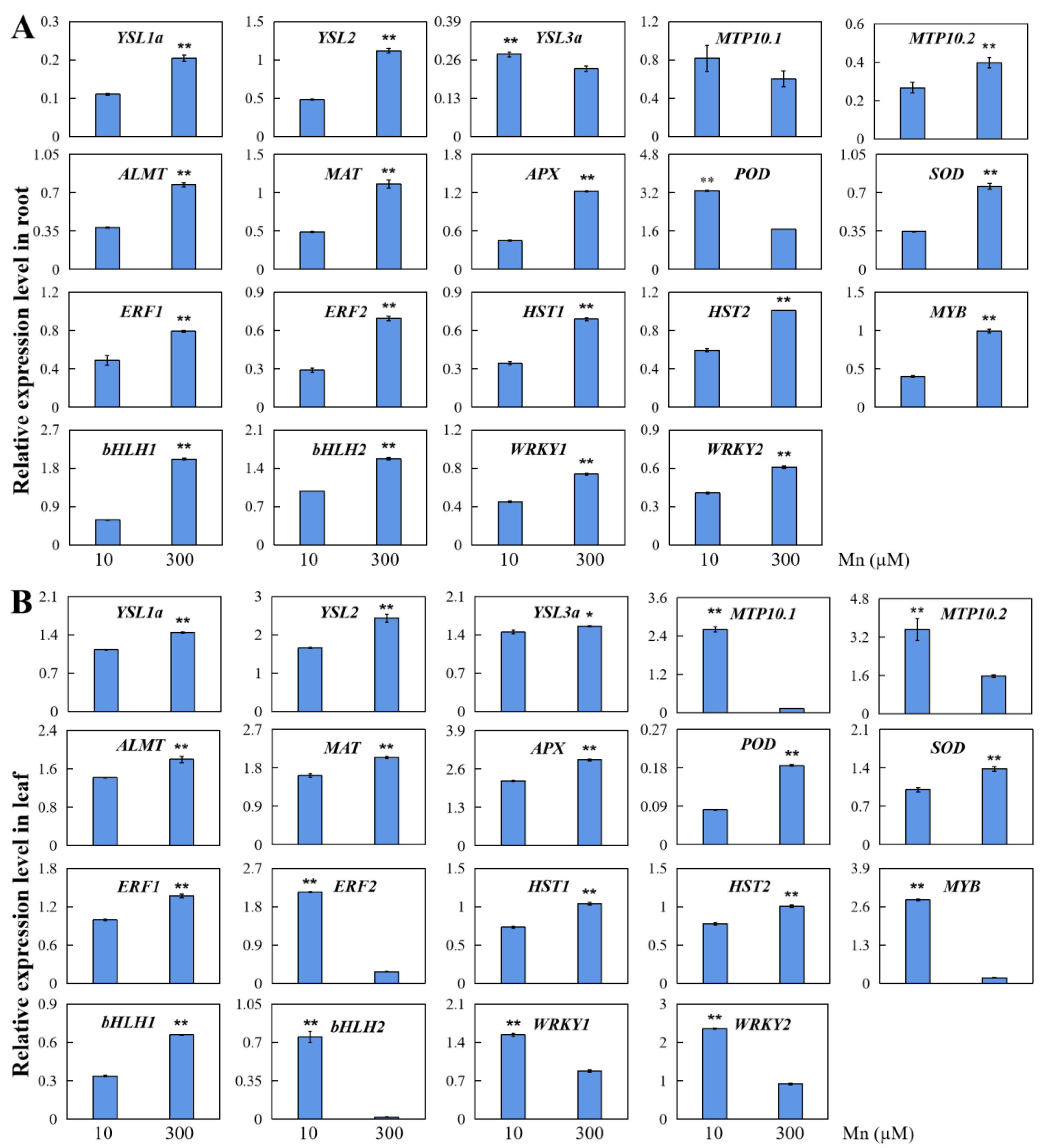
2.10. qRT-PCR Analysis of DEGs
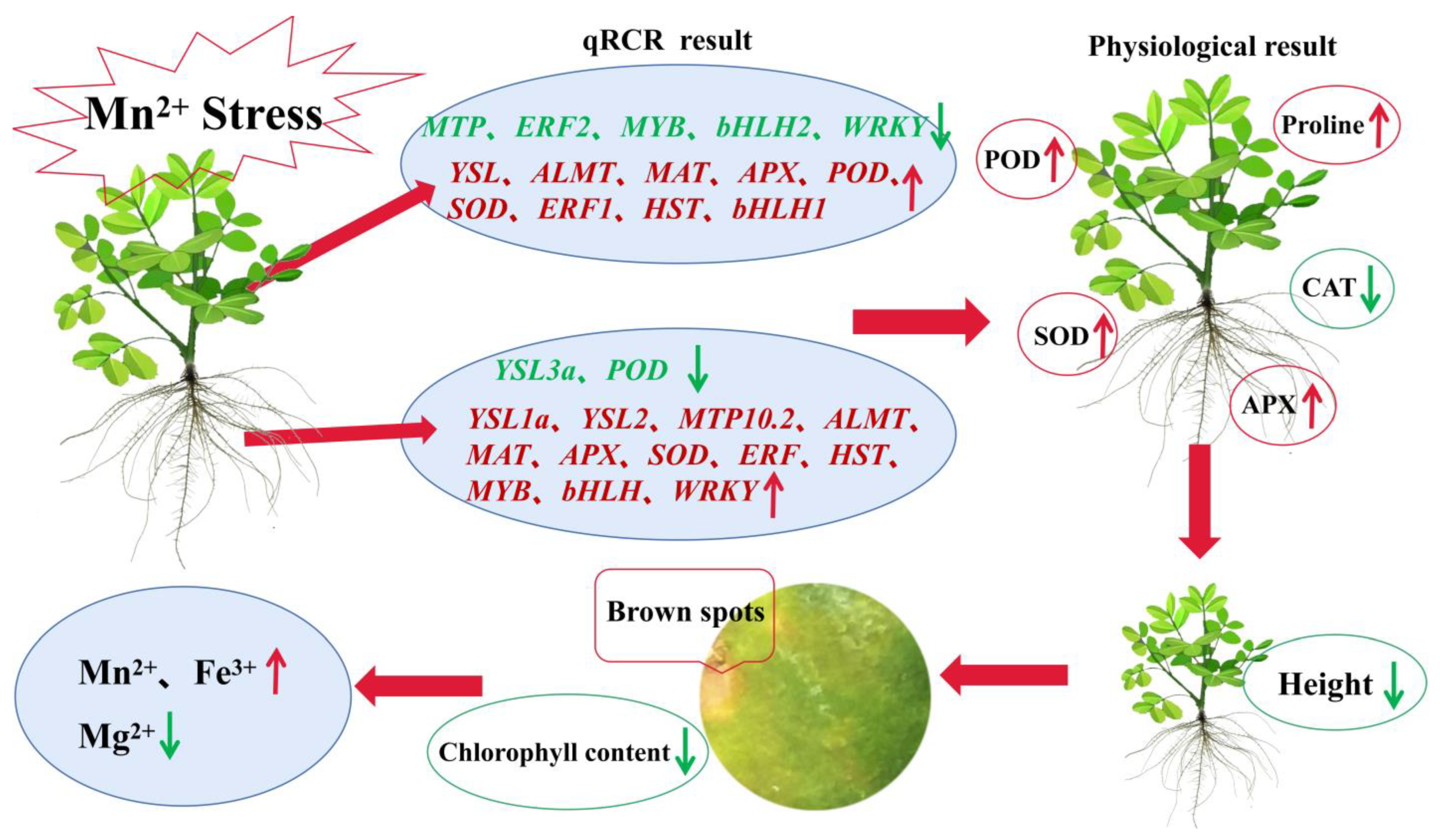
3. Discussion
4. Materials and Methods
4.1. Plant Material Sources and Processing Methods
4.2. Determination of Plant Heights, Fresh and Dry Weights of Peanuts
4.3. Brown Spot Measurement
4.4. Analysis of Morphological Root Parameters
4.5. Malondialdehyde (MDA) Content Assaying
4.6. Soluble Protein Content Assaying
4.7. Proline Content Analysis
4.8. The Enzyme Activities and SPAD Values Measurement
4.9. Concentrations of Metal Ions in the Leaves and Roots
4.10. Preparation of cDNA Libraries and Transcriptomic Sequencing Analysis
4.11. Real-Time Fluorescence Quantitative PCR (qRT-PCR) Testing
4.12. Statistic Evaluation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Li, J.F.; Jia, Y.D.; Dong, R.S.; Huang, R.; Liu, P.D.; Li, X.Y.; Wang, Z.Y.; Liu, G.D.; Chen, Z.J. Advances in the mechanisms of plant tolerance to manganese toxicity. Int. J. Mol. Sci. 2019, 20, 5096. [Google Scholar] [CrossRef]
- Liu, Y.; Xue, Y.B.; Xie, B.X.; Zhu, S.N.; Lu, X.; Liang, C.Y.; Tian, J. Complex gene regulation between young and old soybean leaves in responses to manganese toxicity. Plant Physiol. Bioch. 2020, 155, 231–242. [Google Scholar] [CrossRef]
- Yang, S.; Yi, K.; Chang, M.M.; Ling, G.Z.; Zhao, Z.K.; Li, X.F. Sequestration of Mn into the cell wall contributes to Mn tolerance in sugarcane (Saccharum officinarum L.). Plant Soil 2019, 436, 475–487. [Google Scholar] [CrossRef]
- Shao, J.F.; Yamaji, N.; Shen, R.F.; Ma, J.F. The key to Mn homeostasis in plants: Regulation of Mn transporters. Trends Plant Sci. 2017, 22, 215–224. [Google Scholar] [CrossRef]
- Pan, G.; Zhang, H.P.; Liu, P.; Xiao, Z.H.; Li, X.H.; Liu, W.S. Effects of manganese stress on phenology and biomass allocation in Xanthium strumarium from metalliferous and non-metalliferous sites. Ecotox. Environ. Saf. 2019, 172, 308–316. [Google Scholar] [CrossRef]
- Santos, E.F.; Santini, J.M.K.; Paixão, A.P.; Júnior, E.F.; Lavres, J.; Campos, M.; Reis, A.R. Physiological highlights of manganese toxicity symptoms in soybean plants: Mn toxicity responses. Plant Physiol. Bioch. 2017, 113, 6–19. [Google Scholar] [CrossRef]
- Fecht-Christoffers, M.M.; Führs, H.; Braun, H.P.; Horst, W.J. The role of hydrogen peroxide-producing and hydrogen peroxide-consuming peroxidases in the leaf apoplast of cowpea in manganese tolerance. Plant Physiol. 2006, 140, 1451–1463. [Google Scholar] [CrossRef]
- Chen, Z.J.; Sun, L.L.; Liu, P.D.; Liu, G.D.; Tian, J.; Liao, H. Malate synthesis and secretion mediated by a manganese-enhanced malate dehydrogenase confers superior manganese tolerance in Stylosanthes guianensis. Plant Physiol. 2015, 167, 176–188. [Google Scholar] [CrossRef]
- Jumrani, K.; Bhatia, V.S. Interactive effect of temperature and water stress on physiological and biochemical processes in soybean. Physiol. Mol. Biol. Pla. 2019, 25, 667–681. [Google Scholar] [CrossRef]
- Takeuchi, J.; Fukui, K.; Seto, Y.; Takaoka, Y.; Okamoto, M. Ligand–receptor interactions in plant hormone signaling. Plant J. 2021, 105, 290–306. [Google Scholar] [CrossRef]
- Shafique, F.; Ali, Q.; Saleem, M.Z.; Bhatti, Y.; Zikrea, A.; Malik, D.A. Effect of manganese and chromium toxicity on growth and photosynthetic pigments of maize. Plant Cell Biotechnol. Mol. Bio. 2021, 21, 58–64. [Google Scholar]
- Takagi, D.; Ishiyama, K.; Suganami, M.; Ushijima, T.; Fujii, T.; Tazoe, Y.; Kawasaki, M.; Noguchi, K.; Makino, A. Manganese toxicity disrupts indole acetic acid homeostasis and suppresses the CO2 assimilation reaction in rice leaves. Sci. Rep. 2021, 11, 20922. [Google Scholar] [CrossRef]
- Aprile, A.; De Bellis, L. Editorial for Special Issue “Heavy metals accumulation, toxicity, and detoxification in plants”. Int. J. Mol. Sci. 2020, 21, 4103. [Google Scholar] [CrossRef]
- Liu, Y.; Chen, J.Y.; Li, X.H.; Yang, S.X.; Hu, H.Q.; Xue, Y.B. Effects of manganese toxicity on the growth and gene expression at the seedling stage of soybean. Phyton. Int. J. Exp. Bot. 2022, 91, 975–987. [Google Scholar] [CrossRef]
- Sheng, H.; Zeng, J.; Liu, Y.; Wang, X.; Wang, Y.; Kang, H.; Fan, X.; Sha, L.; Zhang, H.; Zhou, Y. Sulfur mediated alleviation of Mn toxicity in polish wheat relates to regulating Mn allocation and improving antioxidant system. Front. Plant Sci. 2016, 7, 1382. [Google Scholar] [CrossRef]
- Horst, W.J.; Maier, P.; Fecht, M.; Naumann, A.; Wissemeier, A.H. The physiology of manganese toxicity and tolerance in Vigna unguiculata (L.) Walp. J. Plant Nutr. Soil Sci. 1999, 162, 263–274. [Google Scholar] [CrossRef]
- Delhaize, E.; Gruber, B.D.; Pittman, J.K.; White, R.G.; Leung, H.; Miao, Y.; Jiang, L.; Ryan, P.R.; Richardson, A.E. A role for the AtMTP11 gene of Arabidopsis in manganese transport and tolerance. Plant J. 2007, 51, 198–210. [Google Scholar] [CrossRef]
- Chen, Z.; Fujii, Y.; Yamaji, N.; Masuda, S.; Takemoto, Y.; Kamiya, T.; Yusuyin, Y.; Iwasaki, K.; Kato, S.; Maeshima, M.; et al. Mn tolerance in rice is mediated by MTP8.1, a member of the cation diffusion facilitator family. J. Exp. Bot. 2013, 64, 4375–4387. [Google Scholar] [CrossRef]
- Migocka, M.; Papierniak, A.; Maciaszczyk-Dziubinska, E.; Poździk, P.; Posyniak, E.; Garbiec, A.; Filleur, S. Cucumber metal transport protein MTP8 confers increased tolerance to manganese when expressed in yeast and Arabidopsis thaliana. J. Exp. Bot. 2014, 65, 5367–5384. [Google Scholar] [CrossRef]
- De la Luz Mora, M.; Rosas, A.; Ribera, A.; Rengel, Z. Differential tolerance to Mn toxicity in perennial ryegrass genotypes: Involvement of antioxidative enzymes and root exudation of carboxylates. Plant Soil 2009, 320, 79–89. [Google Scholar] [CrossRef]
- Hauck, M.; Paul, A.; Gross, S.; Raubuch, M. Manganese toxicity in epiphytic lichens: Chlorophyll degradation and interaction with iron and phosphorus. Environ. Exp. Bot. 2003, 49, 181–191. [Google Scholar] [CrossRef]
- Dou, C.; Fu, X.; Chen, X.; Shi, J.; Chen, Y. Accumulation and interaction of calcium and manganese in Phytolacca americana. Plant Sci. 2009, 177, 601–606. [Google Scholar] [CrossRef]
- Alejandro, S.; Höller, S.; Meier, B.; Peiter, E. Manganese in plants: From acquisition to subcellular allocation. Front. Plant Sci. 2020, 11, 300. [Google Scholar] [CrossRef] [PubMed]
- He, S.N.; Chen, Y.; Xiang, W.; Chen, X.W.; Wang, X.L.; Chen, Y. Carbon and nitrohgen footprints accounting of peanut and peanut oil production in China. J. Clean Prod. 2021, 291, 125964. [Google Scholar] [CrossRef]
- Boukid, F. Peanut protein-an underutilised by-product with great potential: A review. Int. J. Food Sci. Technol. 2022, 57, 5585–5591. [Google Scholar] [CrossRef]
- Yu, F.M.; Li, Y.; Li, F.R.; Li, C.M.; Liu, K.H. The effects of EDTA on plant growth and manganese (Mn) accumulation in Polygonum pubescens Blume cultured in unexplored soil, mining soil and tailing soil from the pingle Mn mine, China. Ecotox. Environ. Saf. 2019, 173, 235–242. [Google Scholar] [CrossRef]
- Chen, C.; Cao, Q.; Jiang, Q.; Li, J.; Yu, R.G.; Shi, G.R. Comparative transcriptome analysis reveals gene network regulating cadmium uptake and translocation in peanut roots under iron deficiency. BMC Plant Biol. 2019, 19, 35. [Google Scholar] [CrossRef]
- Liu, Y.; Chen, J.Y.; Li, X.H.; Xue, Y.B. Effects of manganese on growth and development of peanut (Arachis hypogaea L.) seedlings. E3S Web Conf. 2021, 245, 03013. [Google Scholar] [CrossRef]
- Cao, Z.Z.; Lin, X.Y.; Yang, Y.J.; Guan, M.Y.; Xu, P.; Chen, M.X. Gene identification and transcriptome analysis of low cadmium accumulation rice mutant (lcd1) in response to cadmium stress using MutMap and RNA-seq. BMC Plant Biol. 2019, 19, 250. [Google Scholar] [CrossRef]
- Rosa-Santos, T.M.; Silva, R.G.D.; Kumar, P.; Kottapalli, P.; Crasto, C.; Kottapalli, K.R.; Franca, S.C.; Zingaretti, S.M. Molecular mechanisms underlying sugarcane response to aluminum stress by RNA-Seq. Int. J. Mol. Sci. 2020, 21, 7934. [Google Scholar] [CrossRef]
- Peng, H.; Gao, J.; Song, X. Identification of heavy metal-responsive genes in radish (Raphanus sativus L.) through RNA-Seq meta-analysis. Sci. Hortic. 2021, 288, 110402. [Google Scholar]
- Wu, F.L.; Huang, H.Y.; Peng, M.Y.; Lai, Y.H.; Ren, Q.Q.; Zhang, J.; Huang, Z.G.; Yang, L.T.; Rensing, C.; Chen, L.S. Adaptive responses of Citrus grandis leaves to copper toxicity revealed by RNA-Seq and physiology. Int. J. Mol. Sci. 2021, 22, 12023. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Wei, Y.; Sun, L.; Tian, J.; Liao, H. Proteomic analysis reveals growth inhibition of soybean roots by manganese toxicity is associated with alteration of cell wall structure and lignification. J. Proteom. 2016, 143, 151–160. [Google Scholar] [CrossRef] [PubMed]
- González, A.; Steffen, K.; Lynch, J. Light and excess manganese implications for oxidative stress in common bean. Plant Physiol. 1998, 118, 493–504. [Google Scholar] [PubMed]
- Shi, Q.H.; Zhu, Z.J.; Xu, M.; Qian, Q.Q.; Yu, J.Q. Effect of excess manganese on the antioxidant system in Cucumis sativus L. under two light intensities. Environ. Exp. Bot. 2006, 58, 197–205. [Google Scholar] [CrossRef]
- Millaleo, R.; Reyes-Díaz, M.; Ivanov, A.; Mora, M.; Alberdi, M. Manganese as essential and toxic element for plants: Transport, accumulation and resistance mechanism. J. Soil Sci. Plant Nutr. 2010, 10, 470–481. [Google Scholar] [CrossRef]
- Chang, J.D.; Huang, S.; Yamaji, N.Y.; Zhang, W.W.; Ma, J.F.; Zhao, F.J. OsNRAMP1 transporter contributes to cadmium and manganese uptake in rice. Plant Cell Environ. 2020, 43, 2476–2491. [Google Scholar] [CrossRef]
- Liu, P.D.; Huang, R.; Hu, X.; Jia, Y.D.; Ki, J.F.; Luo, J.J.; Liu, Q.; Luo, L.J.; Liu, G.D.; Chen, Z.J. Physiological responses and proteomic changes reveal insights into Stylosanthes response to manganese toxicity. BMC Plant Biol. 2019, 19, 212. [Google Scholar] [CrossRef]
- Fecht-Christoffers, M.M.; Braun, H.P.; Lemaitre-Guillier, C.; Van-Dorsselaer, A.; Horst, W.J. Effect of manganese toxicity on the proteome of the leaf apoplast in cowpea. Plant Physiol. 2003, 133, 1935–1946. [Google Scholar] [CrossRef]
- Bot, J.L.; Kirkby, E.A.; Beusicchem, M.L.V. Manganese toxicity in tomato plants: Effects on cation uptake and distribution. J. Plant Nutr. 1990, 13, 513–525. [Google Scholar] [CrossRef]
- Foy, C.D.; Webb, H.W.; Coradetti, C.A. Differential manganese tolerances of cotton genotypes in nutrient solution. J. Plant Nutr. 1995, 18, 685–706. [Google Scholar] [CrossRef]
- Raja, V.; Majeed, U.; Kang, H.; Andrabi, K.I.; John, R. Abiotic stress: Interplay between ROS, hormones and MAPKs. Environ. Exp. Bot. 2017, 137, 142–157. [Google Scholar] [CrossRef]
- Che, Y.; Zhang, N.; Zhu, X.; Li, S.; Wang, S.; Si, H. Enhanced tolerance of the transgenic potato plants overexpressing Cu/Zn superoxide dismutase to low temperature. Sci. Hortic. 2020, 261, 108949. [Google Scholar] [CrossRef]
- Wang, D.; Li, W.X.; Li, D.; Li, L.; Luo, Z.S. Effect of high carbon dioxide treatment on reactive oxygen species accumulation and antioxidant capacity in fresh-cut pear fruit during storage. Sci. Hortic. 2021, 281, 109925. [Google Scholar] [CrossRef]
- Killi, D.; Raschi, A.; Bussotti, F. Lipid peroxidation and chlorophyll fluorescence of photosystem II performance during drought and heat stress is associated with the antioxidant capacities of C3 sunflower and C4 maize varieties. Int. J. Mol. Sci. 2020, 21, 4846. [Google Scholar] [CrossRef] [PubMed]
- Gill, S.S.; Tuteja, N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Bioch. 2010, 48, 909–930. [Google Scholar] [CrossRef]
- Wu, Y.; Gao, H.C.; Zhang, B.X.; Zhang, H.L.; Wang, Q.W.; Liu, X.L.; Luan, X.Y.; Ma, Y.S. Effects of 24-epioleucin lactones on fertility, physiology and cellular ultrastructure of soybean under salinity stress. Chin. Agr. Sci. 2017, 50, 811–821. [Google Scholar]
- Bai, Y.D.; Xiao, S.; Zhang, Z.C.; Zhang, Y.J.; Sun, H.C.; Zhang, K.; Wang, X.D.; Bai, Z.Y.; Li, C.D.; Liu, L.T. Melatonin improves the germination rate of cotton seeds under drought stress by opening pores in the seed coat. Peer J. 2020, 8, e9450. [Google Scholar] [CrossRef]
- Alisofi, S.; Einali, A.; Sangtarash, M.H. Jasmonic acid-induced metabolic responses in bitter melon (Momordica charantia) seedlings under salt stress. J. Hortic. Sci. Biotechnol. 2020, 95, 247–259. [Google Scholar] [CrossRef]
- Yoon, J.Y.; Hamayun, M.; Lee, S.K.; Lee, I.J. Methyl jasmonate alleviated salinity stress in soybean. J. Crop. Sci. Biotechnol. 2009, 12, 63–68. [Google Scholar] [CrossRef]
- Zhu, Q.Y.; Xu, P.X.; Lei, L.L.; Jing, Y.X. Transcriptome analysis reveals decreased accumulation and toxicity of Cd in upland rice inoculated with arbuscular mycorrhizal fungi. Appl. Soil. Ecol. 2022, 177, 104501. [Google Scholar] [CrossRef]
- Ren, Q.Q.; Huang, Z.R.; Huang, W.L.; Huang, W.T.; Chen, H.H.; Yang, L.T.; Ye, X.; Chen, L.S. Physiological and molecular adaptations of Citrus grandis roots to long-term copper excess revealed by physiology, metabolome and transcriptome. Environ. Exp. Bot. 2022, 203, 105049. [Google Scholar] [CrossRef]
- Chen, S.S.; Qi, X.Y.; Feng, J.; Chen, H.J.; Qin, Z.Y.; Wang, H.D.; Deng, Y.M. Biochemistry and transcriptome analyses reveal key genes and pathways involved in high-aluminum stress response and tolerance in hydrangea sepals. Plant Physiol. Biochem. 2022, 185, 268–278. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; Xiao, X.; Ou, Y.; Wu, X.; Xu, G. Root transcriptome analysis on the grape genotypes with contrast translocation pattern of excess manganese from root to shoot. Plant Soil 2015, 387, 49–67. [Google Scholar] [CrossRef]
- Xue, Y.B.; Chen, J.Y.; Li, X.H.; Liu, Y. Transcriptome analysis of soybean leaves response to manganese toxicity. Biotechnol. Biotechnol. Equip. 2021, 35, 1043–1051. [Google Scholar] [CrossRef]
- Huang, H.M.; Zhao, Y.L.; Xu, Z.G.; Zhang, W.; Jiang, K.K. Physiological responses of Broussonetia papyrifera to manganese stress, a candidate plant for phytoremediation. Ecotox. Environ. Saf. 2019, 181, 18–25. [Google Scholar] [CrossRef]
- Ishimaru, Y.; Masuda, H.; Bashir, K.; Inoue, H.; Tsukamoto, T.; Takahashi, M.; Nakanishi, H.; Aoki, N.; Hirose, T.; Ohsugi, R.; et al. Rice metal-nicotianamine transporter, OsYSL2, is required for the long-distance transport of iron and manganese. Plant J. 2010, 62, 379–390. [Google Scholar] [CrossRef]
- Wintz, H.; Fox, T.; Wu, Y.Y.; Feng, V.; Chen, W.; Chang, H.S.; Zhu, T.; Vulpe, C. Expression profiles of Arabidopsis thaliana in mineral deficiencies reveal novel transporters involved in metal homeostasis. J. Biol. Chem. 2003, 278, 47644–47653. [Google Scholar] [CrossRef]
- Wu, Z.; Liang, F.; Hong, B.; Young, J.C.; Sussman, M.R.; Harper, J.F.; Sze, H. An endoplasmic reticulum-bound Ca2+/Mn2+ pump, ECA1, supports plant growth and confers tolerance to Mn2+ stress. Plant Physiol. 2002, 130, 128–137. [Google Scholar] [CrossRef]
- Mills, R.F.; Doherty, M.L.; Lopez-Marques, R.L.; Weimar, T.; Dupree, P.; Palmgren, M.G.; Pittman, J.K.; Williams, L.E. ECA3, a Golgi-localized P2A-type ATPase, plays a crucial role in manganese nutrition in Arabidopsis. Plant Physiol. 2008, 146, 116–128. [Google Scholar] [CrossRef]
- Zhang, M.; Liu, B. Identification of a rice metal tolerance protein OsMTP11 as a manganese transporter. PloS ONE 2017, 12, e0174987. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.Y.; Li, Q.H.; Xu, W.L.; Zhao, H.; Guo, F.; Wang, P.; Wang, Y.; Ni, D.; Wang, M.; Wei, C. Identification of MTP gene family in tea plant (Camellia sinensis L.) and characterization of CsMTP8.2 in manganese toxicity. Ecotox. Environ. Saf. 2020, 202, 110904. [Google Scholar] [CrossRef] [PubMed]
- Goss, M.J.; Carvalho, M.J.G.P.R. Manganese toxicity: The significance of magnesium for the sensitivity of wheat plants. Plant Soil 1992, 139, 91–98. [Google Scholar] [CrossRef]
- Ribera-Fonseca, A.; Inostroza-Blancheteau, C.; Cartes, P.; Rengel, Z.; Mora, M.L. Early induction of Fe-SOD gene expression is involved in tolerance to Mn toxicity in perennial ryegrass. Plant Physiol. Bioch. 2013, 73, 77–82. [Google Scholar] [CrossRef]
- Yamada, K.; Nagano, A.J.; Nishina, M.; Hara-Nishimura, I.; Nishimura, M. Identification of two novel endoplasmic reticulum body-specific integral membrane proteins. Plant Physiol. 2013, 161, 108–120. [Google Scholar] [CrossRef]
- Pornaro, C.; Macolino, S.; Menegon, A.; Richardson, M. Winrhizo technology for measuring morphological traits of bermudagrass stolons. Agron J. 2017, 109, 3007–3010. [Google Scholar] [CrossRef]
- Chen, G.J.; Zheng, D.F.; Feng, N.J.; Zhou, H.; Mu, D.W.; Liu, L.; Zhao, L.M.; Shen, X.F.; Rao, G.H.; Li, T.Z. Effects of exogenous salicylic acid and abscisic acid on growth, photosynthesis and antioxidant system of rice. Chil. J. Agr. Res. 2022, 82, 21–32. [Google Scholar] [CrossRef]
- De Araújo Silva, M.M.; Ferreira, L.T.; De Vasconcelos, F.M.T.; Willadino, L.; Camara, T.R.; Dos Santos, D.Y.A.C.; De Oliveira, A.F.M. Water stress-induced responses in the growth, cuticular wax composition, chloroplast pigments and soluble protein content, and redox metabolism of two genotypes of Ricinus communis L. J. Plant Growth Regul. 2021, 40, 342–352. [Google Scholar] [CrossRef]
- Ghaffari, H.; Tadayon, M.R.; Nadeem, M.; Cheema, M.; Razmjoo, J. Proline-mediated changes in antioxidant enzymatic activities and the physiology of sugar beet under drought stress. Acta Physiol Plant 2019, 41, 23. [Google Scholar] [CrossRef]
- Sadak, M.S.; Abdalla, A.M.; Abd Elhamid, E.M.; Ezzo, M.I. Role of melatonin in improving growth, yield quantity and quality of Moringa oleifera L. plant under drought stress. Bull. Nat. Res. Cent. 2020, 44, 18. [Google Scholar] [CrossRef]
- Rácz, A.; Hideg, É.; Czégény, G. Selective responses of class III plant peroxidase isoforms to environmentally relevant UV-B doses. J. Plant Physiol. 2018, 221, 101–106. [Google Scholar] [CrossRef] [PubMed]
- Ekinci, M.; Ors, S.; Yildirim, E.; Turan, M.; Sahin, U.; Dursun, A.; Kul, R. Determination of physiological indices and some antioxidant enzymes of chard exposed to nitric oxide under drought stress. Russ. J. Plant Physiol. 2020, 67, 740–749. [Google Scholar] [CrossRef]
- Li, C.G.; Li, J.X.; Du, X.H.; Zhang, J.X.; Zou, Y.R.; Liu, Y.D.; Li, Y.; Lin, H.G.; Li, H.; Liu, D.; et al. Chloroplast thylakoidal ascorbate peroxidase, PtotAPX, has enhanced resistance to oxidative stress in Populus tomentosa. Int. J. Mol. Sci. 2022, 23, 3340. [Google Scholar] [CrossRef] [PubMed]
- Uddling, J.; Gelang-Alfredsson, J.; Piikki, K.; Pleijel, H. Evaluating the relationship between leaf chlorophyll concentration and SPAD-502 chlorophyll meter readings. Photosynth. Res. 2007, 91, 37–46. [Google Scholar] [CrossRef]
- Kalogiouri, N.P.; Manousi, N.; Zachariadis, G.A. Determination of the toxic and nutrient element content of almonds, walnuts, hazelnuts and pistachios by ICP-AES. Separations 2021, 8, 28. [Google Scholar] [CrossRef]
- Xue, Y.B.; Zhuang, Q.L.; Zhu, S.N.; Xiao, B.X.; Liang, C.Y.; Liao, H.; Tian, J. Genome wide transcriptome analysis reveals complex regulatory mechanisms underlying phosphate homeostasis in soybean nodules. Int. J. Mol. Sci. 2018, 19, 2924. [Google Scholar] [CrossRef]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef]
- Zhuang, W.; Chen, H.; Yang, M.; Wang, J.; Pandey, M.K.; Zhang, C.; Chang, W.C.; Zhang, L.; Zhang, X.; Tang, R.; et al. The genome of cultivated peanut provides insight into legume karyotypes, polyploid evolution and crop domestication. Nat. Genet. 2019, 51, 865–876. [Google Scholar] [CrossRef]
- Wang, L.; Feng, Z.; Wang, X.; Zhang, X. DEGseq: An R package for identifying differentially expressed genes from RNA-seq data. Bioinform 2010, 26, 136–138. [Google Scholar] [CrossRef]
- Thimm, O.; Bläsing, O.; Gibon, Y.; Nagel, A.; Meyer, S.; Krüger, P.; Selbig, J.; Müller, L.A.; Rhee, S.Y.; Stitt, M. MapMan: A user-driven tool to display genomic data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 2004, 37, 914–939. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.Y.; Piñeros, M.A.; Tian, J.; Yao, Z.F.; Sun, L.L.; Liu, J.P.; Shaff, J.; Coluccio, A.; Kochian, L.V.; Liao, H. Low pH, aluminum, and phosphorus coordinately regulate malate exudation through GmALMT1 to improve soybean adaptation to acid soils. Plant Physiol. 2013, 161, 1347–1361. [Google Scholar] [CrossRef] [PubMed]
- Li, X.X.; Zhao, J.; Tan, Z.Y.; Zeng, R.S.; Liao, H. GmEXPB2, a cell wall β-expansin, affects soybean nodulation through modifying root architecture and promoting nodule formation and development. Plant Physiol. 2015, 169, 2640–2653. [Google Scholar] [CrossRef] [PubMed]

| Parameters of Peanut Development | Concentrations of Mn (μM) | |
|---|---|---|
| 10 | 300 | |
| Height of plant (cm) | 25.10 ± 0.36 ** | 20.93 ± 0.17 |
| SPAD values | 44.27 ± 0.09 ** | 39.53 ± 0.29 |
| Number of brown spots on the fifth leaves | 0 | 60.00 ± 2.45 ** |
| Fresh weights of the shoot (g) | 6.06 ± 0.96 ** | 2.19 ± 0.12 |
| Fresh weights of the root (g) | 1.57 ± 0.35 ** | 0.68 ± 0.02 |
| Shoot dry weights | 0.80 ± 0.13 ** | 0.37 ± 0.03 |
| Root dry weights | 0.16 ± 0.03 * | 0.08 ± 0.00 |
| Parameters of Peanut Root Growth | Concentrations of Mn (μM) | |
|---|---|---|
| 10 | 300 | |
| Average diameter of root (mm) | 0.89 ± 0.03 | 0.88 ± 0.01 |
| Volume of root (cm3) | 184.98 ± 4.88 ** | 16.71 ± 3.39 |
| Surface area of root (cm2) | 1393.15 ± 18.23 ** | 459.97 ± 59.43 |
| Total length of root (cm) | 4239.83 ± 730.85 ** | 1741.37 ± 203.57 |
| Root tip number | 12,789.67 ± 781.01 ** | 6473.67 ± 504.61 |
| Physiological Indices | Leaves | Roots | ||
|---|---|---|---|---|
| 10 μM | 300 μM | 10 μM | 300 μM | |
| Activity of POD (U/g FW) | 1833.33 ± 155.90 ** | 16,500.00 ± 810.09 | 32,500.00 ± 1503.47 | 32,000.00 ± 2215.01 |
| Activity of CAT (U/g FW) | 1787.33 ± 84.98 ** | 1516.67 ± 62.36 | 500.00 ± 40.82 ** | 133.33 ± 23.57 |
| Activity of APX (U/g FW) | 206.67 ± 30.91 | 603.33 ± 26.25 ** | 206.67 ± 4.71 | 376.67 ± 12.47 ** |
| Activity of SOD (U/g FW) | 542.36 ± 15.00 | 832.08 ± 11.60 ** | 319.80 ± 11.60 | 388.97 ± 6.18 ** |
| Content of soluble protein (mg/g FW) | 36,389.47 ± 561.00 * | 33,638.26 ± 816.76 | 15,787.37 ± 843.39 | 17,988.34 ± 154.62 * |
| Content of MDA (μM/g FW) | 0.033 ± 0.002 * | 0.026 ± 0.002 | 0.012 ± 0.001 | 0.014 ± 0.002 |
| Content of proline (μg/g FW) | 66.68 ± 17.02 | 277.55 ± 32.05 ** | 19.24 ± 2.00 | 39.38 ± 6.08 ** |
| Number of Total Expressed Genes | Number of Up-Regulated DEGs | Number of Down-Regulated DEGs | Number of DEGs Identified in Both Roots and Leaves | Number of DEGs Identified Only in Roots or Leaves | |
|---|---|---|---|---|---|
| Leaves | 47,675 | 3566 | 1023 | 310 | 4718 |
| Roots | 52,320 | 542 | 207 |
| Gene ID | log2FoldChange | Description | |
|---|---|---|---|
| Roots | Leaves | ||
| AH11G27280 | 1.02 | - | Oligopeptide transporter |
| AH10G04950 | 1.08 | - | Oligopeptide transporter |
| AH14G01590 | −1.97 | 1.22 | Oligopeptide transporter |
| AH17G01740 | - | −1.57 | Oligopeptide transporter |
| AH11G23790 | - | −2.13 | Oligopeptide transporter |
| AH01G08540 | - | 1.46 | Oligopeptide transporter |
| AH11G00400 | - | 1.02 | Oligopeptide transporter |
| AH14G23400 | 1.78 | 1.71 | Calcium-transporting ATPase |
| AH20G34940 | 1.42 | 4.30 | Calcium-transporting ATPase |
| AH06G11590 | 1.27 | 2.15 | Calcium-transporting ATPase |
| AH16G01740 | 1.19 | 1.67 | Calcium-transporting ATPase |
| AH17G01280 | 1.14 | 1.71 | Calcium-transporting ATPase |
| AH01G07000 | 1.04 | 1.32 | Calcium-transporting ATPase |
| AH12G04160 | - | 4.24 | Calcium-transporting ATPase |
| AH10G27230 | - | 3.18 | Calcium-transporting ATPase |
| AH10G27240 | - | 3.13 | Calcium-transporting ATPase |
| AH07G01100 | - | 2.68 | Calcium-transporting ATPase |
| AH13G12150 | - | 2.47 | Calcium-transporting ATPase |
| AH11G01720 | - | 2.31 | Calcium-transporting ATPase |
| AH19G39830 | - | 2.04 | Calcium-transporting ATPase |
| AH02G03820 | - | 1.66 | Calcium-transporting ATPase |
| AH01G06980 | - | 1.60 | Calcium-transporting ATPase |
| AH15G14410 | - | 1.53 | Calcium-transporting ATPase |
| AH10G27570 | - | 1.11 | Calcium-transporting ATPase |
| AH09G00230 | - | 1.02 | Calcium-transporting ATPase |
| AH15G14430 | - | −2.20 | Calcium-transporting ATPase |
| AH15G32580 | −1.14 | 1.24 | Metal-nicotianamine transporter YSL |
| AH09G18140 | −3.51 | - | Metal-nicotianamine transporter YSL |
| AH17G27180 | - | 1.78 | Metal-nicotianamine transporter YSL |
| AH08G03900 | - | 1.57 | Metal-nicotianamine transporter YSL |
| AH11G01940 | - | 1.42 | Metal-nicotianamine transporter YSL |
| AH05G22970 | - | 1.04 | Metal-nicotianamine transporter YSL |
| AH03G40290 | 2.27 | 4.22 | High-affinity nitrate transporter |
| AH01G16560 | −2.59 | - | High-affinity nitrate transporter |
| AH13G43220 | - | 1.63 | High-affinity nitrate transporter |
| AH06G25880 | - | 1.25 | High-affinity nitrate transporter |
| AH03G23240 | - | −3.41 | High-affinity nitrate transporter |
| AH10G30290 | 2.74 | - | Metal tolerance protein |
| AH13G56980 | 2.53 | - | Metal tolerance protein |
| AH16G14430 | - | 1.40 | Metal tolerance protein |
| AH09G00220 | - | 1.10 | Metal tolerance protein |
| AH13G46720 | 1.88 | - | Potassium transporter |
| AH16G06470 | 1.52 | 2.00 | Potassium transporter |
| AH06G03660 | 1.27 | 2.96 | Potassium transporter |
| AH01G04720 | 1.24 | - | Potassium transporter |
| AH11G03840 | −1.99 | - | Potassium transporter |
| AH06G07520 | - | 4.56 | Potassium transporter |
| AH16G14880 | - | 4.07 | Potassium transporter |
| AH03G40840 | - | 3.23 | Potassium transporter |
| AH10G19740 | - | 2.72 | Potassium transporter |
| AH13G43650 | - | 1.84 | Potassium transporter |
| AH20G23230 | - | 1.42 | Potassium transporter |
| AH13G50440 | - | 1.20 | Potassium transporter |
| AH18G28880 | 1.07 | - | Sulfate transporter |
| AH20G08820 | −1.11 | 4.28 | Sulfate transporter |
| AH10G05870 | −1.20 | - | Sulfate transporter |
| AH20G08800 | −3.04 | - | Sulfate transporter |
| AH06G11450 | - | 2.26 | Sulfate transporter |
| AH10G09900 | - | 1.30 | Sulfate transporter |
| AH14G17090 | - | 1.01 | Sulfate transporter |
| AH17G34960 | - | −1.21 | Sulfate transporter |
| AH06G15690 | - | −1.44 | Sulfate transporter |
| AH09G31000 | 1.66 | - | Aluminum-activated malate transporter |
| AH08G16020 | 1.37 | - | Aluminum-activated malate transporter |
| AH05G32880 | −1.56 | −2.16 | Aluminum-activated malate transporter |
| AH13G14340 | - | 7.25 | Aluminum-activated malate transporter |
| AH06G10950 | - | 1.60 | Aluminum-activated malate transporter |
| AH16G02290 | - | 1.26 | Aluminum-activated malate transporter |
| AH05G03780 | - | −2.05 | Aluminum-activated malate transporter |
| AH01G13250 | - | −2.81 | Aluminum-activated malate transporter |
| AH11G13250 | - | −3.12 | Aluminum-activated malate transporter |
| AH05G36140 | −1.13 | −3.35 | Vacuolar iron transporter |
| AH15G37400 | −1.16 | −2.89 | Vacuolar iron transporter |
| AH13G48510 | −1.58 | - | Vacuolar iron transporter |
| AH03G45850 | −1.78 | - | Vacuolar iron transporter |
| AH20G09060 | −2.07 | - | Vacuolar iron transporter |
| AH03G45860 | −2.18 | - | Vacuolar iron transporter |
| AH19G35130 | −2.49 | - | Vacuolar iron transporter |
| AH10G06170 | −2.60 | - | Vacuolar iron transporter |
| AH13G48520 | −2.67 | - | Vacuolar iron transporter |
| AH13G48530 | −3.08 | - | Vacuolar iron transporter |
| AH04G24840 | - | −1.70 | Vacuolar iron transporter |
| AH05G30820 | 1.05 | −2.03 | Zinc transporter |
| AH13G05680 | −1.45 | - | Zinc transporter |
| AH04G00880 | - | 3.97 | Zinc transporter |
| AH18G03990 | - | 1.19 | Zinc transporter |
| AH17G00620 | - | −2.31 | Zinc transporter |
| AH15G22860 | - | −2.77 | Zinc transporter |
| AH18G31650 | −1.39 | - | Boron transporter |
| AH08G26430 | −1.40 | 3.33 | Boron transporter |
| AH12G28290 | - | 2.05 | Magnesium transporter |
| AH13G08910 | - | 1.21 | Magnesium transporter |
| AH16G38110 | - | 1.13 | Magnesium transporter |
| AH16G35450 | - | −1.22 | Magnesium transporter |
| AH17G04910 | - | 3.91 | Calcium permeable stress-gated cation channel |
| AH00G05400 | - | 3.42 | Calcium permeable stress-gated cation channel |
| AH11G27870 | - | 2.62 | Calcium permeable stress-gated cation channel |
| Gene ID | log2FoldChange | Description | |
|---|---|---|---|
| Root | Leaf | ||
| AH13G38160 | - | 1.28 | L-ascorbate oxidase |
| AH17G04410 | - | 2.7 | L-ascorbate oxidase |
| AH03G34360 | - | 3.01 | L-ascorbate oxidase |
| AH07G17320 | 3.5 | - | Peroxidase |
| AH08G26960 | −1.35 | - | Peroxidase |
| AH00G03280 | - | 2.84 | Peroxidase |
| AH01G05760 | 2.43 | - | Peroxidase |
| AH01G05780 | 1.43 | - | Peroxidase |
| AH20G08730 | 1.32 | - | Peroxidase |
| AH14G25410 | 1.07 | 3.29 | Peroxidase |
| AH14G25430 | −1.04 | - | Peroxidase |
| AH09G31660 | −2.22 | −1.15 | Peroxidase |
| AH19G36370 | −2.53 | - | Peroxidase |
| AH07G12590 | - | 5.9 | Peroxidase |
| AH04G21700 | - | 3.7 | Peroxidase |
| AH20G08720 | - | 3.36 | Peroxidase |
| AH10G05800 | - | 2.75 | Peroxidase |
| AH10G05810 | - | 2.68 | Peroxidase |
| AH04G21680 | - | 1.15 | Peroxidase |
| AH13G48270 | - | −1.78 | Peroxidase |
| AH15G09760 | 1.5 | - | Peroxidase |
| AH05G12980 | 1.3 | - | Peroxidase |
| AH09G08990 | 1.21 | - | Peroxidase |
| AH15G33990 | 1.14 | - | Peroxidase |
| AH20G14810 | 1.09 | - | Peroxidase |
| AH18G05400 | 1.02 | −2.08 | Peroxidase |
| AH08G15110 | −1.07 | - | Peroxidase |
| AH04G09840 | −1.07 | 1.53 | Peroxidase |
| AH14G08440 | −1.1 | 1.95 | Peroxidase |
| AH01G05770 | −1.19 | - | Peroxidase |
| AH10G20050 | −1.29 | - | Peroxidase |
| AH06G20840 | −1.99 | - | Peroxidase |
| AH16G25800 | −2.23 | - | Peroxidase |
| AH14G08450 | - | 6.33 | Peroxidase |
| AH14G08420 | - | 5.79 | Peroxidase |
| AH18G10570 | - | 4.14 | Peroxidase |
| AH06G26990 | - | 3.94 | Peroxidase |
| AH04G09830 | - | 3.37 | Peroxidase |
| AH04G09870 | - | 2.71 | Peroxidase |
| AH17G11990 | - | 2.29 | Peroxidase |
| AH14G08430 | - | 1.96 | Peroxidase |
| AH06G24710 | - | 1.91 | Peroxidase |
| AH01G31200 | - | 1.87 | Peroxidase |
| AH12G38300 | - | 1.63 | Peroxidase |
| AH04G09850 | - | 1.63 | Peroxidase |
| AH16G25780 | - | 1.62 | Peroxidase |
| AH09G19280 | - | 1.48 | Peroxidase |
| AH15G01790 | - | 1.1 | Peroxidase |
| AH11G28810 | - | −1.05 | Peroxidase |
| AH01G21450 | - | −1.15 | Peroxidase |
| AH10G10440 | - | −1.32 | Peroxidase |
| AH00G04650 | - | −1.79 | Peroxidase |
| AH18G07180 | - | −2.49 | Peroxidase |
| AH03G05320 | 1.22 | - | Peroxidase |
| AH03G07350 | - | 3.32 | Peroxidase |
| AH13G09650 | - | 2.47 | Peroxidase |
| AH19G19840 | - | 2.6 | Superoxide dismutase |
| Gene ID | log2FoldChange | Description | |
|---|---|---|---|
| Root | Leaf | ||
| AH19G30930 | - | 1.24 | AP2/ERF and B3 domain-containing transcription factor |
| AH09G30280 | - | 1.03 | AP2/ERF and B3 domain-containing transcription factor |
| AH17G24920 | - | 1.26 | bZIP transcription factor |
| AH02G13610 | - | 1.22 | bZIP transcription factor |
| AH02G08810 | - | 1.82 | Ethylene-responsive transcription factor |
| AH03G24670 | - | 5.29 | Ethylene-responsive transcription factor |
| AH19G42640 | - | 6.77 | Ethylene-responsive transcription factor |
| AH18G34060 | - | 4.82 | Ethylene-responsive transcription factor |
| AH13G35500 | - | 2.19 | Ethylene-responsive transcription factor |
| AH02G05220 | - | −1.23 | Ethylene-responsive transcription factor |
| AH03G39850 | - | 3.87 | Ethylene-responsive transcription factor |
| AH16G07170 | - | 3.34 | Ethylene-responsive transcription factor |
| AH09G10470 | 1.22 | - | Ethylene-responsive transcription factor |
| AH17G30770 | −1.38 | - | Ethylene-responsive transcription factor |
| AH20G26610 | 1.2 | 2.71 | Ethylene-responsive transcription factor |
| AH16G37720 | 1.03 | 1.09 | Ethylene-responsive transcription factor |
| AH20G06980 | - | 1.35 | Ethylene-responsive transcription factor |
| AH05G03100 | - | 1.39 | Ethylene-responsive transcription factor |
| AH09G21370 | - | 1.77 | Ethylene-responsive transcription factor |
| AH08G21930 | - | 2.49 | Ethylene-responsive transcription factor |
| AH10G02570 | - | 2.18 | Ethylene-responsive transcription factor |
| AH20G07000 | - | 2.07 | Ethylene-responsive transcription factor |
| AH11G14950 | - | 2.1 | Ethylene-responsive transcription factor |
| AH05G03090 | - | 1.97 | Ethylene-responsive transcription factor |
| AH01G14550 | - | 1.17 | Ethylene-responsive transcription factor |
| AH03G15340 | - | 3.27 | Ethylene-responsive transcription factor |
| AH13G17810 | - | 3.2 | Ethylene-responsive transcription factor |
| AH10G20310 | - | 3.88 | Ethylene-responsive transcription factor |
| AH13G27750 | - | 4.36 | Ethylene-responsive transcription factor |
| AH16G13670 | - | −1.67 | Ethylene-responsive transcription factor |
| AH09G25330 | - | 2.02 | Ethylene-responsive transcription factor |
| AH08G17790 | - | 2.72 | Ethylene-responsive transcription factor |
| AH13G32380 | - | 2.07 | Ethylene-responsive transcription factor |
| AH12G16570 | - | 3.32 | Ethylene-responsive transcription factor |
| AH02G14060 | - | 2.8 | Ethylene-responsive transcription factor |
| AH10G14340 | - | 5.22 | Ethylene-responsive transcription factor |
| AH20G18990 | - | 5.12 | Ethylene-responsive transcription factor |
| AH10G01270 | - | 1.11 | Ethylene-responsive transcription factor |
| AH06G04120 | - | 6.22 | Ethylene-responsive transcription factor |
| AH17G18750 | - | 2.06 | Ethylene-responsive transcription factor |
| AH20G13550 | - | 1.39 | Ethylene-responsive transcription factor |
| AH03G01980 | - | −1.4 | Ethylene-responsive transcription factor |
| AH00G01740 | - | −3.06 | Ethylene-responsive transcription factor |
| AH19G32730 | - | 1.65 | GATA transcription factor |
| AH09G34390 | - | 1.64 | GATA transcription factor |
| AH01G26750 | - | 1.38 | GATA transcription factor |
| AH12G17550 | - | −1.08 | GATA transcription factor |
| AH01G21520 | - | 2.2 | Heat stress transcription factor |
| AH06G11540 | - | 2.31 | Heat stress transcription factor |
| AH13G40070 | - | 3.18 | Heat stress transcription factor |
| AH03G36820 | - | 3.21 | Heat stress transcription factor |
| AH15G35830 | - | 2.7 | Heat stress transcription factor |
| AH05G34690 | - | 2.26 | Heat stress transcription factor |
| AH05G17170 | - | 1.21 | Heat stress transcription factor |
| AH06G02170 | - | 1.31 | Heat stress transcription factor |
| AH05G34050 | - | 1.27 | MADS-box transcription factor |
| AH01G14370 | 4.18 | - | Myb family transcription factor |
| AH14G23270 | - | −1.95 | Myb family transcription factor |
| AH12G25410 | - | −2.45 | Myb family transcription factor |
| AH14G07590 | - | 1.61 | Myb family transcription factor |
| AH13G01940 | - | −1.4 | Myb family transcription factor |
| AH14G23270 | −2.11 | - | Myb family transcription factor |
| AH04G20450 | - | −2.16 | Myb family transcription factor |
| AH02G22970 | - | −1.89 | Myb family transcription factor |
| AH01G14370 | - | 2 | Myb family transcription factor |
| AH13G54280 | - | −1.38 | Myb family transcription factor |
| AH19G15410 | −4.5 | - | Myb family transcription factor |
| AH04G18490 | - | −2.38 | Myb family transcription factor |
| AH06G04320 | - | −2.25 | Myb family transcription factor |
| AH03G21480 | - | 1.72 | Myb family transcription factor |
| AH08G29090 | - | 4.62 | Myb family transcription factor |
| AH12G03980 | - | 3.02 | Myb family transcription factor |
| AH14G44850 | - | −1.8 | Myb family transcription factor |
| AH16G39650 | - | 1.59 | Myb family transcription factor |
| AH08G03280 | - | 3.94 | Myb family transcription factor |
| AH17G26480 | - | 5.78 | Myb family transcription factor |
| AH16G42280 | - | 2.5 | Myb family transcription factor |
| AH03G42580 | - | 1.16 | NAC transcription factor |
| AH08G11600 | - | −1.73 | Nuclear transcription factor |
| AH05G27340 | - | 1.32 | BEE transcription factor |
| AH03G21460 | - | 1.39 | BEE transcription factor |
| AH08G08420 | −3.78 | - | bHLH transcription factor |
| AH16G10910 | −1.12 | - | bHLH transcription factor |
| AH09G15020 | −1.67 | - | bHLH transcription factor |
| AH03G03430 | - | −1.13 | bHLH transcription factor |
| AH08G25020 | - | 1.32 | bHLH transcription factor |
| AH18G29680 | - | 1.68 | bHLH transcription factor |
| AH11G35750 | - | 5.31 | bHLH transcription factor |
| AH01G22060 | - | 4.16 | bHLH transcription factor |
| AH03G05080 | - | 1.58 | bHLH transcription factor |
| AH02G04370 | - | −2.12 | bHLH transcription factor |
| AH12G04850 | - | −1.99 | bHLH transcription factor |
| AH05G39020 | - | 1.01 | bHLH transcription factor |
| AH18G02090 | - | −1.03 | bHLH transcription factor |
| AH17G15960 | - | −2.33 | bHLH transcription factor |
| AH07G20060 | - | 1.29 | bHLH transcription factor |
| AH17G17690 | - | 1.36 | bHLH transcription factor |
| AH05G16400 | - | 5.21 | CPC transcription factor |
| AH15G06520 | - | 5.2 | CPC transcription factor |
| AH17G07910 | - | 1.2 | DIVARICATA transcription factor |
| AH09G29640 | - | 1.06 | DIVARICATA transcription factor |
| AH15G18860 | - | −1.41 | FAMA transcription factor |
| AH16G03850 | - | 3.04 | HBP-1b transcription factor |
| AH02G16860 | - | 1.68 | HBP-1b transcription factor |
| AH12G19950 | - | 4.38 | HBP-1b transcription factor |
| AH17G10210 | −2.85 | - | KAN transcription factor |
| AH16G05200 | - | 1.45 | ORG transcription factor |
| AH19G38650 | - | 1.85 | PERIANTHIA transcription factor |
| AH20G33360 | 1.28 | - | TCP transcription factor |
| AH14G42340 | - | 6.33 | TCP transcription factor |
| AH19G37380 | - | 1.11 | TCP transcription factor |
| AH03G34200 | - | −1.01 | TGA transcription factor |
| AH20G00510 | - | −1.45 | UNE transcription factor |
| AH08G30400 | - | −1.8 | UNE transcription factor |
| AH14G41690 | 3.89 | - | Trihelix transcription factor |
| AH12G24650 | - | −2.2 | Trihelix transcription factor |
| AH09G16910 | - | −1.56 | Trihelix transcription factor |
| AH07G03020 | 1.44 | - | WRKY transcription factor |
| AH01G21930 | - | 1.58 | WRKY transcription factor |
| AH13G34700 | - | 5.27 | WRKY transcription factor |
| AH08G28680 | 1.86 | 2.74 | WRKY transcription factor |
| AH12G03520 | 2.21 | 2.73 | WRKY transcription factor |
| AH16G13340 | 2.05 | 4.06 | WRKY transcription factor |
| AH06G25830 | 1.1 | 2.83 | WRKY transcription factor |
| AH03G28760 | 1.57 | 3.18 | WRKY transcription factor |
| AH13G32420 | 1.44 | 2.71 | WRKY transcription factor |
| AH08G09100 | 1.4 | 2.93 | WRKY transcription factor |
| AH03G21920 | - | 1.23 | WRKY transcription factor |
| AH13G25080 | - | 1.18 | WRKY transcription factor |
| AH08G25500 | - | 2.38 | WRKY transcription factor |
| AH18G30430 | - | 2 | WRKY transcription factor |
| AH06G09380 | - | 3.71 | WRKY transcription factor |
| AH16G32150 | - | 2.65 | WRKY transcription factor |
| AH19G01540 | - | 2.17 | WRKY transcription factor |
| AH09G00690 | - | 2.31 | WRKY transcription factor |
| AH17G33510 | - | 2.27 | WRKY transcription factor |
| AH13G37760 | - | 4.33 | WRKY transcription factor |
| AH03G33940 | - | 4.02 | WRKY transcription factor |
| AH08G19200 | - | 1.47 | WRKY transcription factor |
| AH10G17080 | - | 3.06 | WRKY transcription factor |
| AH20G22960 | - | 2.1 | WRKY transcription factor |
| AH17G03120 | - | 5.7 | WRKY transcription factor |
| AH06G24570 | - | 1.53 | WRKY transcription factor |
| AH03G30560 | - | 3.58 | WRKY transcription factor |
| AH16G30280 | - | 1.28 | WRKY transcription factor |
| AH01G28930 | - | 1.48 | WRKY transcription factor |
| AH11G28210 | - | 1.66 | WRKY transcription factor |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Y.; Zhao, M.; Chen, J.; Yang, S.; Chen, J.; Xue, Y. Comparative Transcriptome Analysis Reveals Complex Physiological Response and Gene Regulation in Peanut Roots and Leaves under Manganese Toxicity Stress. Int. J. Mol. Sci. 2023, 24, 1161. https://doi.org/10.3390/ijms24021161
Liu Y, Zhao M, Chen J, Yang S, Chen J, Xue Y. Comparative Transcriptome Analysis Reveals Complex Physiological Response and Gene Regulation in Peanut Roots and Leaves under Manganese Toxicity Stress. International Journal of Molecular Sciences. 2023; 24(2):1161. https://doi.org/10.3390/ijms24021161
Chicago/Turabian StyleLiu, Ying, Min Zhao, Jingye Chen, Shaoxia Yang, Jianping Chen, and Yingbin Xue. 2023. "Comparative Transcriptome Analysis Reveals Complex Physiological Response and Gene Regulation in Peanut Roots and Leaves under Manganese Toxicity Stress" International Journal of Molecular Sciences 24, no. 2: 1161. https://doi.org/10.3390/ijms24021161
APA StyleLiu, Y., Zhao, M., Chen, J., Yang, S., Chen, J., & Xue, Y. (2023). Comparative Transcriptome Analysis Reveals Complex Physiological Response and Gene Regulation in Peanut Roots and Leaves under Manganese Toxicity Stress. International Journal of Molecular Sciences, 24(2), 1161. https://doi.org/10.3390/ijms24021161




