Diaphragm Fatigue in SMNΔ7 Mice and Its Molecular Determinants: An Underestimated Issue
Abstract
1. Introduction
2. Results
2.1. Ex Vivo Functional Analysis
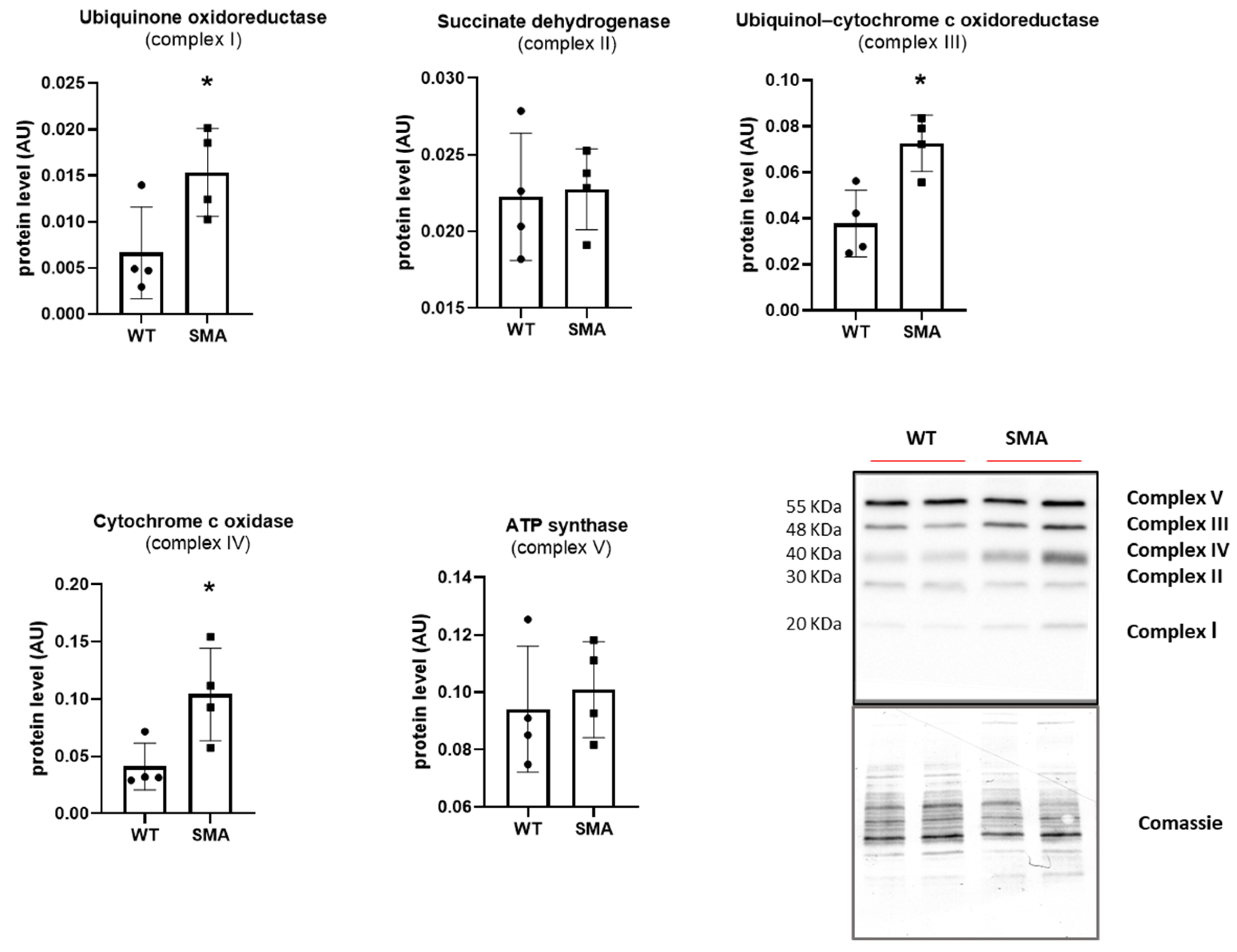
2.2. Energy Imbalance and Oxidative Metabolism
2.3. Autophagy and Mitophagy
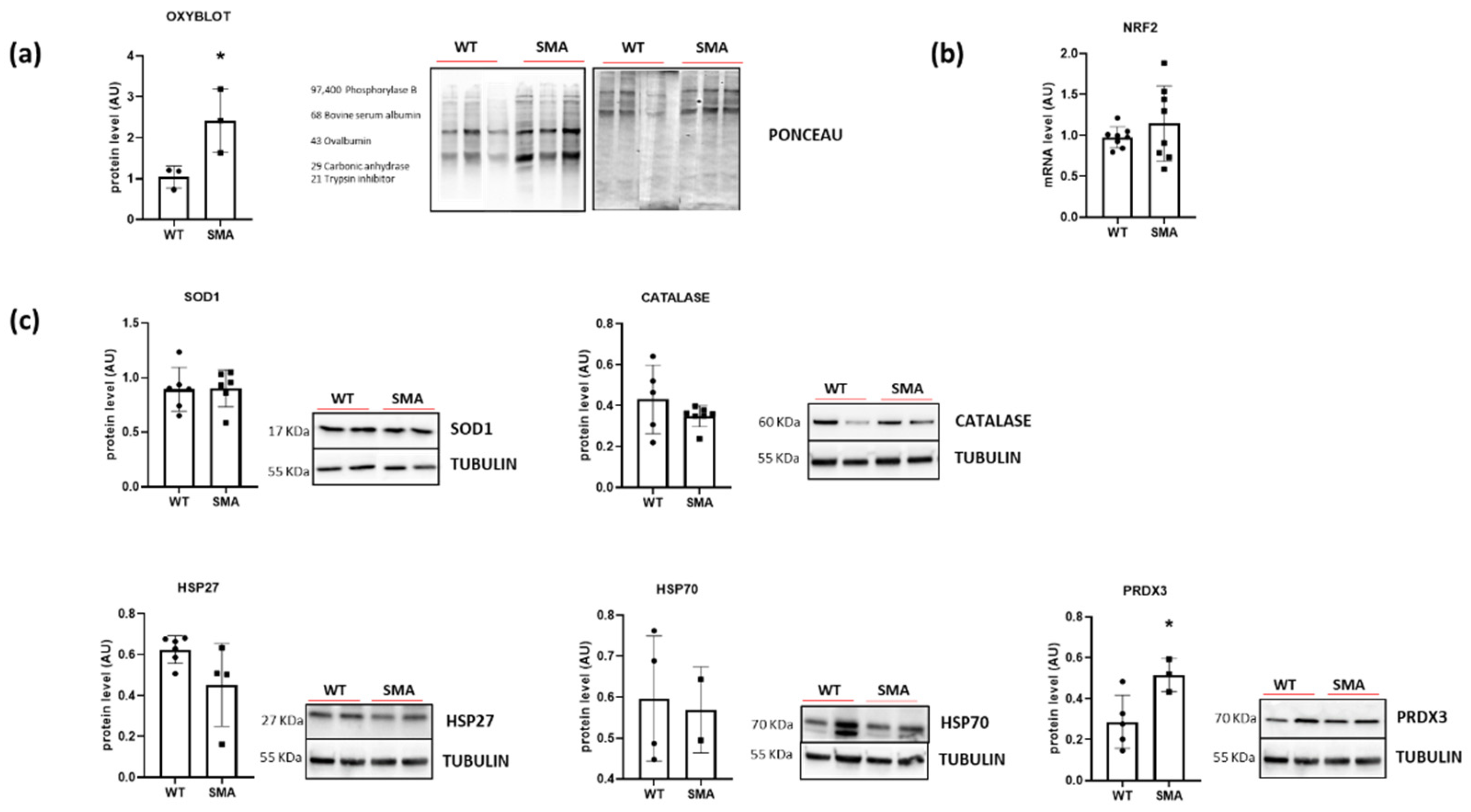
2.4. Redox Imbalance
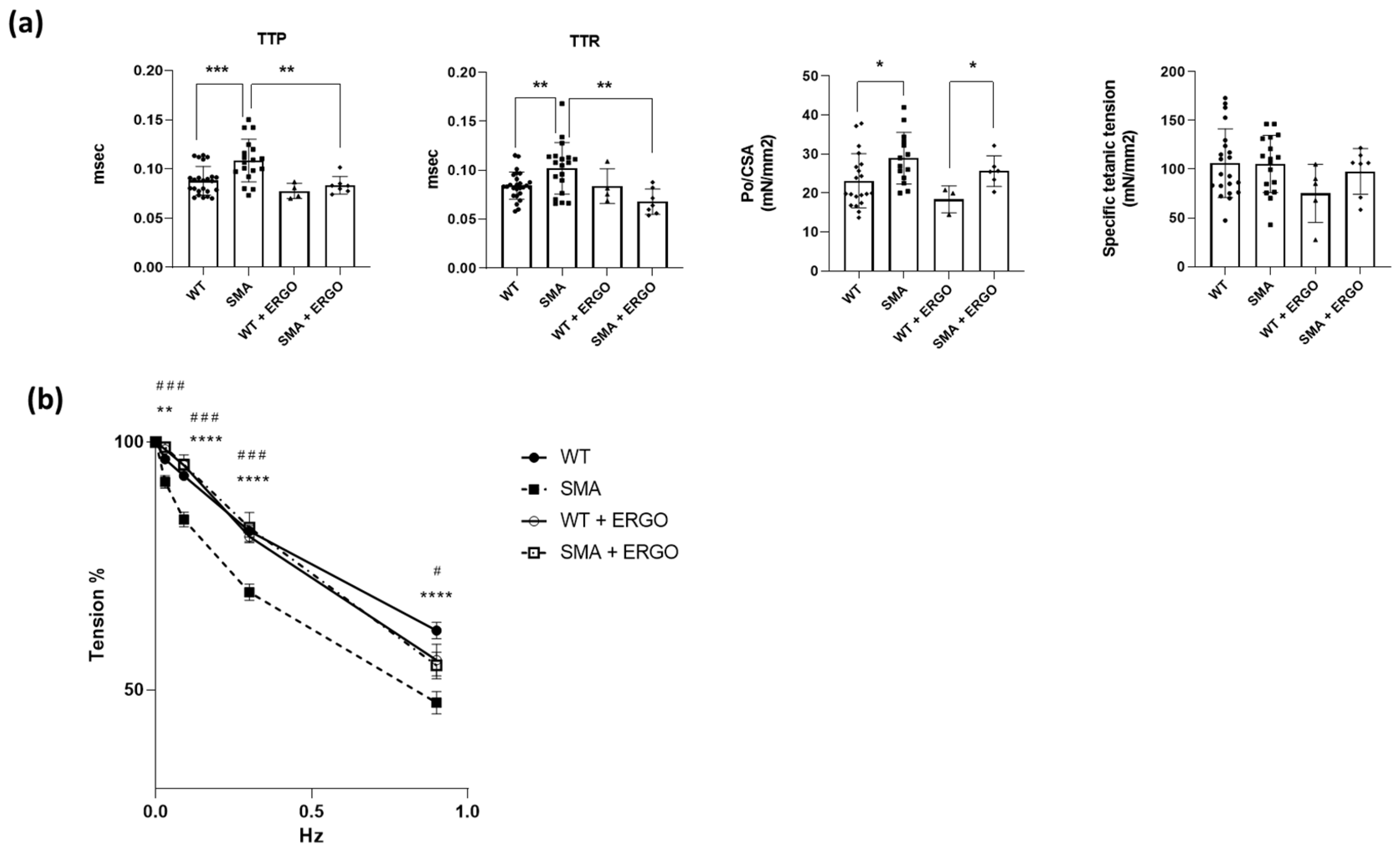
2.5. Effect of Exposure to the Antioxidant Molecule Ergothioneine
3. Discussion
3.1. SMA Diaphragm Exhibits Reduced Fatigue Resistance
3.2. Energy Balance in SMA Diaphragm
3.3. Oxidative Stress in SMA Diaphragm
4. Materials and Methods
4.1. Animals
- Homozygous for both transgenes and heterozygous for the null allele (7SMN+/+; SMN2 +/+; SMN+/−—50%). These mice do not show SMA phenotype and are used as breeders.
- Homozygous for both transgenes and heterozygous for the null allele (7SMN+/+; SMN2 +/+; SMN+/+—25%). These mice do not show SMA phenotype and are used as control mice (WTs).
- Homozygous for both transgenes and heterozygous for the null allele (7SMN+/+; SMN2 +/+; SMN−/−—25%). These mice show SMA phenotype (SMAs).
4.2. Genotyping
4.3. Ex Vivo Functional Analysis
4.4. OXYBLOT Analysis
4.5. Gene Expression Analysis
4.6. Western Blot Analysis
4.7. Oxygraph-2k for High-Resolution Respirometry (O2k HRR)
4.8. Citrate Synthase Activity
4.9. Electron Microscopy (EM)
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schroth, M.K. Special considerations in the respiratory management of spinal muscular atrophy. Pediatrics 2009, 123 (Suppl. 4), S245–S249. [Google Scholar] [CrossRef] [PubMed]
- Prior, T.W. Spinal muscular atrophy: A time for screening. Curr. Opin. Pediatr. 2010, 22, 696–702. [Google Scholar] [CrossRef]
- Lefebvre, S.; Bürglen, L.; Reboullet, S.; Clermont, O.; Burlet, P.; Viollet, L.; Benichou, B.; Cruaud, C.; Millasseau, P.; Zeviani, M. Identification and characterization of a spinal muscular atrophy-determining gene. Cell 1995, 80, 155–165. [Google Scholar] [CrossRef]
- Lorson, C.L.; Hahnen, E.; Androphy, E.J.; Wirth, B. A single nucleotide in the SMN gene regulates splicing and is responsible for spinal muscular atrophy. Proc. Natl. Acad. Sci. USA 1999, 96, 6307–6311. [Google Scholar] [CrossRef]
- Yeo, C.J.J.; Darras, B.T. Overturning the Paradigm of Spinal Muscular Atrophy as Just a Motor Neuron Disease. Pediatr. Neurol. 2020, 109, 12–19. [Google Scholar] [CrossRef]
- Nash, L.A.; Burns, J.K.; Chardon, J.W.; Kothary, R.; Parks, R.J. Spinal Muscular Atrophy: More than a Disease of Motor Neurons? Curr. Mol. Med. 2016, 16, 779–792. [Google Scholar] [CrossRef] [PubMed]
- Simone, C.; Ramirez, A.; Bucchia, M.; Rinchetti, P.; Rideout, H.; Papadimitriou, D.; Re, D.B.; Corti, S. Is spinal muscular atrophy a disease of the motor neurons only: Pathogenesis and therapeutic implications? Cell Mol. Life Sci. 2016, 73, 1003–1020. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.K.; Jha, N.N.; Feng, Z.; Faleiro, M.R.; Chiriboga, C.A.; Wei-Lapierre, L.; Dirksen, R.T.; Ko, C.P.; Monani, U.R. Muscle-specific SMN reduction reveals motor neuron-independent disease in spinal muscular atrophy models. J. Clin. Investig. 2020, 130, 1271–1287. [Google Scholar] [CrossRef]
- Lee, Y.I.; Mikesh, M.; Smith, I.; Rimer, M.; Thompson, W. Muscles in a mouse model of spinal muscular atrophy show profound defects in neuromuscular development even in the absence of failure in neuromuscular transmission or loss of motor neurons. Dev. Biol. 2011, 356, 432–444. [Google Scholar] [CrossRef]
- Ripolone, M.; Ronchi, D.; Violano, R.; Vallejo, D.; Fagiolari, G.; Barca, E.; Lucchini, V.; Colombo, I.; Villa, L.; Berardinelli, A.; et al. Impaired Muscle Mitochondrial Biogenesis and Myogenesis in Spinal Muscular Atrophy. JAMA Neurol. 2015, 72, 666–675. [Google Scholar] [CrossRef]
- James, R.; Chaytow, H.; Ledahawsky, L.M.; Gillingwater, T.H. Revisiting the role of mitochondria in spinal muscular atrophy. Cell Mol. Life Sci. 2021, 78, 4785–4804. [Google Scholar] [CrossRef]
- Chemello, F.; Pozzobon, M.; Tsansizi, L.I.; Varanita, T.; Quintana-Cabrera, R.; Bonesso, D.; Piccoli, M.; Lanfranchi, G.; Giacomello, M.; Scorrano, L.; et al. Dysfunctional mitochondria accumulate in a skeletal muscle knockout model of Smn1, the causal gene of spinal muscular atrophy. Cell Death Dis. 2023, 14, 162. [Google Scholar] [CrossRef] [PubMed]
- Durmus, H.; Yilmaz, R.; Gulsen-Parman, Y.; Oflazer-Serdaroglu, P.; Cuttini, M.; Dursun, M.; Deymeer, F. Muscle magnetic resonance imaging in spinal muscular atrophy type 3: Selective and progressive involvement. Muscle Nerve 2017, 55, 651–656. [Google Scholar] [CrossRef] [PubMed]
- LoMauro, A.; Aliverti, A.; Mastella, C.; Arnoldi, M.T.; Banfi, P.; Baranello, G. Spontaneous Breathing Pattern as Respiratory Functional Outcome in Children with Spinal Muscular Atrophy (SMA). PLoS ONE 2016, 11, e0165818. [Google Scholar] [CrossRef] [PubMed]
- Michaud, M.; Arnoux, T.; Bielli, S.; Durand, E.; Rotrou, Y.; Jablonka, S.; Robert, F.; Giraudon-Paoli, M.; Riessland, M.; Mattei, M.G.; et al. Neuromuscular defects and breathing disorders in a new mouse model of spinal muscular atrophy. Neurobiol. Dis. 2010, 38, 125–135. [Google Scholar] [CrossRef]
- Fauroux, B.; Griffon, L.; Amaddeo, A.; Stremler, N.; Mazenq, J.; Khirani, S.; Baravalle-Einaudi, M. Respiratory management of children with spinal muscular atrophy (SMA). Arch. Pediatr. 2020, 27, 7S29–7S34. [Google Scholar] [CrossRef]
- Kariya, S.; Park, G.H.; Maeno-Hikichi, Y.; Leykekhman, O.; Lutz, C.; Arkovitz, M.S.; Landmesser, L.T.; Monani, U.R. Reduced SMN protein impairs maturation of the neuromuscular junctions in mouse models of spinal muscular atrophy. Hum. Mol. Genet. 2008, 17, 2552–2569. [Google Scholar] [CrossRef]
- Voigt, T.; Neve, A.; Schümperli, D. The craniosacral progression of muscle development influences the emergence of neuromuscular junction alterations in a severe murine model for spinal muscular atrophy. Neuropathol. Appl. Neurobiol. 2014, 40, 416–434. [Google Scholar] [CrossRef]
- Neve, A.; Trüb, J.; Saxena, S.; Schümperli, D. Central and peripheral defects in motor units of the diaphragm of spinal muscular atrophy mice. Mol. Cell Neurosci. 2016, 70, 30–41. [Google Scholar] [CrossRef]
- Le, T.T.; Pham, L.T.; Butchbach, M.E.; Zhang, H.L.; Monani, U.R.; Coovert, D.D.; Gavrilina, T.O.; Xing, L.; Bassell, G.J.; Burghes, A.H. SMNDelta7, the major product of the centromeric survival motor neuron (SMN2) gene, extends survival in mice with spinal muscular atrophy and associates with full-length SMN. Hum. Mol. Genet. 2005, 14, 845–857. [Google Scholar] [CrossRef]
- Butchbach, M.E.; Edwards, J.D.; Burghes, A.H. Abnormal motor phenotype in the SMNDelta7 mouse model of spinal muscular atrophy. Neurobiol. Dis. 2007, 27, 207–219. [Google Scholar] [CrossRef]
- Butchbach, M.E.; Rose, F.F.; Rhoades, S.; Marston, J.; McCrone, J.T.; Sinnott, R.; Lorson, C.L. Effect of diet on the survival and phenotype of a mouse model for spinal muscular atrophy. Biochem. Biophys. Res. Commun. 2010, 391, 835–840. [Google Scholar] [CrossRef] [PubMed]
- Qaisar, R.; Pharaoh, G.; Bhaskaran, S.; Xu, H.; Ranjit, R.; Bian, J.; Ahn, B.; Georgescu, C.; Wren, J.D.; Van Remmen, H. Restoration of Sarcoplasmic Reticulum Ca. Int. J. Mol. Sci. 2020, 22, 37. [Google Scholar] [CrossRef] [PubMed]
- Allen, D.G.; Lamb, G.D.; Westerblad, H. Skeletal muscle fatigue: Cellular mechanisms. Physiol. Rev. 2008, 88, 287–332. [Google Scholar] [CrossRef] [PubMed]
- Kong, L.; Wang, X.; Choe, D.W.; Polley, M.; Burnett, B.G.; Bosch-Marcé, M.; Griffin, J.W.; Rich, M.M.; Sumner, C.J. Impaired synaptic vesicle release and immaturity of neuromuscular junctions in spinal muscular atrophy mice. J. Neurosci. 2009, 29, 842–851. [Google Scholar] [CrossRef]
- Torres-Benito, L.; Neher, M.F.; Cano, R.; Ruiz, R.; Tabares, L. SMN requirement for synaptic vesicle, active zone and microtubule postnatal organization in motor nerve terminals. PLoS ONE 2011, 6, e26164. [Google Scholar] [CrossRef]
- Acsadi, G.; Lee, I.; Li, X.; Khaidakov, M.; Pecinova, A.; Parker, G.C.; Hüttemann, M. Mitochondrial dysfunction in a neural cell model of spinal muscular atrophy. J. Neurosci. Res. 2009, 87, 2748–2756. [Google Scholar] [CrossRef]
- Chan, D.C. Mitochondrial Dynamics and Its Involvement in Disease. Annu. Rev. Pathol. 2020, 15, 235–259. [Google Scholar] [CrossRef]
- Favaro, G.; Romanello, V.; Varanita, T.; Andrea Desbats, M.; Morbidoni, V.; Tezze, C.; Albiero, M.; Canato, M.; Gherardi, G.; De Stefani, D.; et al. DRP1-mediated mitochondrial shape controls calcium homeostasis and muscle mass. Nat. Commun. 2019, 10, 2576. [Google Scholar] [CrossRef]
- Xie, L.; Shi, F.; Li, Y.; Li, W.; Yu, X.; Zhao, L.; Zhou, M.; Hu, J.; Luo, X.; Tang, M.; et al. Drp1-dependent remodeling of mitochondrial morphology triggered by EBV-LMP1 increases cisplatin resistance. Signal Transduct. Target. Ther. 2020, 5, 56. [Google Scholar] [CrossRef]
- Loh, J.K.; Lin, C.C.; Yang, M.C.; Chou, C.H.; Chen, W.S.; Hong, M.C.; Cho, C.L.; Hsu, C.M.; Cheng, J.T.; Chou, A.K.; et al. GSKIP- and GSK3-mediated anchoring strengthens cAMP/PKA/Drp1 axis signaling in the regulation of mitochondrial elongation. Biochim. Biophys. Acta 2015, 1853, 1796–1807. [Google Scholar] [CrossRef][Green Version]
- Gomes, L.C.; Di Benedetto, G.; Scorrano, L. During autophagy mitochondria elongate, are spared from degradation and sustain cell viability. Nat. Cell Biol. 2011, 13, 589–598. [Google Scholar] [CrossRef] [PubMed]
- Navratil, M.; Terman, A.; Arriaga, E.A. Giant mitochondria do not fuse and exchange their contents with normal mitochondria. Exp. Cell Res. 2008, 314, 164–172. [Google Scholar] [CrossRef] [PubMed]
- Leduc-Gaudet, J.P.; Picard, M.; St-Jean Pelletier, F.; Sgarioto, N.; Auger, M.J.; Vallée, J.; Robitaille, R.; St-Pierre, D.H.; Gouspillou, G. Mitochondrial morphology is altered in atrophied skeletal muscle of aged mice. Oncotarget 2015, 6, 17923–17937. [Google Scholar] [CrossRef] [PubMed]
- Rambold, A.S.; Kostelecky, B.; Elia, N.; Lippincott-Schwartz, J. Tubular network formation protects mitochondria from autophagosomal degradation during nutrient starvation. Proc. Natl. Acad. Sci. USA 2011, 108, 10190–10195. [Google Scholar] [CrossRef] [PubMed]
- Cantó-Santos, J.; Grau-Junyent, J.M.; Garrabou, G. The Impact of Mitochondrial Deficiencies in Neuromuscular Diseases. Antioxidants 2020, 9, 964. [Google Scholar] [CrossRef] [PubMed]
- Marra, F.; Lunetti, P.; Curcio, R.; Lasorsa, F.M.; Capobianco, L.; Porcelli, V.; Dolce, V.; Fiermonte, G.; Scarcia, P. An Overview of Mitochondrial Protein Defects in Neuromuscular Diseases. Biomolecules 2021, 11, 1633. [Google Scholar] [CrossRef]
- Boncompagni, S.; Rossi, A.E.; Micaroni, M.; Beznoussenko, G.V.; Polishchuk, R.S.; Dirksen, R.T.; Protasi, F. Mitochondria are linked to calcium stores in striated muscle by developmentally regulated tethering structures. Mol. Biol. Cell 2009, 20, 1058–1067. [Google Scholar] [CrossRef]
- Bolaños, P.; Guillen, A.; Rojas, H.; Boncompagni, S.; Caputo, C. The use of CalciumOrange-5N as a specific marker of mitochondrial Ca2+ in mouse skeletal muscle fibers. Pflugers Arch. 2008, 455, 721–731. [Google Scholar] [CrossRef]
- Rossi, A.E.; Boncompagni, S.; Dirksen, R.T. Sarcoplasmic reticulum-mitochondrial symbiosis: Bidirectional signaling in skeletal muscle. Exerc. Sport. Sci. Rev. 2009, 37, 29–35. [Google Scholar] [CrossRef]
- Rossi, A.E.; Boncompagni, S.; Wei, L.; Protasi, F.; Dirksen, R.T. Differential impact of mitochondrial positioning on mitochondrial Ca(2+) uptake and Ca(2+) spark suppression in skeletal muscle. Am. J. Physiol. Cell Physiol. 2011, 301, C1128–C1139. [Google Scholar] [CrossRef] [PubMed]
- Filomeni, G.; De Zio, D.; Cecconi, F. Oxidative stress and autophagy: The clash between damage and metabolic needs. Cell Death Differ. 2015, 22, 377–388. [Google Scholar] [CrossRef]
- Brand, M.D.; Buckingham, J.A.; Esteves, T.C.; Green, K.; Lambert, A.J.; Miwa, S.; Murphy, M.P.; Pakay, J.L.; Talbot, D.A.; Echtay, K.S. Mitochondrial superoxide and aging: Uncoupling-protein activity and superoxide production. Biochem. Soc. Symp. 2004, 71, 203–213. [Google Scholar] [CrossRef]
- Wonsey, D.R.; Zeller, K.I.; Dang, C.V. The c-Myc target gene PRDX3 is required for mitochondrial homeostasis and neoplastic transformation. Proc. Natl. Acad. Sci. USA 2002, 99, 6649–6654. [Google Scholar] [CrossRef]
- Powers, S.K.; Jackson, M.J. Exercise-induced oxidative stress: Cellular mechanisms and impact on muscle force production. Physiol. Rev. 2008, 88, 1243–1276. [Google Scholar] [CrossRef] [PubMed]
- Gruber, J.; Fong, S.; Chen, C.B.; Yoong, S.; Pastorin, G.; Schaffer, S.; Cheah, I.; Halliwell, B. Mitochondria-targeted antioxidants and metabolic modulators as pharmacological interventions to slow ageing. Biotechnol. Adv. 2013, 31, 563–592. [Google Scholar] [CrossRef]
- Kerley, R.N.; McCarthy, C.; Kell, D.B.; Kenny, L.C. The potential therapeutic effects of ergothioneine in pre-eclampsia. Free Radic. Biol. Med. 2018, 117, 145–157. [Google Scholar] [CrossRef]
- Rossi, R.; Bottinelli, R.; Sorrentino, V.; Reggiani, C. Response to caffeine and ryanodine receptor isoforms in mouse skeletal muscles. Am. J. Physiol. Cell Physiol. 2001, 281, C585–C594. [Google Scholar] [CrossRef]
- Careccia, G.; Saclier, M.; Tirone, M.; Ruggieri, E.; Principi, E.; Raffaghello, L.; Torchio, S.; Recchia, D.; Canepari, M.; Gorzanelli, A.; et al. Rebalancing expression of HMGB1 redox isoforms to counteract muscular dystrophy. Sci. Transl. Med. 2021, 13, eaay8416. [Google Scholar] [CrossRef]
- Mahmoodzadeh, S.; Koch, K.; Schriever, C.; Xu, J.; Steinecker, M.; Leber, J.; Dworatzek, E.; Purfürst, B.; Kunz, S.; Recchia, D.; et al. Age-related decline in murine heart and skeletal muscle performance is attenuated by reduced Ahnak1 expression. J. Cachexia Sarcopenia Muscle 2021, 12, 1249–1265. [Google Scholar] [CrossRef]
- Halliwell, B.; Cheah, I.K.; Tang, R.M.Y. Ergothioneine—A diet-derived antioxidant with therapeutic potential. FEBS Lett. 2018, 592, 3357–3366. [Google Scholar] [CrossRef] [PubMed]
- Borodina, I.; Kenny, L.C.; McCarthy, C.M.; Paramasivan, K.; Pretorius, E.; Roberts, T.J.; van der Hoek, S.A.; Kell, D.B. The biology of ergothioneine, an antioxidant nutraceutical. Nutr. Res. Rev. 2020, 33, 190–217. [Google Scholar] [CrossRef] [PubMed]
- Roda, E.; Priori, E.C.; Ratto, D.; De Luca, F.; Di Iorio, C.; Angelone, P.; Locatelli, C.A.; Desiderio, A.; Goppa, L.; Savino, E.; et al. Neuroprotective Metabolites of. Int. J. Mol. Sci. 2021, 22, 6379. [Google Scholar] [CrossRef] [PubMed]
- Perry, C.G.; Kane, D.A.; Lin, C.T.; Kozy, R.; Cathey, B.L.; Lark, D.S.; Kane, C.L.; Brophy, P.M.; Gavin, T.P.; Anderson, E.J.; et al. Inhibiting myosin-ATPase reveals a dynamic range of mitochondrial respiratory control in skeletal muscle. Biochem. J. 2011, 437, 215–222. [Google Scholar] [CrossRef]
- Zuccarelli, L.; Baldassarre, G.; Magnesa, B.; Degano, C.; Comelli, M.; Gasparini, M.; Manferdelli, G.; Marzorati, M.; Mavelli, I.; Pilotto, A.; et al. Peripheral impairments of oxidative metabolism after a 10-day bed rest are upstream of mitochondrial respiration. J. Physiol. 2021, 599, 4813–4829. [Google Scholar] [CrossRef]
- Makrecka-Kuka, M.; Krumschnabel, G.; Gnaiger, E. High-Resolution Respirometry for Simultaneous Measurement of Oxygen and Hydrogen Peroxide Fluxes in Permeabilized Cells, Tissue Homogenate and Isolated Mitochondria. Biomolecules 2015, 5, 1319–1338. [Google Scholar] [CrossRef]
- Larsen, S.; Nielsen, J.; Hansen, C.N.; Nielsen, L.B.; Wibrand, F.; Stride, N.; Schroder, H.D.; Boushel, R.; Helge, J.W.; Dela, F.; et al. Biomarkers of mitochondrial content in skeletal muscle of healthy young human subjects. J. Physiol. 2012, 590, 3349–3360. [Google Scholar] [CrossRef]





| SMN1 WT FORWARD | 5′CTCCGGGATATTGGGATTG 3′ |
| SMN1 WT REVERSE | 5′ TTTCTTCTGGCTGTGCCTTT 3′ |
| SMN1 MUTANT REVERSE | 5′ GGTAACGCCAGGGTTTTCC 3′ |
| SMN2 WT FORWARD | 5′ CTGACCTACCAGGGATGAGG 3′ |
| SMN2 TRANSGENE | 5′ GGTCTGTTCTACAGCCACAGC 3′ |
| SMN2 WT REVERSE | 5′CCCAGGTGGTTTATAGACTCAGA 3′ |
| SMN TRANSGENE 01 | 5′ TCCATTTCCTTCTGGACCAC 3′ |
| SMN TRANSGENE 02 | 5′ ACCCATTCCACTTCCTTTTT 3′ |
| SMN POSITIVE CTRL FORWARD | 5′ CAAATGTTGCTTGTCTGGTG 3′ |
| SMN POSITIVE CTRL REVERSE | 5′ GTCAGTCGAGTGCACAGTTT 3′ |
| Forward Primer | Reverse Primer | |
|---|---|---|
| SERCA1 | TGTTTGTCCTATTTCGGGGTG | AATCCGCACAAGCAGGTCTTC |
| SERCA2 | GAGAACGCTCACACAAAGACC | CAATTCGTTGGAGCCCCAT |
| PGC1α | ACCCCAGAGTCACCAAATGA | CGAAGCCTTGAAAGGGTTATC |
| SIRT1 | CCGCGGATAGGTCCATATACT | AACAATCTGCCACAGCGTCA |
| NRF1 | GCACCTTTGGAGAATGTGGT | CTGAGCCTGGGTCATTTTGT |
| NRF2 | TTCTTTCAGCAGCATCCTCTCCAC | ACAGCCTTCAATAGTCCCGTCCAG |
| BNIP3 | TTCCACTAGCACCTTCTGATGA | GAACACCGCATTTACAGAACAA |
| Primary Antibody | Species | Dilution | Supplier | Catalog Number |
|---|---|---|---|---|
| p-AMPK | Rabbit | 1:1000 | Cell Signaling | #4188 |
| AMPK | Rabbit | 1:1000 | Cell Signaling | #2532 |
| PGC1α | Rabbit | 1:1000 | Abcam | Ab54481 |
| TOM20 | Rabbit | 1:1000 | Santa Cruz | sc-11415 |
| OPA1 | Mouse | 1:3000 | Abcam | Ab157457 |
| MNF1 | Mouse | 1:1000 | Abcam | Ab57602 |
| MNF2 | Rabbit | 1:1000 | Abcam | Ab50843 |
| FIS1 | Rabbit | 1:1000 | Abcam | Ab71498 |
| DRP1 pSer616 | Rabbit | 1:1000 | Cell Signaling | #3455 |
| DRP1 pSer637 | Rabbit | 1:1000 | Cell Signaling | #4867 |
| DRP1 | Rabbit | 1:1000 | Cell Signaling | #8570 |
| SOD1 | Rabbit | 1:1000 | Abcam | Ab16831 |
| Catalase | Rabbit | 1:1000 | Abcam | Ab52477 |
| Hsp27 | Mouse | 1:1000 | Abcam | Ab2790 |
| Hsp70 | Mouse | 1:1000 | Abcam | Ab47455 |
| PRDX3 | Rabbit | 1:1000 | Abcam | Ab73349 |
| LC3B | Rabbit | 1:1000 | Sigma | L7543 |
| p62 | Rabbit | 1:2000 | Cell Signaling | #5114 |
| PINK1 | Rabbit | 1:1000 | Invitrogen | #PA1-16604 |
| PARKIN | Mouse | 1:2000 | Invitrogen | #39-0900 |
| Tubulin | Mouse | 1:2000 | Sigma | T6199 |
| OXPHOS | Mouse | 1:1000 | Abcam | Ab110413 |
| SERCA1 | Rabbit | 1:1000 | Cell Signaling | #12293 |
| SERCA2 | Rabbit | 1:1000 | Cell Signaling | #9580 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cadile, F.; Recchia, D.; Ansaldo, M.; Rossi, P.; Rastelli, G.; Boncompagni, S.; Brocca, L.; Pellegrino, M.A.; Canepari, M. Diaphragm Fatigue in SMNΔ7 Mice and Its Molecular Determinants: An Underestimated Issue. Int. J. Mol. Sci. 2023, 24, 14953. https://doi.org/10.3390/ijms241914953
Cadile F, Recchia D, Ansaldo M, Rossi P, Rastelli G, Boncompagni S, Brocca L, Pellegrino MA, Canepari M. Diaphragm Fatigue in SMNΔ7 Mice and Its Molecular Determinants: An Underestimated Issue. International Journal of Molecular Sciences. 2023; 24(19):14953. https://doi.org/10.3390/ijms241914953
Chicago/Turabian StyleCadile, Francesca, Deborah Recchia, Massimiliano Ansaldo, Paola Rossi, Giorgia Rastelli, Simona Boncompagni, Lorenza Brocca, Maria Antonietta Pellegrino, and Monica Canepari. 2023. "Diaphragm Fatigue in SMNΔ7 Mice and Its Molecular Determinants: An Underestimated Issue" International Journal of Molecular Sciences 24, no. 19: 14953. https://doi.org/10.3390/ijms241914953
APA StyleCadile, F., Recchia, D., Ansaldo, M., Rossi, P., Rastelli, G., Boncompagni, S., Brocca, L., Pellegrino, M. A., & Canepari, M. (2023). Diaphragm Fatigue in SMNΔ7 Mice and Its Molecular Determinants: An Underestimated Issue. International Journal of Molecular Sciences, 24(19), 14953. https://doi.org/10.3390/ijms241914953







