Nonspecific Amyloid Aggregation of Chicken Smooth-Muscle Titin: In Vitro Investigations
Abstract
1. Introduction
2. Results
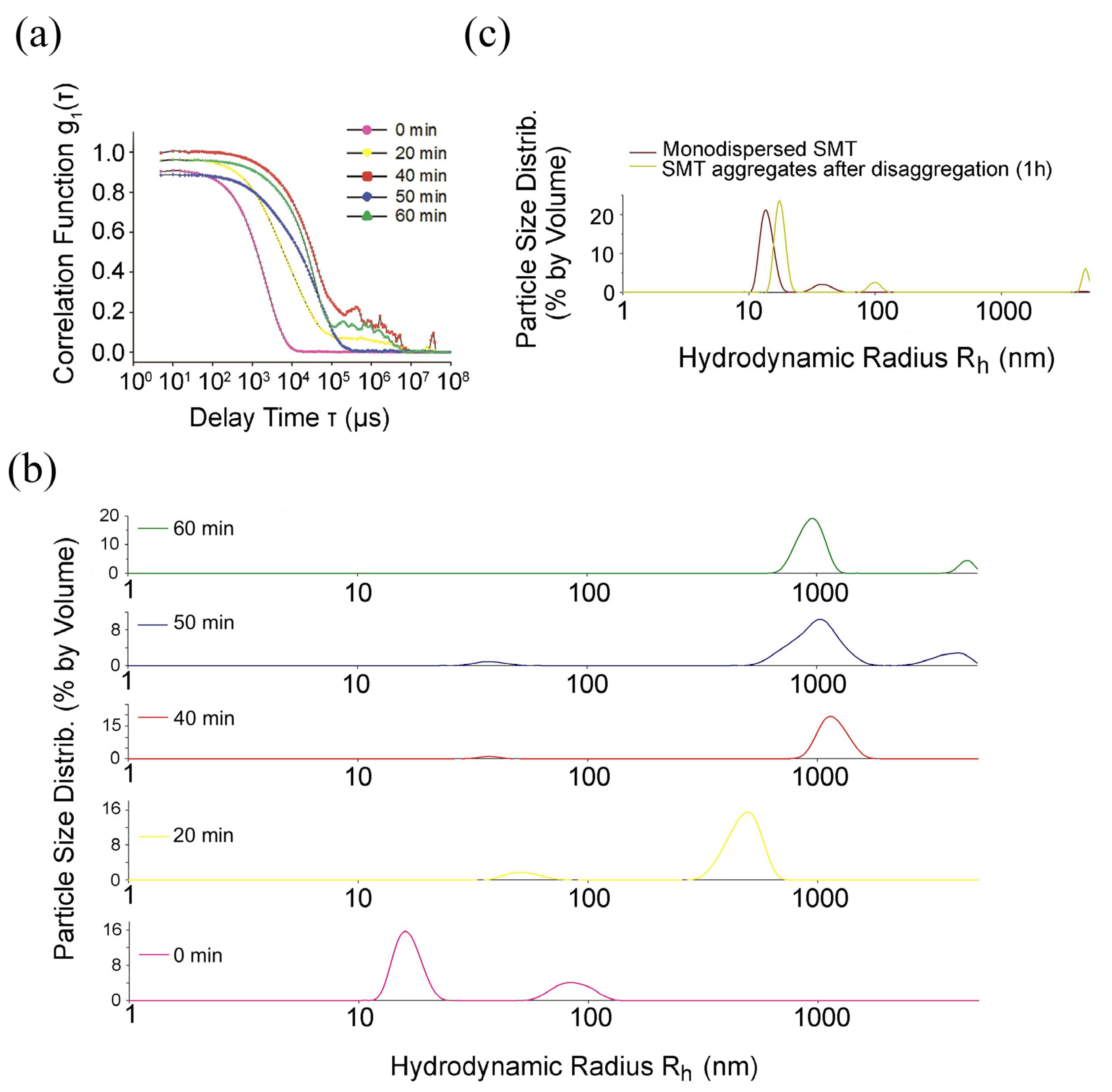
2.1. DLS Analysis of SMT Aggregations
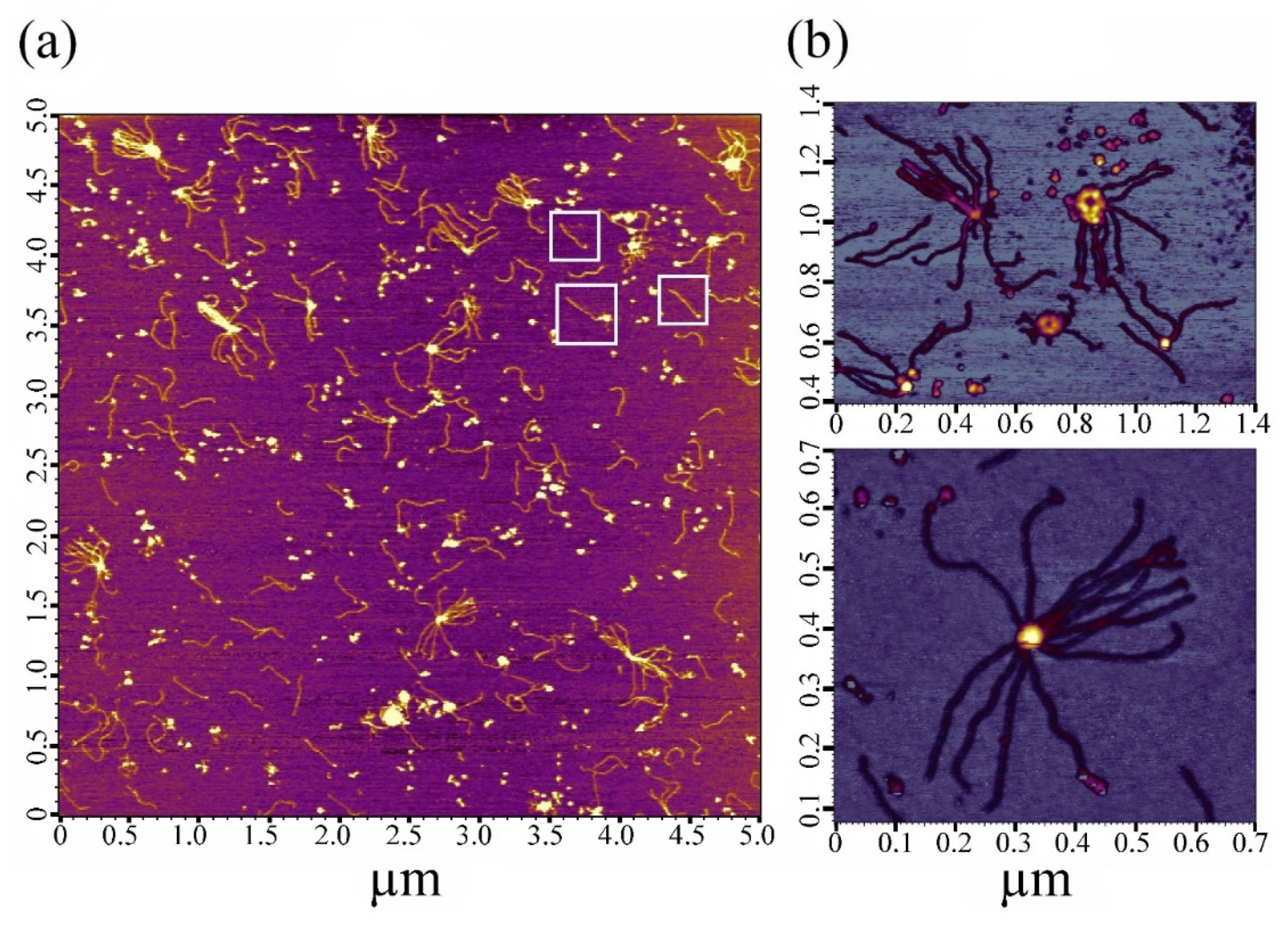
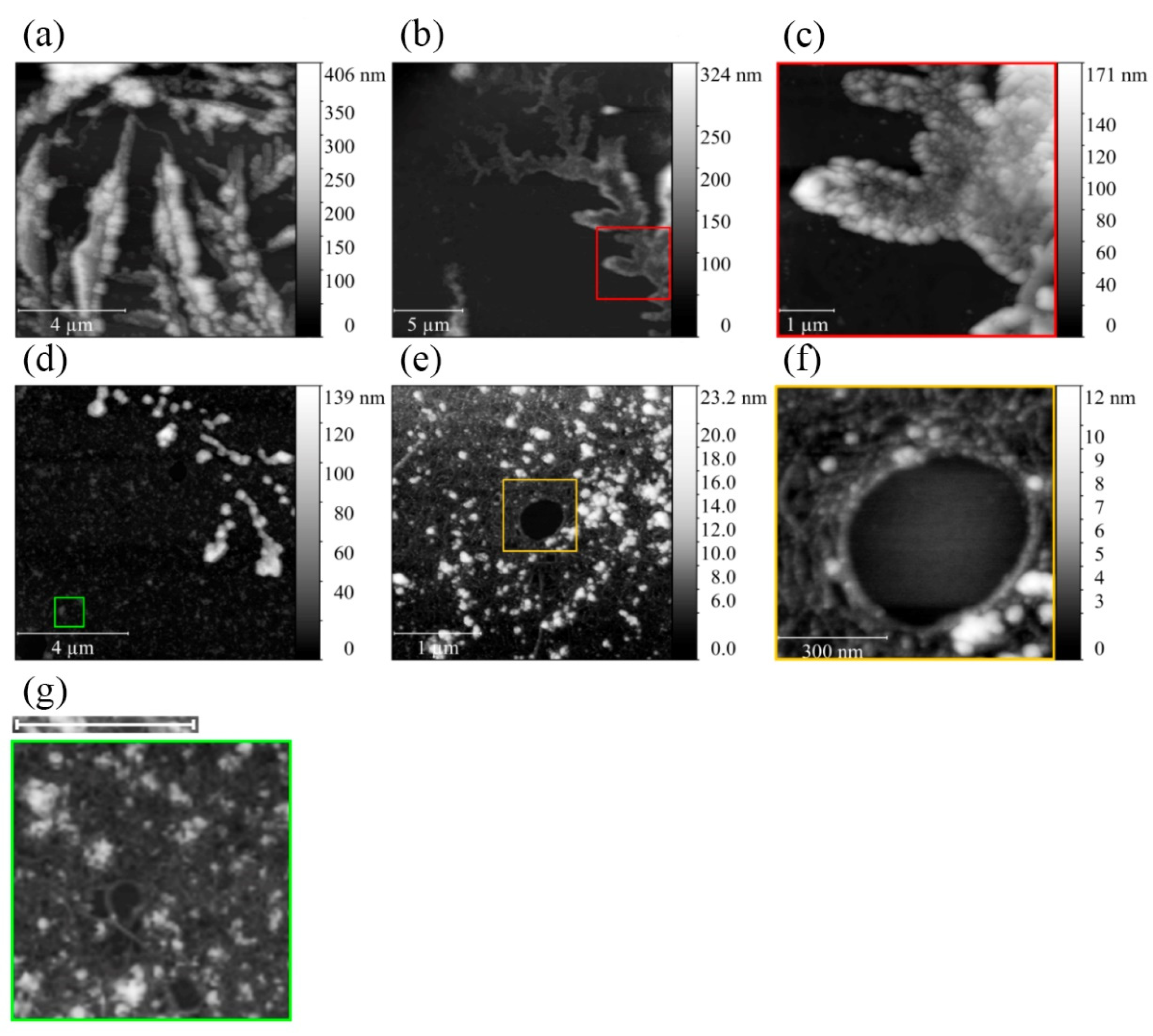
2.2. Electron and Atomic-Force Microscopy of SMTHMW Aggregates
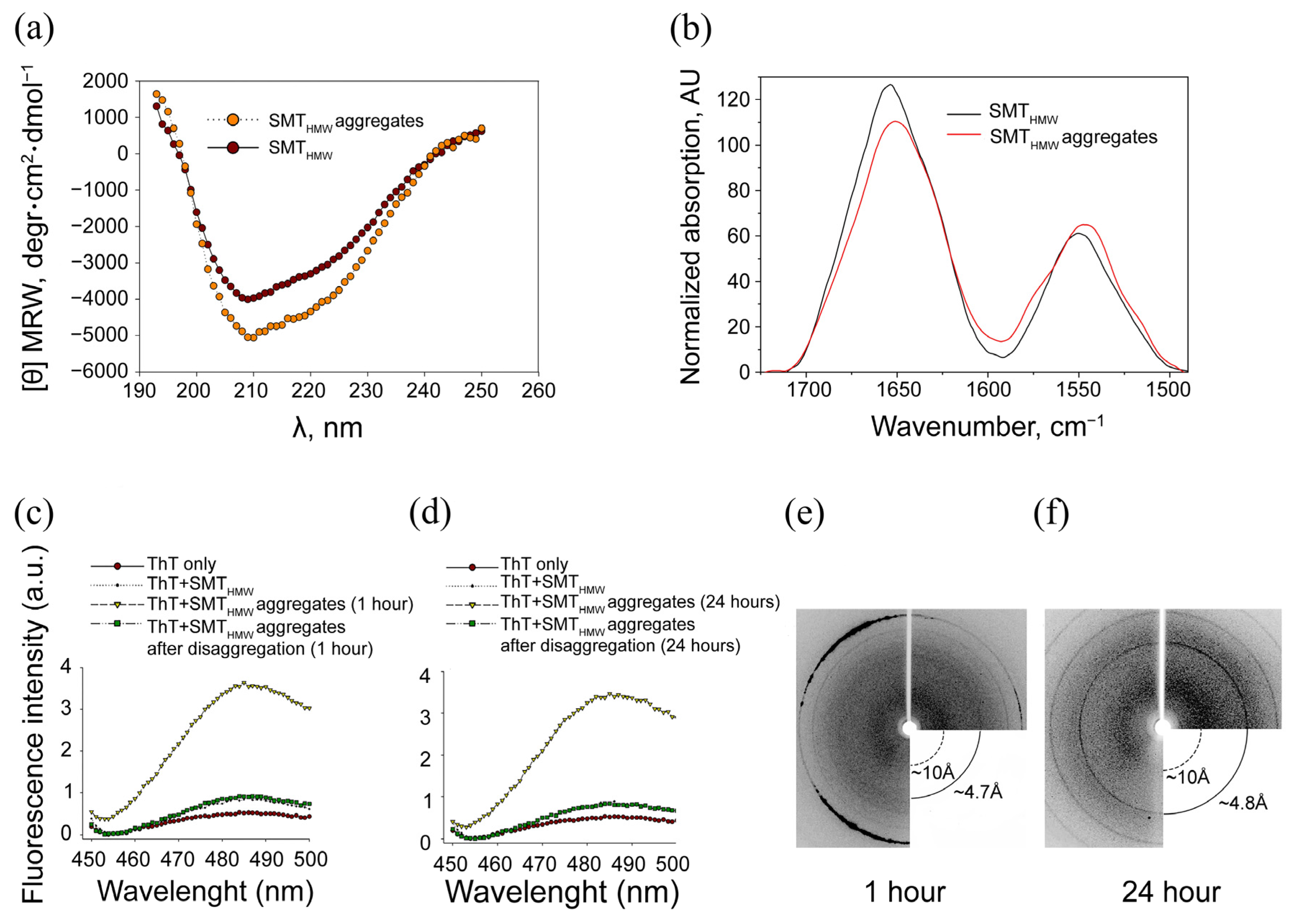
2.3. Circular Dichroism
2.4. Fourier-Transform Infrared Spectroscopy
2.5. Association of SMTHMW Aggregates with Thioflavin T
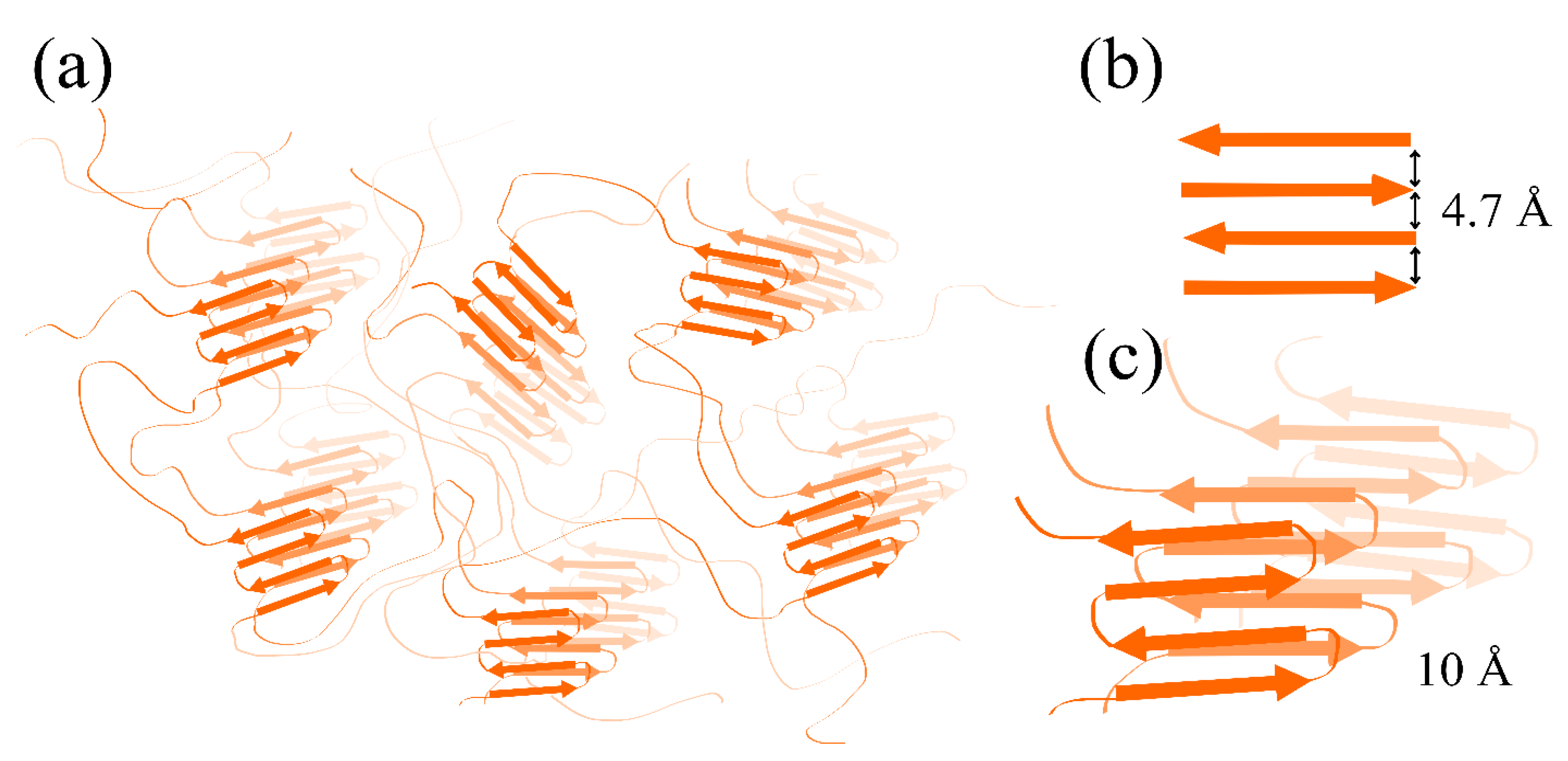
2.6. X-ray Diffraction of SMTHMW Aggregates
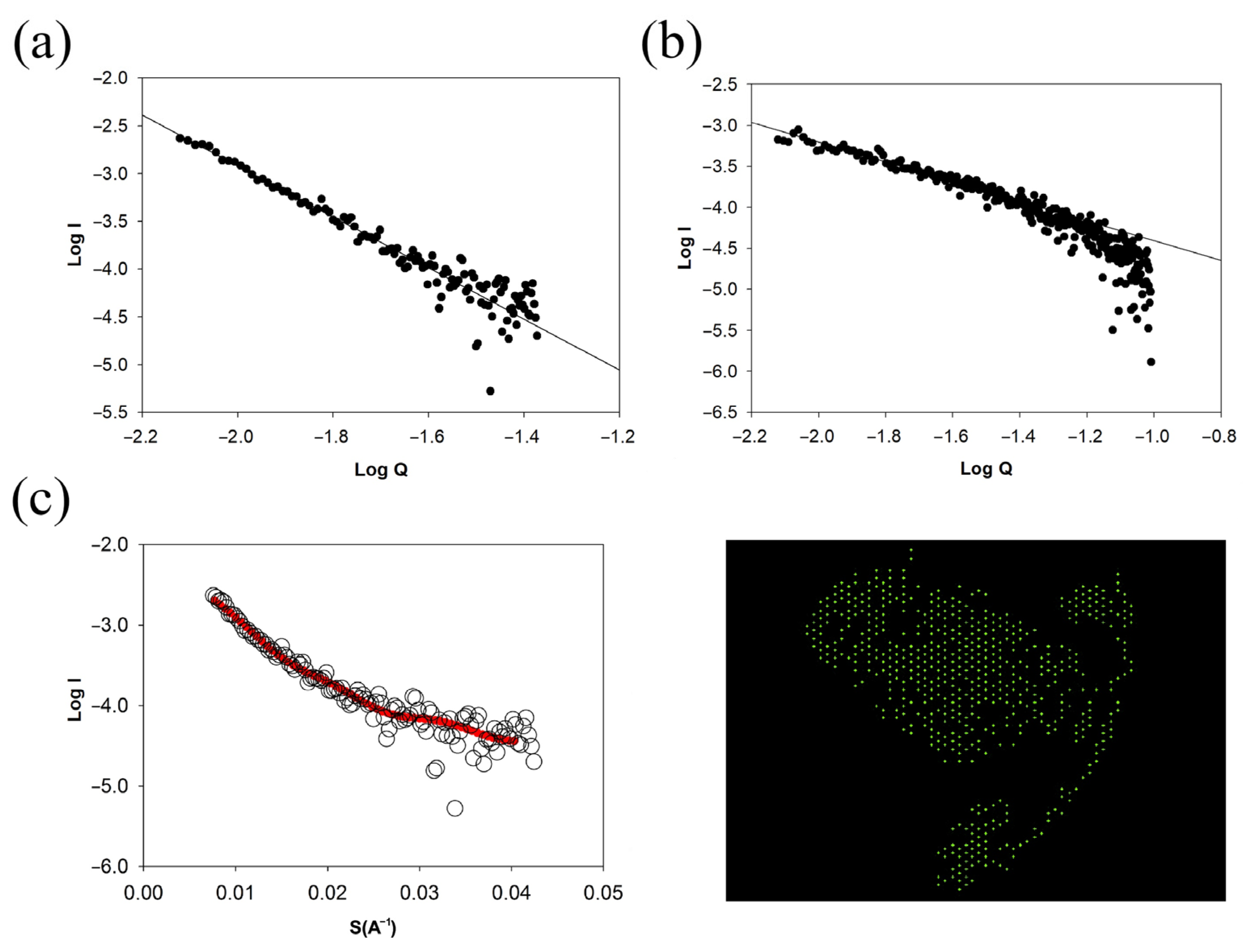
2.7. Small-Angle X-ray Scattering (SAXS)
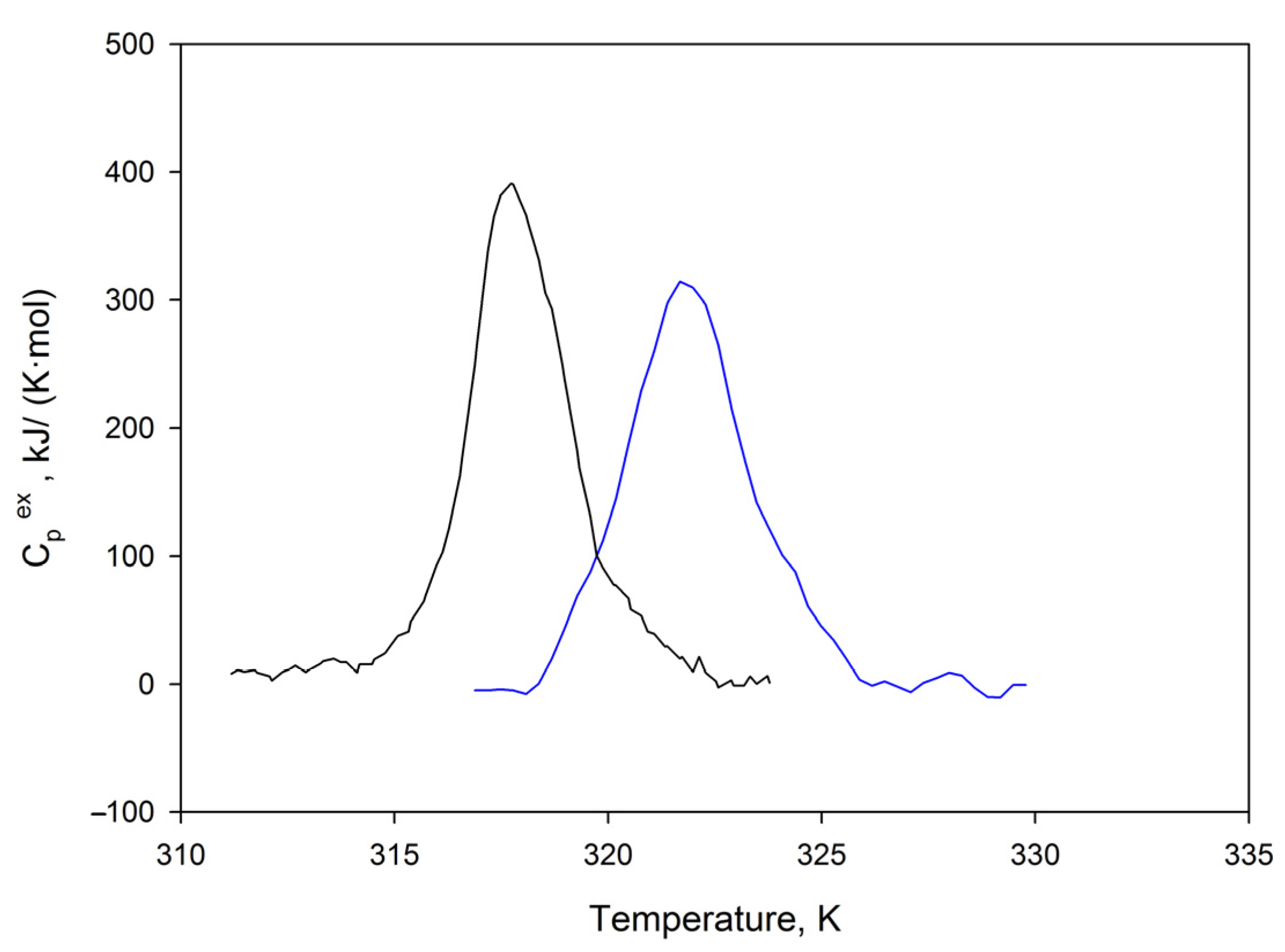
2.8. Differential Scanning Calorimetry of SMTHMW
2.9. SMT Amino Acid Sequence Identity
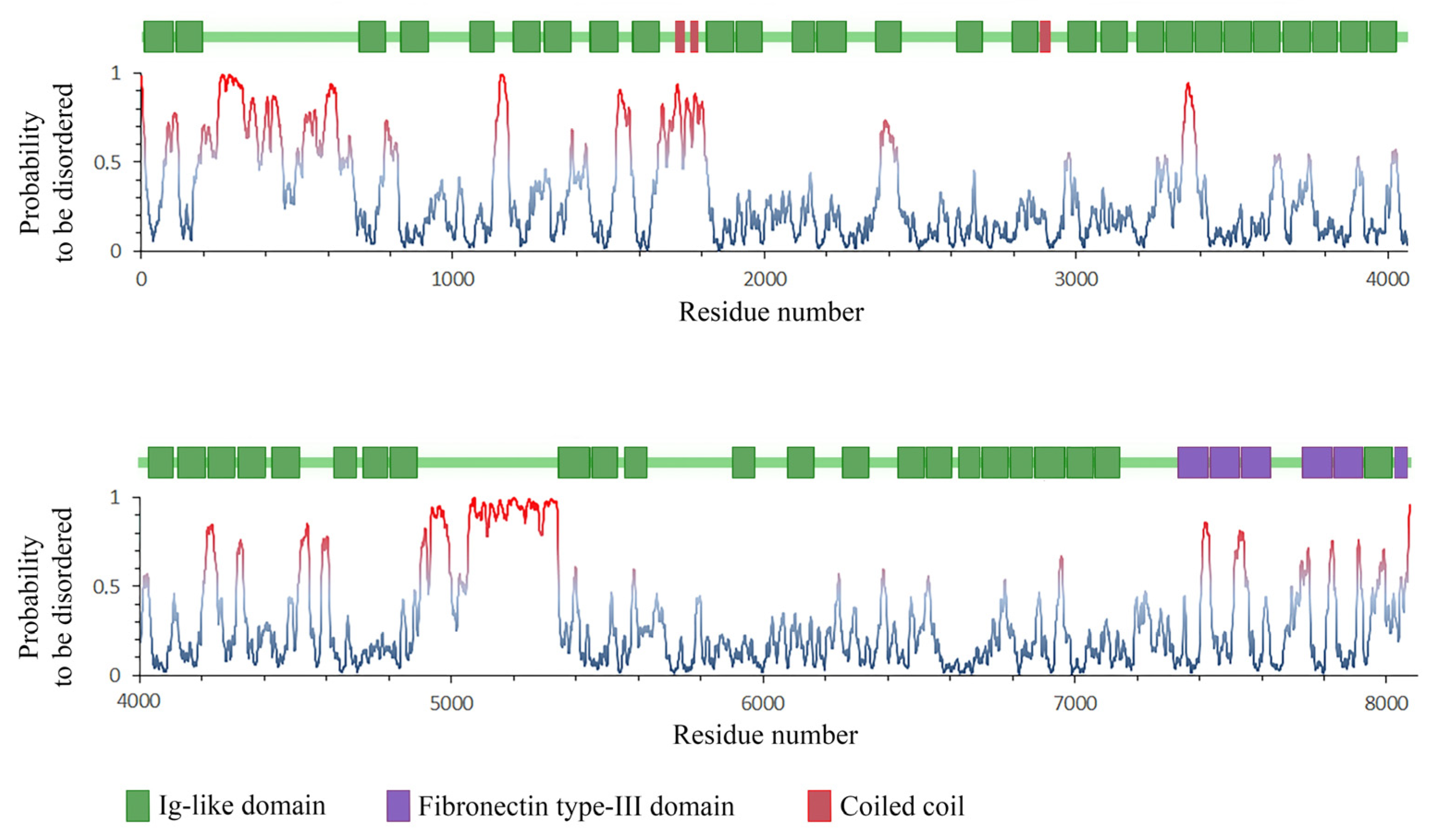
2.10. Calculation of Unstructured Areas in the SMT Molecule
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Purification of Chicken Gizzard SMT
5.2. SDS-PAGE and Mass Spectrometry Analysis of Titin
5.3. Conditions for the Formation of SMT Aggregates
5.4. Dynamic Light Scattering Experiments
5.5. Transmission Electron Microscopy
5.6. Atomic-Force Microscopy
5.7. Circular Dichroism
5.8. Fourier-Transform Infrared Spectroscopy
5.9. Fluorescence Analysis with Thioflavin T
5.10. X-ray Diffraction
5.11. Small Angle X-ray Scattering
5.12. Differential Scanning Calorimetry
5.13. Calculation of the Identity of the Amino Acid Sequence and Disordered Regions in the SMTHMW Molecule
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Tsytlonok, M.; Craig, P.O.; Sivertsson, E.; Serquera, D.; Perrett, S.; Best, R.B.; Wolynes, P.G.; Itzhaki, L.S. Complex energy landscape of a giant repeat protein. Structure 2013, 21, 1954–1965. [Google Scholar] [CrossRef] [PubMed]
- Tian, P.; Best, R.B. Best structural determinants of misfolding in multidomain proteins. PLoS Comput. Biol. 2016, 12, e1004933. [Google Scholar] [CrossRef]
- Gershenson, A.; Gierasch, L.M.; Pastore, A.; Radford, S.E. Energy landscapes of functional proteins are inherently risky. Nat. Chem. Biol. 2014, 10, 884–891. [Google Scholar] [CrossRef] [PubMed]
- Dobson, C.M. Protein folding and misfolding. Nature 2003, 426, 884–890. [Google Scholar] [CrossRef]
- Stefani, M.; Rigacci, S. Protein folding and aggregation into amyloid: The interference by natural phenolic compounds. Int. J. Mol. Sci. 2013, 14, 12411–12457. [Google Scholar] [CrossRef]
- Ittner, L.M.; Ke, Y.D.; Delerue, F.; Bi, M.; Gladbach, A.; van Eersel, J.; Wölfing, H.; Chieng, B.C.; Christie, M.J.; Napier, I.A.; et al. Dendritic function of tau mediates amyloid-beta toxicity in Alzheimer’s disease mouse models. Cell 2010, 142, 387–397. [Google Scholar] [CrossRef]
- Haass, C.; Mandelkow, E. Fyn-tau-amyloid: A toxic triad. Cell 2010, 142, 356–358. [Google Scholar] [CrossRef]
- Marshall, K.E.; Marchante, R.; Xue, W.F.; Serpell, L.C. The relationship between amyloid structure and cytotoxicity. Prion 2014, 8, 192–196. [Google Scholar] [CrossRef]
- Bucciantini, M.; Giannoni, E.; Chiti, F.; Baroni, F.; Formigli, L.; Zurdo, J.; Taddei, N.; Ramponi, G.; Dobson, C.M.; Stefani, M. Inherent toxicity of aggregates implies a common mechanism for protein misfolding diseases. Nature 2002, 416, 507–511. [Google Scholar] [CrossRef]
- Sengupta, U.; Nilson, A.N.; Kayed, R. The role of amyloid-β oligomers in toxicity, propagation, and immunotherapy. EBioMedicine 2016, 6, 42–49. [Google Scholar] [CrossRef]
- Knowles, T.P.; Vendruscolo, M.; Dobson, C.M. The amyloid state and its association with protein misfolding diseases. Nat. Rev. Mol. Cell Biol. 2014, 15, 384–396. [Google Scholar] [CrossRef]
- Dobson, C.M. Experimental investigation of protein folding and misfolding. Methods 2004, 34, 4–14. [Google Scholar] [CrossRef]
- Buxbaum, J.N.; Linke, R.P. A molecular history of the amyloidosis. J. Mol. Biol. 2000, 421, 142–159. [Google Scholar] [CrossRef] [PubMed]
- Dobson, C.M. Principles of protein folding, misfolding and aggregation. Semin. Cell Dev. Biol. 2004, 15, 3–16. [Google Scholar] [CrossRef]
- Sunde, M.; Serpell, L.C.; Bartlam, M.; Fraser, P.E.; Pepys, M.B.; Blake, C.C. Common core structure of amyloid fibrils by synchrotron X-ray diffraction. J. Mol. Biol. 1997, 273, 729–739. [Google Scholar] [CrossRef]
- Nelson, R.; Eisenberg, D. Recent atomic models of amyloid fibril structure. Curr. Opin. Struct. Biol. 2006, 16, 260–265. [Google Scholar] [CrossRef]
- Olsen, A.; Jonsson, A.; Normark, S. Fibronectin binding mediated by a novel class of surface organelles on Escherichia coli. Nature 1989, 338, 652–655. [Google Scholar] [CrossRef]
- Römling, U.; Bian, Z.; Hammar, M.; Sierralta, W.D.; Normark, S. Curli fibers are highly conserved between Salmonella typhimurium and Escherichia coli with respect to open structure and regulation. J. Bacteriol. 1998, 180, 722–731. [Google Scholar] [CrossRef]
- Otzen, D.; Nielsen, P.H. We find them here, we find them there: Functional bacterial amyloid. Cell Mol. Life Sci. 2008, 65, 910–927. [Google Scholar] [CrossRef]
- Claessen, D.; Rink, R.; de Jong, W.; Siebring, J.; de Vreugd, P.; Boersma, F.G.; Dijkhuizen, L.; Wosten, H.A. A novel class of secreted hydrophobic proteins is involved in aerial hyphae formation in Streptomyces coelicolor by forming amyloid-like fibrils. Genes Dev. 2003, 17, 1714–1726. [Google Scholar] [CrossRef]
- Si, K.; Lindquist, S.L.; Kandel, E.R. A neuronal isoform of the aplysia CPEB has prion-like properties. Cell 2003, 115, 879–891. [Google Scholar] [CrossRef]
- Iconomidou, V.A.; Vriend, G.; Hamodrakas, S.J. Amyloids protect the silkmoth oocyte and embryo. FEBS Lett. 2000, 479, 141–145. [Google Scholar] [CrossRef]
- Iconomidou, V.A.; Chryssikos, G.D.; Giorius, V.; Galarius, A.S.; Cordopatis, P.; Hoenger, A.; Hamodrakas, S.J. Amyloid fibril formation propensity is inherent into the hexapeptide tandemly repeating sequence of the central domain of silkmoth chorion proteins of the A-family. J. Struct. Biol. 2006, 156, 480–488. [Google Scholar] [CrossRef]
- Podrabsky, J.E.; Carpenter, J.F.; Hand, S.C. Survival of water stress in annual fish embryos: Dehydration avoidance and egg envelope amyloid fibers. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2001, 280, R123–R131. [Google Scholar] [CrossRef]
- Fowler, D.M.; Koulov, A.V.; Alory-Jost, C.; Marks, M.S.; Balch, W.E.; Kelly, J.W. Functional amyloid formation within mammalian tissue. PLoS Biol. 2005, 4, e6. [Google Scholar] [CrossRef]
- Berson, J.F.; Theos, A.C.; Harper, D.C.; Tenza, D.; Raposo, G.; Marks, M.S. Proprotein convertase cleavage liberates a fibrillogenic fragment of a resident glycoprotein to initiate melanosome biogenesis. J. Cell. Biol. 2003, 161, 521–533. [Google Scholar] [CrossRef]
- Antonets, K.S.; Belousov, M.V.; Sulatskaya, A.I.; Belousova, M.E.; Kosolapova, A.O.; Sulatsky, M.I.; Andreeva, E.A.; Zykin, P.A.; Malovichko, Y.V.; Shtark, O.Y.; et al. Accumulation of storage proteins in plant seeds is mediated by amyloid formation. PLoS Biol. 2020, 18, e3000564. [Google Scholar] [CrossRef]
- Wang, K.; McClure, J.; Tu, A. Titin: Major myofibrillar components of striated muscle. Proc. Natl. Acad. Sci. USA 1979, 76, 3698–3702. [Google Scholar] [CrossRef]
- Maruyama, K.; Kimura, S.; Ohashi, K.; Kuwano, Y. Connectin, an elastic protein of muscle. Identification of “titin” with connectin. J. Biochem. 1981, 89, 701–709. [Google Scholar] [CrossRef]
- Koretz, J.F.; Irving, T.C.; Wang, K. Filamentous aggregates of native titin and binding of C-protein and AMP-deaminase. Arch. Biochem. Biophys. 1993, 304, 305–309. [Google Scholar] [CrossRef]
- Kellermayer, M.S.; Bustamante, C.; Granzier, H.L. Mechanics and structure of titin oligomers explored with atomic force microscopy. Biochim. Biophys. Acta 2003, 1604, 105–114. [Google Scholar] [CrossRef] [PubMed]
- Wright, C.F.; Teichmann, S.A.; Clarke, J.; Dobson, C.M. The importance of sequence diversity in the aggregation and evolution of proteins. Nature 2005, 438, 878–881. [Google Scholar] [CrossRef] [PubMed]
- Borgia, A.; Kemplen, K.R.; Borgia, M.B.; Soranno, A.; Shammas, S.; Wunderlich, B.; Nettels, D.; Best, R.B.; Clarke, J.; Schuler, B. Transient misfolding dominates multidomain protein folding. Nat. Commun. 2015, 6, 8861. [Google Scholar] [CrossRef]
- Bobylev, A.G.; Galzitskaya, O.V.; Fadeev, R.S.; Bobyleva, L.G.; Yurshenas, D.A.; Molochkov, N.V.; Dovidchenko, N.V.; Selivanova, O.M.; Penkov, N.V.; Podlubnaya, Z.A.; et al. Smooth muscle titin forms in vitro amyloid aggregates. Biosci. Rep. 2016, 36, e00334. [Google Scholar] [CrossRef]
- Yakupova, E.I.; Vikhlyantsev, I.M.; Bobyleva, L.G.; Penkov, N.V.; Timchenko, A.A.; Timchenko, M.A.; Enin, G.A.; Khutzian, S.S.; Selivanova, O.M.; Bobylev, A.G. Different amyloid aggregation of smooth muscles titin in vitro. J. Biomol. Struct. Dyn. 2018, 36, 2237–2248. [Google Scholar] [CrossRef] [PubMed]
- Bobylev, A.G.; Fadeev, R.S.; Bobyleva, L.G.; Kobyakova, M.I.; Shlyapnikov, Y.M.; Popov, D.V.; Vikhlyantsev, I.M. Amyloid aggregates of smooth-muscle titin impair cell adhesion. Int. J. Mol. Sci. 2021, 22, 4579. [Google Scholar] [CrossRef] [PubMed]
- Hill, S.E.; Robinson, J.; Matthews, G.; Muschol, M. Amyloid protofibrils of lysozyme nucleate and grow via oligomer fusion. Biophys. J. 2009, 96, 3781–3790. [Google Scholar] [CrossRef]
- Tskhovrebova, L.; Trinick, J. Titin: Properties and family relationships. Nat. Rev. Mol. Cell Biol. 2003, 4, 679–689. [Google Scholar] [CrossRef]
- Vikhlyantsev, I.M.; Okuneva, A.D.; Shumilina, U.V.; Salmov, N.N.; Bobylev, A.G.; Molochkov, N.V.; Podlubnaya, Z.A. Method for isolation of intact titin (connectin) molecules from mammalian cardiac muscle. Biochemistry 2013, 78, 455–462. [Google Scholar] [CrossRef]
- Yang, H.; Yang, S.; Kong, J.; Dong, A.; Yu, S. Obtaining information about protein secondary structures in aqueous solution using Fourier transform IR spectroscopy. Nat. Protoc. 2015, 10, 382–396. [Google Scholar] [CrossRef] [PubMed]
- Svergun, D.I. Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys. J. 1999, 76, 2879–2886. [Google Scholar] [CrossRef]
- Noguchi, H.; Takemori, S.; Kajiwara, J.; Kimura, M.; Maruyama, K.; Kimura, S. Chicken breast muscle connectin: Passive tension and I-band region primary structure. J. Mol. Biol. 2007, 370, 213–219. [Google Scholar] [CrossRef] [PubMed]
- Lobanov, M.Y.; Galzitskaya, O.V. The Ising model for prediction of disordered residues from protein sequence alone. Phys. Biol. 2011, 8, 35004. [Google Scholar] [CrossRef] [PubMed]
- Lobanov, M.Y.; Sokolovskiy, I.V.; Galzitskaya, O.V. IsUnstruct: Prediction of the residue status to be ordered or disordered in the protein chain by a method based on the Ising model. J. Biomol. Struct. Dyn. 2013, 31, 1034–1043. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Walter, J. Phosphorylation of amyloid beta (Aβ) peptides—A trigger for formation of toxic aggregates in Alzheimer’s disease. Aging 2011, 3, 803–812. [Google Scholar] [CrossRef]
- Bobyleva, L.G.; Shumeyko, S.A.; Yakupova, E.I.; Surin, A.K.; Galzitskaya, O.V.; Kihara, H.; Timchenko, A.A.; Timchenko, M.A.; Penkov, N.V.; Nikulin, A.D.; et al. Myosin binding protein-C forms amyloid-like aggregates in vitro. Int. J. Mol. Sci. 2021, 22, 731. [Google Scholar] [CrossRef]
- Pan, K.M.; Baldwin, M.; Nguyen, J.; Gasset, M.; Serban, A.; Groth, D.; Mehlhorn, I.; Huang, Z.; Fletterick, R.J.; Cohen, F.E. Conversion of alpha-helices into beta-sheets features in the formation of the scrapie prion proteins. Proc. Natl. Acad. Sci. USA 1993, 90, 10962–10966. [Google Scholar] [CrossRef]
- Sun, Y.; Kakinen, A.; Xing, Y.; Faridi, P.; Nandakumar, A.; Purcell, A.W.; Davis, T.P.; Ke, P.C.; Ding, F. Amyloid self-assembly of hIAPP8-20 via the accumulation of helical oligomers, α-helix to β-sheet transition, and formation of β-barrel intermediates. Small 2019, 15, e1805166. [Google Scholar] [CrossRef]
- Ji, W.; Yuan, C.; Chakraborty, P.; Gilead, S.; Yan, X.; Gazit, E. Stoichiometry-controlled secondary structure transition of amyloid-derived supramolecular dipeptide co-assemblies. Commun. Chem. 2019, 2, 65. [Google Scholar] [CrossRef]
- Ulamec, S.M.; Radford, S.E. Spot the Difference: Function versus toxicity in amyloid fibrils. Trends Biochem. Sci. 2020, 45, 635–636. [Google Scholar] [CrossRef]
- Eckels, E.C.; Haldar, S.; Tapia-Rojo, R.; Rivas-Pardo, J.A.; Fernández, J.M. The mechanical power of titin folding. Cell Rep. 2019, 27, 1836–1847.e4. [Google Scholar] [CrossRef]
- Rivas-Pardo, J.A.; Eckels, E.C.; Popa, I.; Kosuri, P.; Linke, W.A.; Fernández, J.M. Work done by titin protein folding assists muscle contraction. Cell Rep. 2016, 14, 1339–1347. [Google Scholar] [CrossRef]
- Rousseau, F.; Serrano, L.; Schymkowitz, J.W. How evolutionary pressure against protein aggregation shaped chaperone specificity. J. Mol. Biol. 2006, 355, 1037–1047. [Google Scholar] [CrossRef]
- Goldschmidt, L.; Teng, P.K.; Riek, R.; Eisenberg, D. Identifying the amylome, proteins capable of forming amyloid-like fibrils. Proc. Natl. Acad. Sci. USA 2010, 107, 3487–3492. [Google Scholar] [CrossRef]
- Azuaga, A.I.; Dobson, C.M.; Mateo, P.L.; Conejero-Lara, F. Unfolding and aggregation during the thermal denaturation of streptokinase. Eur. J. Biochem. 2002, 269, 4121–4133. [Google Scholar] [CrossRef]
- Rezaei, H.; Choiset, Y.; Eghiaian, F.; Treguer, E.; Mentre, P.; Debey, P.; Grosclaude, J.; Haertle, T. Amyloidogenic unfolding intermediates differentiate sheep prion protein variants. J. Mol. Biol. 2002, 322, 799–814. [Google Scholar] [CrossRef]
- Baxa, U.; Ross, P.D.; Wickner, R.B.; Steven, A.C. The N-terminal prion domain of Ure2p converts from an unfolded to a thermally resistant conformation upon filament formation. J. Mol. Biol. 2004, 339, 259–264. [Google Scholar] [CrossRef]
- Morel, B.; Casares, S.; Conejero-Lara, F. A single mutation induces amyloid aggregation in the α-spectrin SH3 domain: Analysis of the early stages of fibril formation. J. Mol. Biol. 2006, 356, 453–468. [Google Scholar] [CrossRef]
- Morel, B.; Varela, L.; Conejero-Lara, F. The thermodynamic stability of amyloid fibrils studied by differential scanning calorimetry. J. Phys. Chem. B. 2010, 114, 4010–4019. [Google Scholar] [CrossRef]
- Oberhauser, A.F.; Marszalek, P.E.; Carrion-Vazquez, M.; Fernandez, J.M. Single protein misfolding events captured by atomic force microscopy. Nat. Struct. Biol. 1999, 6, 1025–1028. [Google Scholar] [CrossRef]
- Trinick, J.; Knight, P.; Whiting, A. Purification and properties of native titin. J. Mol. Biol. 1984, 180, 331–356. [Google Scholar] [CrossRef]
- Fritz, J.D.; Swartz, D.R.; Greaser, M.L. Factors affecting polyacrylamide gel electrophoresis and electroblotting of high-molecular-weight myofibrillar proteins. Anal. Biochem. 1989, 180, 205–210. [Google Scholar] [CrossRef]
- Makhnovskii, P.A.; Zgoda, V.G.; Bokov, R.O.; Shagimardanova, E.I.; Gazizova, G.R.; Gusev, O.A.; Lysenko, E.A.; Kolpakov, F.A.; Vinogradova, O.L.; Popov, D.V. Regulation of proteins in human skeletal muscle: The role of transcription. Sci. Rep. 2020, 10, 3514. [Google Scholar] [CrossRef]
- Wiśniewski, J.R.; Zougman, A.; Nagaraj, N.; Mann, M. Universal sample preparation method for proteome analysis. Nat. Methods 2009, 6, 359–362. [Google Scholar] [CrossRef]
- Sreerama, N.; Woody, R.W. Estimation of protein secondary structure from circular dichroism spectra: Comparison of CONTIN, SELCON, and CDSSTR methods with an expanded reference set. Anal. Biochem. 2000, 287, 252–260. [Google Scholar] [CrossRef]
- Venyaminov, S.; Prendergast, F.G. Water (H2O and D2O) molar absorptivity in the 1000–4000 cm−1 range and quantitative infrared spectroscopy of aqueous solutions. Anal. Biochem. 1997, 248, 234–245. [Google Scholar] [CrossRef]
- Feigin, L.A.; Svergun, D.A. Structure Analysis by Small-angle X-ray and Neutron Scattering; Springer: Berlin/Heidelberg, Germany; Plenum Press: New York, NY, USA, 1987. [Google Scholar]
- Senin, A.A.; Potekhin, S.A.; Tiktopulo, E.I.; Filomonov, V.V. Differential scanning microcalorimetr SCAL-1. J. Therm. Anal. Cal. 2000, 62, 153–160. [Google Scholar] [CrossRef]
- Privalov, P.L.; Potekhin, S.A. Scanning microcalorimetry in studying temperature-induced changes in proteins. Methods Enzymol. 1986, 131, 4–51. [Google Scholar] [CrossRef]

| Preparation | α-Helices, % | β-Folds, % | Turns, % | Disordered Structure, % |
|---|---|---|---|---|
| Molecular SMTHMW | 26 ± 1.0 | 25 ± 2.0 | 11.7 ± 0.6 | 32 ± 2.0 |
| Aggregated SMTHMW | 24.6 ± 1.6 | 26 ± 2.0 | 10.7 ± 0.4 | 34 ± 2.0 |
| Protein | Domain 1 (for Pair Comparison) | Number of Domains in the Group | Average Length of Domain | Domain 2 (for Pair Comparison) | (Id) Mean Identity, % | Side Standard Dispersion/Deviation of Identity, % | N0, Number of Domain Pairs with Zero Identity | N, Total Number of Domain Pairs | N0/N·100, % |
|---|---|---|---|---|---|---|---|---|---|
| Chicken titin | Ig-like | 64 | 81.4 | Ig-like | 20% | 11% | 647 | 4032 | 16% |
| Ig-like | 64 | 81.4 | FnIII-like | 3% | 4% | 223 | 384 | 58% | |
| Ig-like | 64 | 81.4 | none | 2% | 6% | 526 | 704 | 75% | |
| FnIII-like | 6 | 84.3 | FnIII-like | 33% | 7% | 0 | 30 | 0% | |
| FnIII-like | 6 | 84.3 | none | 2% | 2% | 43 | 66 | 65% | |
| none | 11 | 104.2 | none | 3% | 5% | 59 | 110 | 54% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bobylev, A.G.; Yakupova, E.I.; Bobyleva, L.G.; Molochkov, N.V.; Timchenko, A.A.; Timchenko, M.A.; Kihara, H.; Nikulin, A.D.; Gabdulkhakov, A.G.; Melnik, T.N.; et al. Nonspecific Amyloid Aggregation of Chicken Smooth-Muscle Titin: In Vitro Investigations. Int. J. Mol. Sci. 2023, 24, 1056. https://doi.org/10.3390/ijms24021056
Bobylev AG, Yakupova EI, Bobyleva LG, Molochkov NV, Timchenko AA, Timchenko MA, Kihara H, Nikulin AD, Gabdulkhakov AG, Melnik TN, et al. Nonspecific Amyloid Aggregation of Chicken Smooth-Muscle Titin: In Vitro Investigations. International Journal of Molecular Sciences. 2023; 24(2):1056. https://doi.org/10.3390/ijms24021056
Chicago/Turabian StyleBobylev, Alexander G., Elmira I. Yakupova, Liya G. Bobyleva, Nikolay V. Molochkov, Alexander A. Timchenko, Maria A. Timchenko, Hiroshi Kihara, Alexey D. Nikulin, Azat G. Gabdulkhakov, Tatiana N. Melnik, and et al. 2023. "Nonspecific Amyloid Aggregation of Chicken Smooth-Muscle Titin: In Vitro Investigations" International Journal of Molecular Sciences 24, no. 2: 1056. https://doi.org/10.3390/ijms24021056
APA StyleBobylev, A. G., Yakupova, E. I., Bobyleva, L. G., Molochkov, N. V., Timchenko, A. A., Timchenko, M. A., Kihara, H., Nikulin, A. D., Gabdulkhakov, A. G., Melnik, T. N., Penkov, N. V., Lobanov, M. Y., Kazakov, A. S., Kellermayer, M., Mártonfalvi, Z., Galzitskaya, O. V., & Vikhlyantsev, I. M. (2023). Nonspecific Amyloid Aggregation of Chicken Smooth-Muscle Titin: In Vitro Investigations. International Journal of Molecular Sciences, 24(2), 1056. https://doi.org/10.3390/ijms24021056











