Crystal Structure of DNA Replication Protein SsbA Complexed with the Anticancer Drug 5-Fluorouracil
Abstract
1. Introduction
2. Results
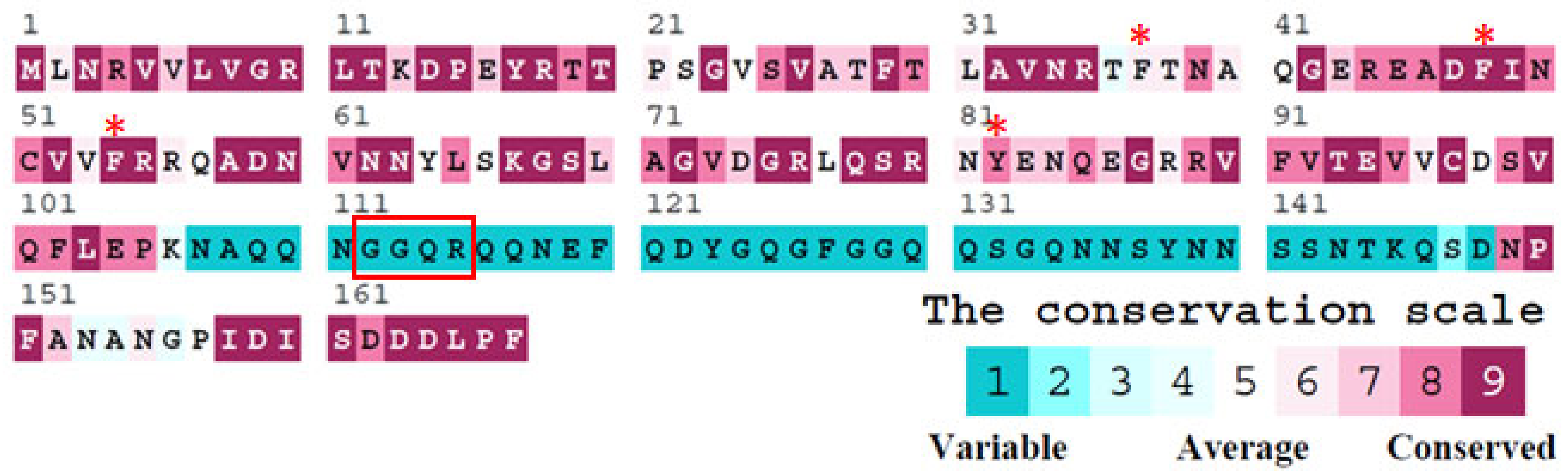
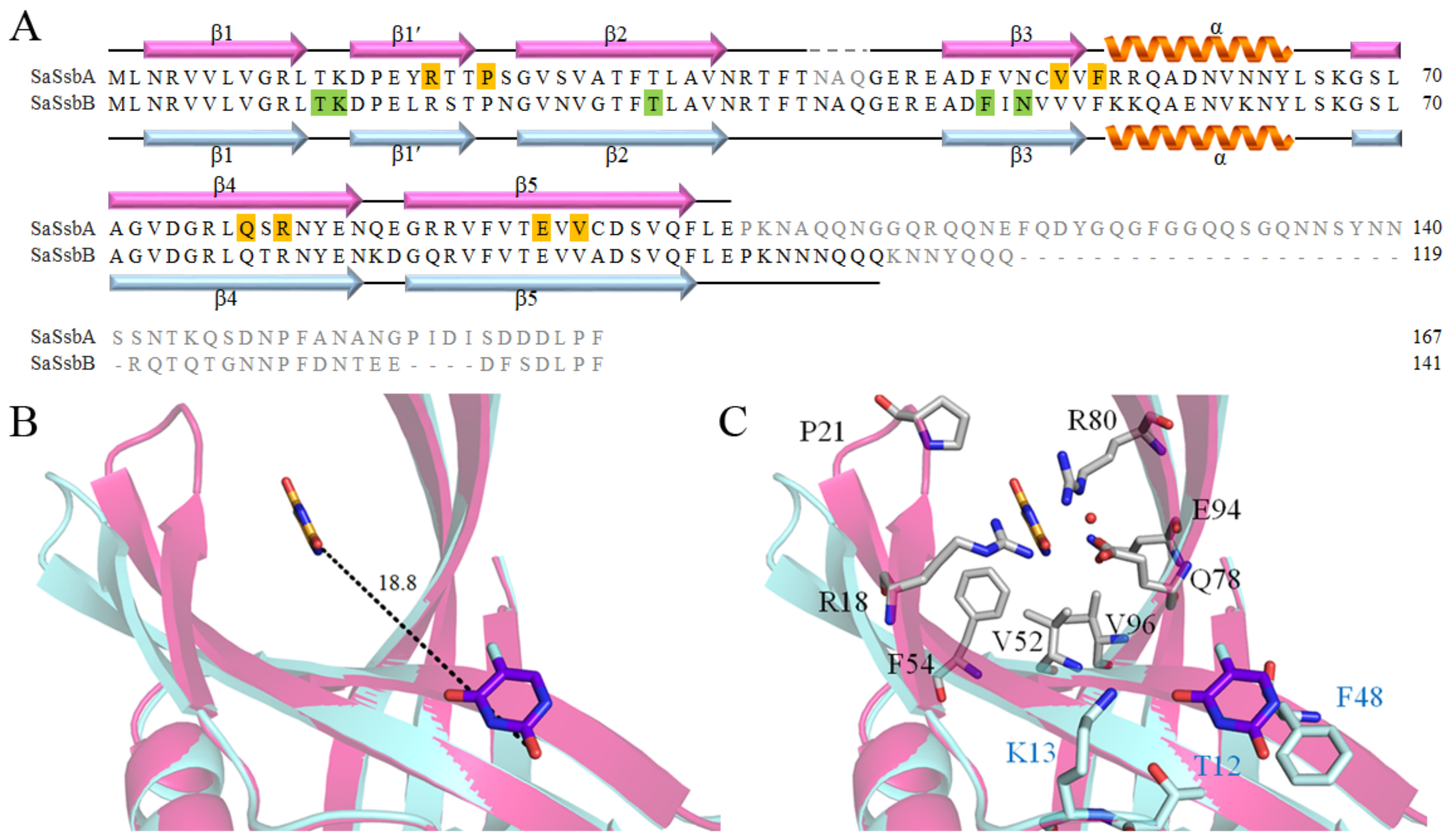
2.1. Sequence Analysis of SaSsbA
2.2. Crystallization of the Glycerol-Bound SaSsbA and the SaSsbA-5-FU Complex
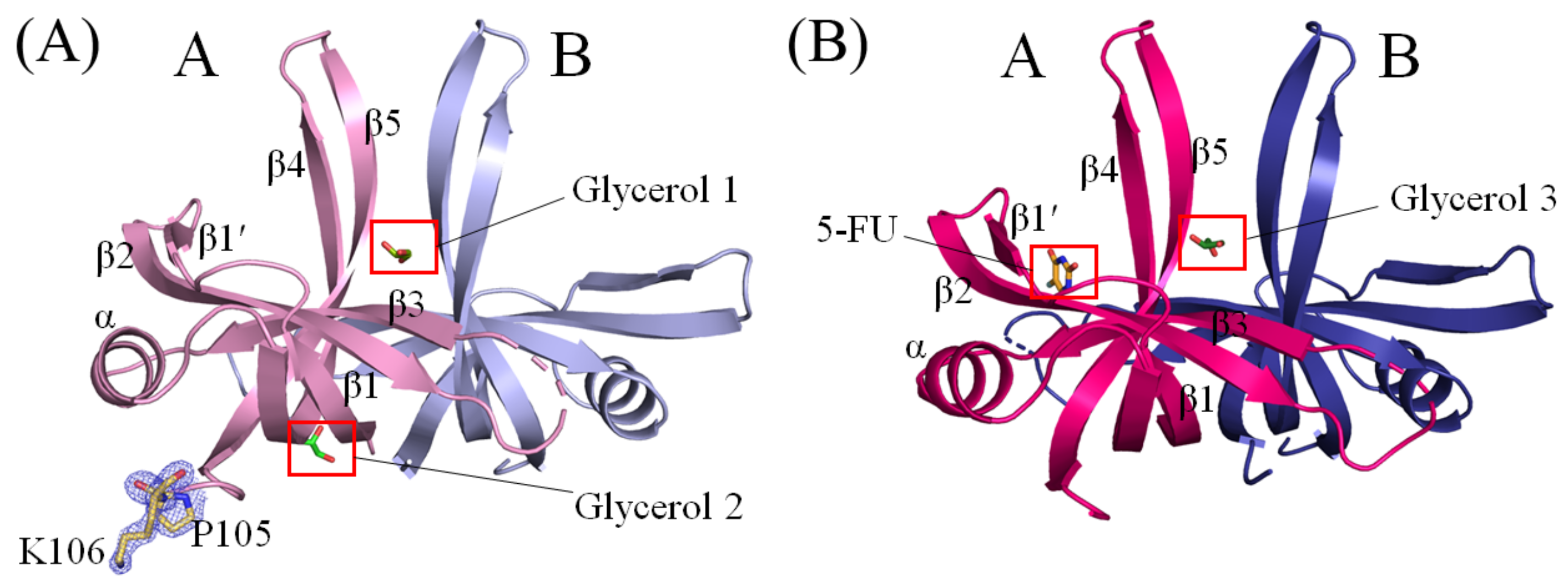
2.3. Crystal Structure of Glycerol-Bound SaSsbA
2.4. Crystal Structure of SaSsbA Complexed with 5-FU
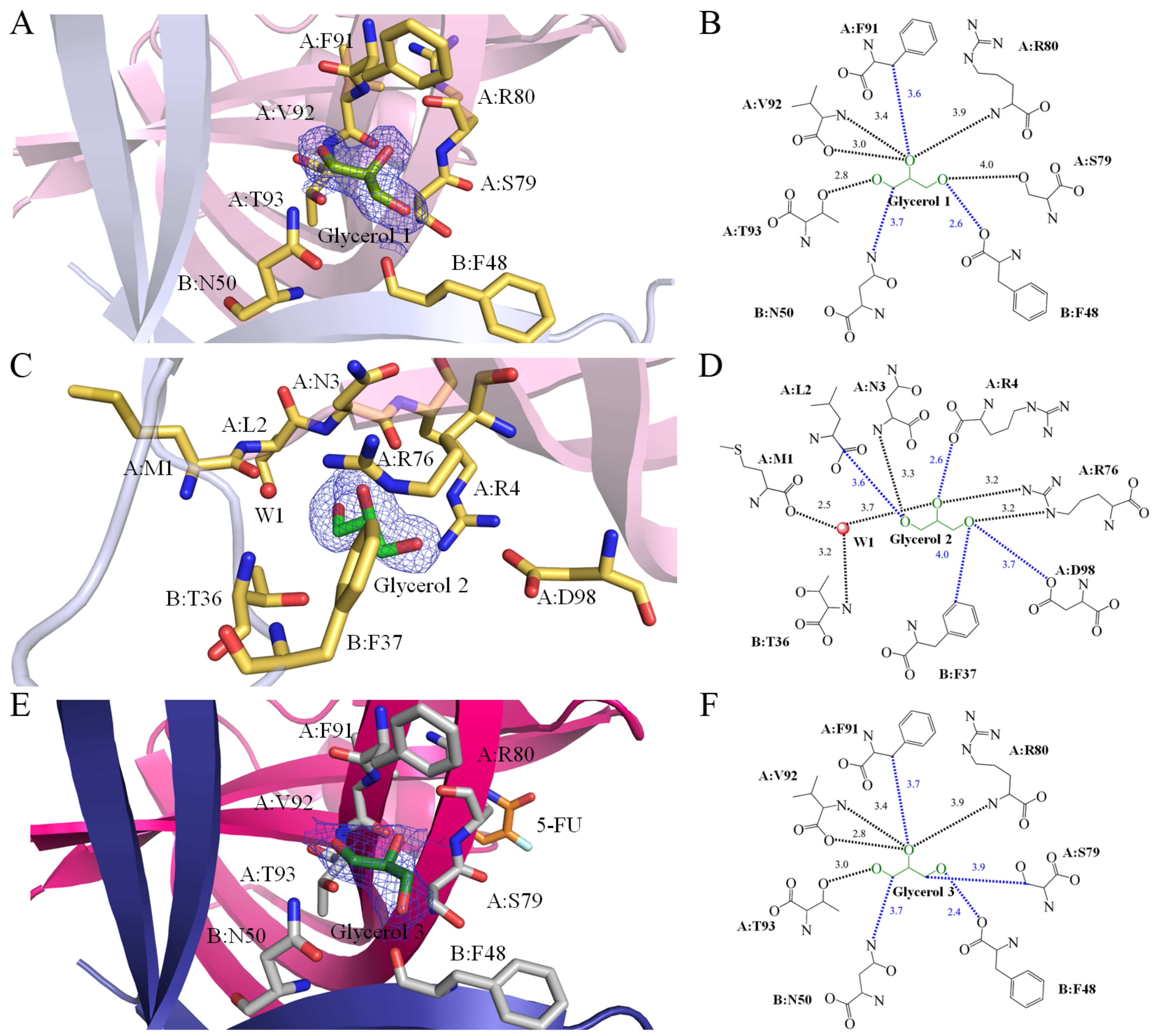
2.5. Glycerol 1 Binding Mode of SaSsbA
2.6. Glycerol 2 Binding Mode of SaSsbA
2.7. Glycerol 3 Binding Mode of SaSsbA
2.8. 5-FU Binding Mode of SaSsbA
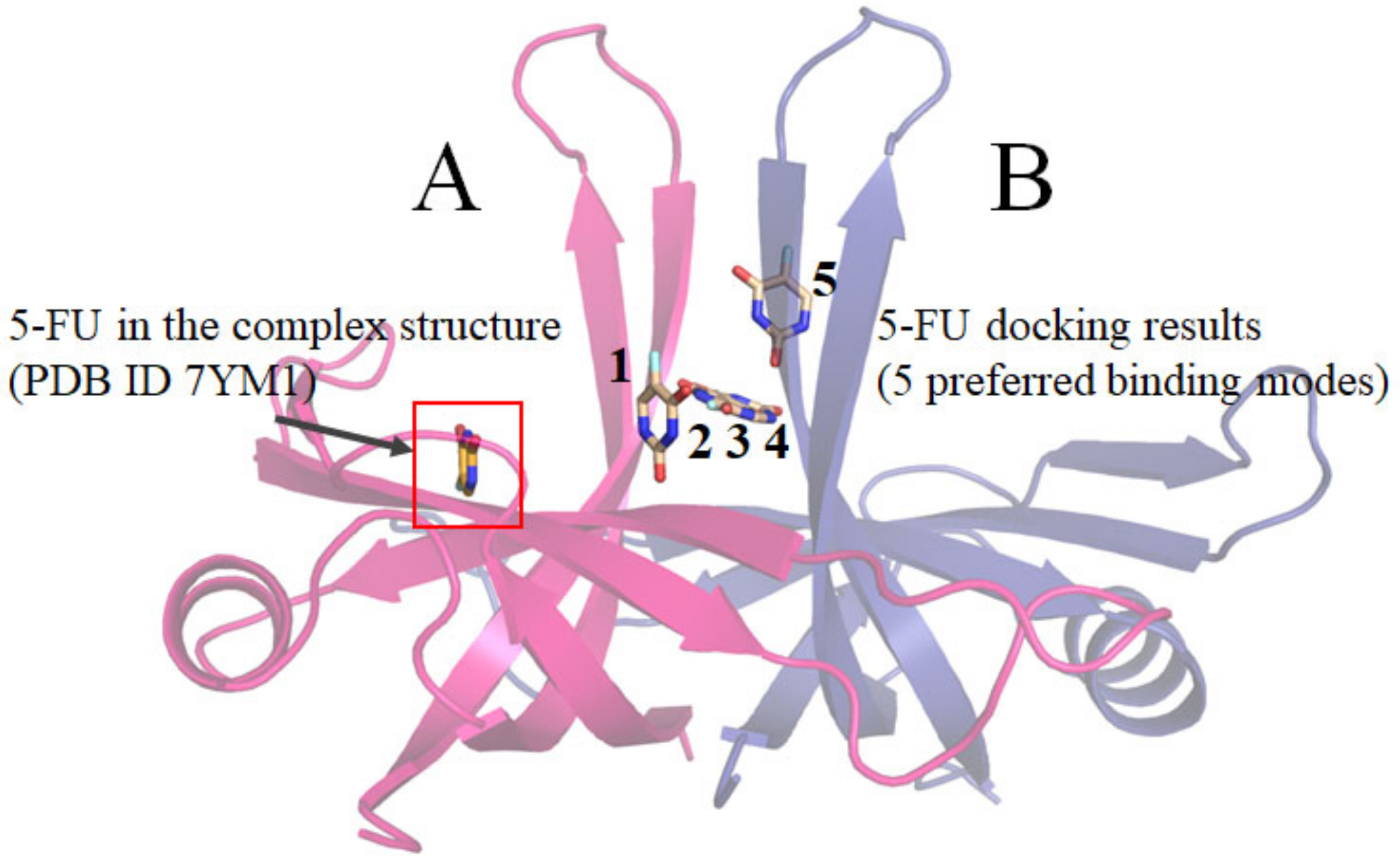
2.9. Comparative Analysis of 5-FU Binding Sites in Different Binding States of SaSsbA
2.10. Comparative Structural Analysis of 5-FU Binding Sites in the 5-FU-Bound and Unbound States of SaSsbA
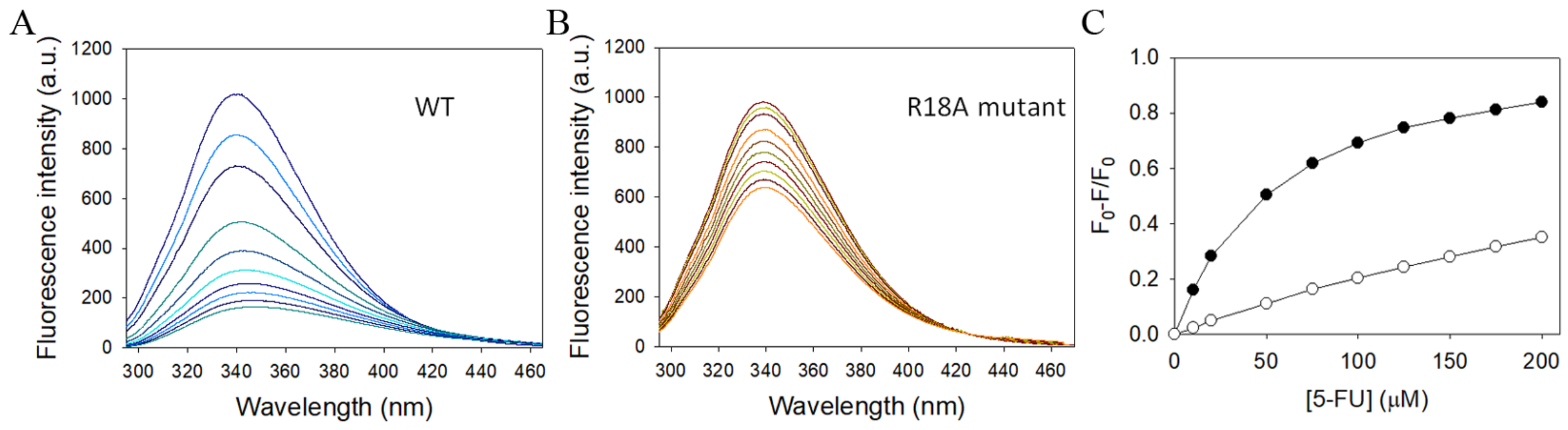
2.11. Structure-Based Mutational Analysis
2.12. Distinct 5-FU Binding Modes in SaSsbA and SaSsbB
3. Discussion
4. Materials and Methods
4.1. Protein Expression and Purification
4.2. Site-Directed Mutagenesis
4.3. Crystallization Experiments
4.4. X-ray Diffraction Data and Structure Determination
4.5. Fluorescence Quenching
4.6. MOE-Dock Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sessler, J.L.; Lawrence, C.M.; Jayawickramarajah, J. Molecular recognition via base-pairing. Chem. Soc. Rev. 2007, 36, 314–325. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Thompson, C.B. Metabolic regulation of cell growth and proliferation. Nat. Rev. Mol. Cell Biol. 2019, 20, 436–450. [Google Scholar] [CrossRef] [PubMed]
- Fung, S.K.; Lok, A.S. Drug insight: Nucleoside and nucleotide analog inhibitors for hepatitis B. Nat. Clin. Pract. Gastroenterol. Hepatol. 2004, 1, 90–97. [Google Scholar] [CrossRef] [PubMed]
- Longley, D.B.; Harkin, D.P.; Johnston, P.G. 5-fluorouracil: Mechanisms of action and clinical strategies. Nat. Rev. Cancer 2003, 3, 330–338. [Google Scholar] [CrossRef]
- Peters, H.L.; Ku, T.C.; Seley-Radtke, K.L. Flexibility as a Strategy in Nucleoside Antiviral Drug Design. Curr. Med. Chem. 2015, 22, 3910–3921. [Google Scholar] [CrossRef]
- Yssel, A.E.J.; Vanderleyden, J.; Steenackers, H.P. Repurposing of nucleoside- and nucleobase-derivative drugs as antibiotics and biofilm inhibitors. J. Antimicrob Chemother. 2017, 72, 2156–2170. [Google Scholar] [CrossRef]
- Ku, S.K.; Baek, M.C.; Bae, J.S. Anti-inflammatory effects of methylthiouracil in vitro and in vivo. Toxicol. Appl. Pharmacol. 2015, 288, 374–386. [Google Scholar] [CrossRef]
- Alvarez, P.; Marchal, J.A.; Boulaiz, H.; Carrillo, E.; Velez, C.; Rodriguez-Serrano, F.; Melguizo, C.; Prados, J.; Madeddu, R.; Aranega, A. 5-Fluorouracil derivatives: A patent review. Expert Opin. Ther. Pat. 2012, 22, 107–123. [Google Scholar] [CrossRef]
- Wilson, P.M.; Danenberg, P.V.; Johnston, P.G.; Lenz, H.J.; Ladner, R.D. Standing the test of time: Targeting thymidylate biosynthesis in cancer therapy. Nat. Rev. Clin. Oncol. 2014, 11, 282–298. [Google Scholar] [CrossRef]
- Huang, Y.H.; Ning, Z.J.; Huang, C.Y. Crystal structure of dihydropyrimidinase in complex with anticancer drug 5-fluorouracil. Biochem. Biophys. Res. Commun. 2019, 519, 160–165. [Google Scholar] [CrossRef]
- Huang, C.Y. Structure, catalytic mechanism, posttranslational lysine carbamylation, and inhibition of dihydropyrimidinases. Adv. Protein Chem. Struct. Biol. 2020, 122, 63–96. [Google Scholar] [PubMed]
- Van Kuilenburg, A.B.; Meinsma, R.; Zonnenberg, B.A.; Zoetekouw, L.; Baas, F.; Matsuda, K.; Tamaki, N.; van Gennip, A.H. Dihydropyrimidinase deficiency and severe 5-fluorouracil toxicity. Clin. Cancer Res. 2003, 9, 4363–4367. [Google Scholar]
- Van Kuilenburg, A.B.; Dobritzsch, D.; Meijer, J.; Meinsma, R.; Benoist, J.F.; Assmann, B.; Schubert, S.; Hoffmann, G.F.; Duran, M.; de Vries, M.C.; et al. Dihydropyrimidinase deficiency: Phenotype, genotype and structural consequences in 17 patients. Biochim. Biophys. Acta 2010, 1802, 639–648. [Google Scholar] [CrossRef] [PubMed]
- Lin, E.S.; Huang, Y.H.; Yang, P.C.; Peng, W.F.; Huang, C.Y. Complexed Crystal Structure of the Dihydroorotase Domain of Human CAD Protein with the Anticancer Drug 5-Fluorouracil. Biomolecules 2023, 13, 149. [Google Scholar] [CrossRef] [PubMed]
- Sillo, T.O.; Beggs, A.D.; Middleton, G.; Akingboye, A. The Gut Microbiome, Microsatellite Status and the Response to Immunotherapy in Colorectal Cancer. Int. J. Mol. Sci. 2023, 24, 5767. [Google Scholar] [CrossRef]
- Jaye, K.; Chang, D.; Li, C.G.; Bhuyan, D.J. Gut Metabolites and Breast Cancer: The Continuum of Dysbiosis, Breast Cancer Risk, and Potential Breast Cancer Therapy. Int. J. Mol. Sci. 2022, 23, 9490. [Google Scholar] [CrossRef]
- Alexander, J.L.; Wilson, I.D.; Teare, J.; Marchesi, J.R.; Nicholson, J.K.; Kinross, J.M. Gut microbiota modulation of chemotherapy efficacy and toxicity. Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 356–365. [Google Scholar] [CrossRef]
- Nakayama, H.; Kinouchi, T.; Kataoka, K.; Akimoto, S.; Matsuda, Y.; Ohnishi, Y. Intestinal anaerobic bacteria hydrolyse sorivudine, producing the high blood concentration of 5-(E)-(2-bromovinyl)uracil that increases the level and toxicity of 5-fluorouracil. Pharmacogenetics 1997, 7, 35–43. [Google Scholar] [CrossRef]
- Narendra, G.; Choudhary, S.; Raju, B.; Verma, H.; Silakari, O. Role of Genetic Polymorphisms in Drug-Metabolizing Enzyme-Mediated Toxicity and Pharmacokinetic Resistance to Anti-Cancer Agents: A Review on the Pharmacogenomics Aspect. Clin. Pharmacokinet. 2022, 61, 1495–1517. [Google Scholar] [CrossRef]
- Schneider, J.J.; Galettis, P.; Martin, J.H. Overcoming barriers to implementing precision dosing with 5-fluorouracil and capecitabine. Br. J. Clin. Pharmacol. 2021, 87, 317–325. [Google Scholar] [CrossRef]
- Gmeiner, W.H. A narrative review of genetic factors affecting fluoropyrimidine toxicity. Precis. Cancer Med. 2021, 4, 38. [Google Scholar] [CrossRef] [PubMed]
- Kobuchi, S.; Ito, Y. Application of Pharmacometrics of 5-Fluorouracil to Personalized Medicine: A Tool for Predicting Pharmacokinetic-Pharmacodynamic/Toxicodynamic Responses. Anticancer Res. 2020, 40, 6585–6597. [Google Scholar] [CrossRef] [PubMed]
- Jiricny, J. The multifaceted mismatch-repair system. Nat. Rev. Mol. Cell Biol. 2006, 7, 335–346. [Google Scholar] [CrossRef]
- Reyes-Lamothe, R.; Sherratt, D.J. The bacterial cell cycle, chromosome inheritance and cell growth. Nat. Rev. Microbiol. 2019, 17, 467–478. [Google Scholar] [CrossRef]
- Papamichos-Chronakis, M.; Peterson, C.L. Chromatin and the genome integrity network. Nat. Rev. Genet. 2013, 14, 62–75. [Google Scholar] [CrossRef] [PubMed]
- Cox, M.M.; Goodman, M.F.; Kreuzer, K.N.; Sherratt, D.J.; Sandler, S.J.; Marians, K.J. The importance of repairing stalled replication forks. Nature 2000, 404, 37–41. [Google Scholar] [CrossRef]
- Meyer, R.R.; Laine, P.S. The single-stranded DNA-binding protein of Escherichia coli. Microbiol. Rev. 1990, 54, 342–380. [Google Scholar] [CrossRef]
- Croft, L.V.; Bolderson, E.; Adams, M.N.; El-Kamand, S.; Kariawasam, R.; Cubeddu, L.; Gamsjaeger, R.; Richard, D.J. Human single-stranded DNA binding protein 1 (hSSB1, OBFC2B), a critical component of the DNA damage response. Semin. Cell Dev. Biol. 2019, 86, 121–128. [Google Scholar] [CrossRef] [PubMed]
- Shereda, R.D.; Kozlov, A.G.; Lohman, T.M.; Cox, M.M.; Keck, J.L. SSB as an organizer/mobilizer of genome maintenance complexes. Crit. Rev. Biochem. Mol. Biol. 2008, 43, 289–318. [Google Scholar] [CrossRef]
- Richard, D.J.; Bolderson, E.; Cubeddu, L.; Wadsworth, R.I.; Savage, K.; Sharma, G.G.; Nicolette, M.L.; Tsvetanov, S.; McIlwraith, M.J.; Pandita, R.K.; et al. Single-stranded DNA-binding protein hSSB1 is critical for genomic stability. Nature 2008, 453, 677–681. [Google Scholar] [CrossRef]
- Lohman, T.M.; Ferrari, M.E. Escherichia coli single-stranded DNA-binding protein: Multiple DNA-binding modes and cooperativities. Annu. Rev. Biochem. 1994, 63, 527–570. [Google Scholar] [CrossRef] [PubMed]
- Antony, E.; Lohman, T.M. Dynamics of E. coli single stranded DNA binding (SSB) protein-DNA complexes. Semin. Cell Dev. Biol. 2019, 86, 102–111. [Google Scholar] [CrossRef] [PubMed]
- Byrne, B.M.; Oakley, G.G. Replication protein A, the laxative that keeps DNA regular: The importance of RPA phosphorylation in maintaining genome stability. Semin. Cell Dev. Biol. 2019, 86, 112–120. [Google Scholar] [CrossRef]
- Iftode, C.; Daniely, Y.; Borowiec, J.A. Replication protein A (RPA): The eukaryotic SSB. Crit. Rev. Biochem. Mol. Biol. 1999, 34, 141–180. [Google Scholar] [CrossRef] [PubMed]
- Dickey, T.H.; Altschuler, S.E.; Wuttke, D.S. Single-stranded DNA-binding proteins: Multiple domains for multiple functions. Structure 2013, 21, 1074–1084. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Curth, U.; Urbanke, C.; Kang, C. Crystal structure of human mitochondrial single-stranded DNA binding protein at 2.4 A resolution. Nat. Struct. Biol. 1997, 4, 153–157. [Google Scholar] [CrossRef]
- Murzin, A.G. OB(oligonucleotide/oligosaccharide binding)-fold: Common structural and functional solution for non-homologous sequences. EMBO J. 1993, 12, 861–867. [Google Scholar] [CrossRef] [PubMed]
- Bianco, P.R. The mechanism of action of the SSB interactome reveals it is the first OB-fold family of genome guardians in prokaryotes. Protein Sci. 2021, 30, 1757–1775. [Google Scholar] [CrossRef]
- Bianco, P.R. The tale of SSB. Prog. Biophys. Mol. Biol. 2017, 127, 111–118. [Google Scholar] [CrossRef]
- Lin, E.S.; Luo, R.H.; Huang, C.Y. A Complexed Crystal Structure of a Single-Stranded DNA-Binding Protein with Quercetin and the Structural Basis of Flavonol Inhibition Specificity. Int. J. Mol. Sci. 2022, 23, 588. [Google Scholar] [CrossRef]
- Lin, E.S.; Huang, Y.H.; Luo, R.H.; Basharat, Z.; Huang, C.Y. Crystal Structure of an SSB Protein from Salmonella enterica and Its Inhibition by Flavanonol Taxifolin. Int. J. Mol. Sci. 2022, 23, 4399. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.Y. Crystal structure of SSB complexed with inhibitor myricetin. Biochem. Biophys. Res. Commun. 2018, 504, 704–708. [Google Scholar] [CrossRef]
- Glanzer, J.G.; Endres, J.L.; Byrne, B.M.; Liu, S.; Bayles, K.W.; Oakley, G.G. Identification of inhibitors for single-stranded DNA-binding proteins in eubacteria. J. Antimicrob. Chemother. 2016, 71, 3432–3440. [Google Scholar] [CrossRef]
- Liu, H.W.; Chiang, W.Y.; Huang, Y.H.; Huang, C.Y. The Inhibitory Effects and Cytotoxic Activities of the Stem Extract of Sarracenia purpurea against Melanoma Cells and the SsbA Protein. Plants 2022, 11, 3164. [Google Scholar] [CrossRef]
- Huang, Y.H.; Guan, H.H.; Chen, C.J.; Huang, C.Y. Staphylococcus aureus single-stranded DNA-binding protein SsbA can bind but cannot stimulate PriA helicase. PLoS ONE 2017, 12, e0182060. [Google Scholar] [CrossRef]
- Chen, K.L.; Cheng, J.H.; Lin, C.Y.; Huang, Y.H.; Huang, C.Y. Characterization of single-stranded DNA-binding protein SsbB from Staphylococcus aureus: SsbB cannot stimulate PriA helicase. RSC Adv. 2018, 8, 28367–28375. [Google Scholar] [CrossRef]
- Huang, Y.H.; Huang, C.Y. SAAV2152 is a single-stranded DNA binding protein: The third SSB in Staphylococcus aureus. Oncotarget 2018, 9, 20239–20254. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lin, E.S.; Huang, Y.H.; Chung, J.C.; Su, H.H.; Huang, C.Y. The Inhibitory Effects and Cytotoxic Activities of the Stem Extract of Nepenthes miranda against Single-Stranded DNA-Binding Protein and Oral Carcinoma Cells. Plants 2023, 12, 2188. [Google Scholar] [CrossRef] [PubMed]
- Lin, E.S.; Huang, C.Y. Crystal structure of the single-stranded DNA-binding protein SsbB in complex with the anticancer drug 5-fluorouracil: Extension of the 5-fluorouracil interactome to include the oligonucleotide/oligosaccharide-binding fold protein. Biochem. Biophys. Res. Commun. 2021, 534, 41–46. [Google Scholar] [CrossRef]
- Curreri, A.R.; Ansfield, F.J.; Mc, I.F.; Waisman, H.A.; Heidelberger, C. Clinical studies with 5-fluorouracil. Cancer Res. 1958, 18, 478–484. [Google Scholar]
- Eidinoff, M.L.; Knoll, J.E.; Klein, D. Effect of 5-fluorouracil on the incorporation of precursors into nucleic acid pyrimidines. Arch. Biochem. Biophys. 1957, 71, 274–275. [Google Scholar] [CrossRef]
- Raghunathan, S.; Kozlov, A.G.; Lohman, T.M.; Waksman, G. Structure of the DNA binding domain of E. coli SSB bound to ssDNA. Nat. Struct. Biol. 2000, 7, 648–652. [Google Scholar] [CrossRef] [PubMed]
- Lin, E.S.; Huang, Y.H.; Huang, C.Y. Characterization of the Chimeric PriB-SSBc Protein. Int. J. Mol. Sci. 2021, 22, 10854. [Google Scholar] [CrossRef] [PubMed]
- Lindner, C.; Nijland, R.; van Hartskamp, M.; Bron, S.; Hamoen, L.W.; Kuipers, O.P. Differential expression of two paralogous genes of Bacillus subtilis encoding single-stranded DNA binding protein. J. Bacteriol. 2004, 186, 1097–1105. [Google Scholar] [CrossRef]
- Huang, C.Y.; Hsu, C.H.; Sun, Y.J.; Wu, H.N.; Hsiao, C.D. Complexed crystal structure of replication restart primosome protein PriB reveals a novel single-stranded DNA-binding mode. Nucleic Acids Res. 2006, 34, 3878–3886. [Google Scholar] [CrossRef] [PubMed]
- Savvides, S.N.; Raghunathan, S.; Futterer, K.; Kozlov, A.G.; Lohman, T.M.; Waksman, G. The C-terminal domain of full-length E. coli SSB is disordered even when bound to DNA. Protein Sci. 2004, 13, 1942–1947. [Google Scholar] [CrossRef]
- Huang, Y.H.; Lin, E.S.; Huang, C.Y. Complexed crystal structure of SSB reveals a novel single-stranded DNA binding mode (SSB)3:1: Phe60 is not crucial for defining binding paths. Biochem. Biophys. Res. Commun. 2019, 520, 353–358. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.H.; Chen, I.C.; Huang, C.Y. Characterization of an SSB-dT25 complex: Structural insights into the S-shaped ssDNA binding conformation. RSC Adv. 2019, 9, 40388–40396. [Google Scholar] [CrossRef]
- Paradzik, T.; Ivic, N.; Filic, Z.; Manjasetty, B.A.; Herron, P.; Luic, M.; Vujaklija, D. Structure-function relationships of two paralogous single-stranded DNA-binding proteins from Streptomyces coelicolor: Implication of SsbB in chromosome segregation during sporulation. Nucleic Acids Res. 2013, 41, 3659–3672. [Google Scholar] [CrossRef]
- Adasme, M.F.; Linnemann, K.L.; Bolz, S.N.; Kaiser, F.; Salentin, S.; Haupt, V.J.; Schroeder, M. PLIP 2021: Expanding the scope of the protein-ligand interaction profiler to DNA and RNA. Nucleic Acids Res. 2021, 49, W530–W534. [Google Scholar] [CrossRef]
- Torres, R.; Carrasco, B.; Alonso, J.C. Bacillus subtilis RadA/Sms-Mediated Nascent Lagging-Strand Unwinding at Stalled or Reversed Forks Is a Two-Step Process: RadA/Sms Assists RecA Nucleation, and RecA Loads RadA/Sms. Int. J. Mol. Sci. 2023, 24, 4536. [Google Scholar] [CrossRef] [PubMed]
- Ramos, C.; Hernández-Tamayo, R.; López-Sanz, M.; Carrasco, B.; Serrano, E.; Alonso, J.C.; Graumann, P.L.; Ayora, S. The RecD2 helicase balances RecA activities. Nucleic Acids Res. 2022, 50, 3432–3444. [Google Scholar] [CrossRef]
- Natarajan, R.; Anthoni Samy, H.N.; Sivaperuman, A.; Subramani, A. Structure-Activity Relationships of Pyrimidine Derivatives and their Biological Activity—A Review. Med. Chem. 2022, 19, 10–30. [Google Scholar] [PubMed]
- Mafi, A.; Rezaee, M.; Hedayati, N.; Hogan, S.D.; Reiter, R.J.; Aarabi, M.H.; Asemi, Z. Melatonin and 5-fluorouracil combination chemotherapy: Opportunities and efficacy in cancer therapy. Cell Commun. Signal 2023, 21, 33. [Google Scholar] [CrossRef]
- Huang, Y.H.; Chiang, W.Y.; Chen, P.J.; Lin, E.S.; Huang, C.Y. Anticancer and Antioxidant Activities of the Root Extract of the Carnivorous Pitcher Plant Sarracenia purpurea. Plants 2022, 11, 1668. [Google Scholar] [CrossRef] [PubMed]
- Guan, H.H.; Huang, Y.H.; Lin, E.S.; Chen, C.J.; Huang, C.Y. Plumbagin, a Natural Product with Potent Anticancer Activities, Binds to and Inhibits Dihydroorotase, a Key Enzyme in Pyrimidine Biosynthesis. Int. J. Mol. Sci. 2021, 22, 6861. [Google Scholar] [CrossRef] [PubMed]
- Wyatt, M.D.; Wilson, D.M., 3rd. Participation of DNA repair in the response to 5-fluorouracil. Cell Mol. Life Sci. 2009, 66, 788–799. [Google Scholar] [CrossRef]
- Malik, Z.; Parveen, R.; Zahiruddin, S.; Gaurav; Husain, S.A.; Ahmad, S. HPTLC Stability Indicating Analytical Method of Andrographolide and 5-fluorouracil with Network Pharmacology Analysis against Cancer. Comb. Chem. High Throughput Screen 2023. ahead of print. [Google Scholar] [CrossRef]
- Yang, H.; Jeffrey, P.D.; Miller, J.; Kinnucan, E.; Sun, Y.; Thoma, N.H.; Zheng, N.; Chen, P.L.; Lee, W.H.; Pavletich, N.P. BRCA2 function in DNA binding and recombination from a BRCA2-DSS1-ssDNA structure. Science 2002, 297, 1837–1848. [Google Scholar] [CrossRef]
- Brosey, C.A.; Yan, C.; Tsutakawa, S.E.; Heller, W.T.; Rambo, R.P.; Tainer, J.A.; Ivanov, I.; Chazin, W.J. A new structural framework for integrating replication protein A into DNA processing machinery. Nucleic Acids Res. 2013, 41, 2313–2327. [Google Scholar] [CrossRef]
- Suksombat, S.; Khafizov, R.; Kozlov, A.G.; Lohman, T.M.; Chemla, Y.R. Structural dynamics of E. coli single-stranded DNA binding protein reveal DNA wrapping and unwrapping pathways. eLife 2015, 4, e08193. [Google Scholar] [CrossRef] [PubMed]
- Zhou, R.; Kozlov, A.G.; Roy, R.; Zhang, J.; Korolev, S.; Lohman, T.M.; Ha, T. SSB functions as a sliding platform that migrates on DNA via reptation. Cell 2011, 146, 222–232. [Google Scholar] [CrossRef]
- Scholz, C.; Knorr, S.; Hamacher, K.; Schmidt, B. DOCKTITE-a highly versatile step-by-step workflow for covalent docking and virtual screening in the molecular operating environment. J. Chem. Inf. Model. 2015, 55, 398–406. [Google Scholar] [CrossRef] [PubMed]
- Dubiel, K.; Myers, A.R.; Kozlov, A.G.; Yang, O.; Zhang, J.; Ha, T.; Lohman, T.M.; Keck, J.L. Structural Mechanisms of Cooperative DNA Binding by Bacterial Single-Stranded DNA-Binding Proteins. J. Mol. Biol. 2019, 431, 178–195. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.H.; Lo, Y.H.; Huang, W.; Huang, C.Y. Crystal structure and DNA-binding mode of Klebsiella pneumoniae primosomal PriB protein. Genes Cells 2012, 17, 837–849. [Google Scholar] [CrossRef] [PubMed]
- Windgassen, T.A.; Wessel, S.R.; Bhattacharyya, B.; Keck, J.L. Mechanisms of bacterial DNA replication restart. Nucleic Acids Res 2018, 46, 504–519. [Google Scholar] [CrossRef]
- Bruand, C.; Ehrlich, S.D.; Janniere, L. Primosome assembly site in Bacillus subtilis. EMBO J. 1995, 14, 2642–2650. [Google Scholar] [CrossRef]
- Otwinowski, Z.; Minor, W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Methods Enzymol. 1997, 276, 307–326. [Google Scholar]
- Kabsch, W. XDS. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 125–132. [Google Scholar] [CrossRef]
- Terwilliger, T.C.; Adams, P.D.; Read, R.J.; McCoy, A.J.; Moriarty, N.W.; Grosse-Kunstleve, R.W.; Afonine, P.V.; Zwart, P.H.; Hung, L.W. Decision-making in structure solution using Bayesian estimates of map quality: The PHENIX AutoSol wizard. Acta Crystallogr. D Biol. Crystallogr. 2009, 65, 582–601. [Google Scholar] [CrossRef]
- Lebedev, A.A.; Young, P.; Isupov, M.N.; Moroz, O.V.; Vagin, A.A.; Murshudov, G.N. JLigand: A graphical tool for the CCP4 template-restraint library. Acta Crystallogr. D Biol. Crystallogr. 2012, 68, 431–440. [Google Scholar] [CrossRef]
- Terwilliger, T.C.; Grosse-Kunstleve, R.W.; Afonine, P.V.; Moriarty, N.W.; Zwart, P.H.; Hung, L.W.; Read, R.J.; Adams, P.D. Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr. D Biol. Crystallogr. 2008, 64, 61–69. [Google Scholar] [CrossRef] [PubMed]
- Winn, M.D.; Ballard, C.C.; Cowtan, K.D.; Dodson, E.J.; Emsley, P.; Evans, P.R.; Keegan, R.M.; Krissinel, E.B.; Leslie, A.G.; McCoy, A.; et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 2011, 67, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Chen, V.B.; Arendall, W.B., 3rd; Headd, J.J.; Keedy, D.A.; Immormino, R.M.; Kapral, G.J.; Murray, L.W.; Richardson, J.S.; Richardson, D.C. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 12–21. [Google Scholar] [CrossRef] [PubMed]
- Peng, W.F.; Huang, C.Y. Allantoinase and dihydroorotase binding and inhibition by flavonols and the substrates of cyclic amidohydrolases. Biochimie 2014, 101, 113–122. [Google Scholar] [CrossRef]
- Huang, Y.H.; Lien, Y.; Chen, J.H.; Lin, E.S.; Huang, C.Y. Identification and characterization of dihydropyrimidinase inhibited by plumbagin isolated from Nepenthes miranda extract. Biochimie 2020, 171–172, 124–135. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.Y. Inhibition of a putative dihydropyrimidinase from Pseudomonas aeruginosa PAO1 by flavonoids and substrates of cyclic amidohydrolases. PLoS ONE 2015, 10, e0127634. [Google Scholar] [CrossRef][Green Version]
- Carrasco, B.; Seco, E.M.; López-Sanz, M.; Alonso, J.C.; Ayora, S. Bacillus subtilis RarA modulates replication restart. Nucleic Acids Res. 2018, 46, 7206–7220. [Google Scholar] [CrossRef]
- Par, S.; Vaides, S.; VanderVere-Carozza, P.S.; Pawelczak, K.S.; Stewart, J.; Turchi, J.J. OB-Folds and Genome Maintenance: Targeting Protein-DNA Interactions for Cancer Therapy. Cancers 2021, 13, 3346. [Google Scholar] [CrossRef]


| Data Collection | ||
|---|---|---|
| Crystal | SaSsbA–5-FU complex | Glycerol-bound SaSsbA |
| Wavelength (Å) | 1 | 1 |
| Resolution (Å) | 27.8–2.36 | 27.9–1.80 |
| Space group | P41212 | P41212 |
| Cell dimension a, b, c (Å) β (°) | 88.09, 88.09, 57.78 90.00 | 88.22, 88.22, 58.00 90.00 |
| Redundancy | 7.8 (7.9) | 9.4 (9.4) |
| Completeness (%) | 99.9 (100.0) | 100.0 (100.0) |
| <I/σI> | 31.6 (8.2) | 35.7 (4.4) |
| CC1/2 | 0.997 (0.974) | 0.989 (0.955) |
| Refinement | ||
| No. of reflections | 9797 | 21,823 |
| Rwork/Rfree | 0.203/0.244 | 0.199/0.224 |
| No. of atoms | ||
| Protein | 199 | 200 |
| Water | 52 | 151 |
| 5-FU | 1 | 0 |
| Glycerol | 1 | 2 |
| r.m.s deviations | ||
| Bond lengths (Å) | 0.008 | 0.007 |
| Bond angles (°) | 1.06 | 1.05 |
| Ramachandran plot | ||
| Favored (%) | 98.43 | 97.40 |
| Allowed (%) | 1.57 | 2.60 |
| Outliers (%) | 0 | 0 |
| PDB entry | 7YM1 | 8GW5 |
| Oligonucleotide | Primer |
|---|---|
| SaSsbA-R18A-N | GAAAGATCCGGAATACGCAACCACTCCCTC |
| SaSsbA-R18A-C | ACACCTGAGGGAGTGGTTGCGTATTCCGGA |
| SaSsbA | λmax (nm) | λem Shift (nm) | Quenching (%) | Kd Value (µM) |
|---|---|---|---|---|
| SaSsbA WT | 339 | 347 | 83.81 | 55.9 ± 0.7 |
| SaSsbA-R18A | 339 | 340 | 34.99 | 497.6 ± 13.5 |
| Mode | S Score | Receptor Residue | Interaction | Distance (Å) | E (kcal/mol) |
|---|---|---|---|---|---|
| 1 | −4.2104 | Phe 48 (A) | H-donor | 2.96 | −5.6 |
| Thr 93 (A) | H-donor | 2.98 | −1.9 | ||
| Asn 50 (A) | H-acceptor | 3.08 | −1.1 | ||
| 2 | −4.1638 | Arg 80 (A) | H-donor | 3.42 | −0.9 |
| Val 92 (A) | H-donor | 3.03 | −1.2 | ||
| Asn 81 (A) | H-acceptor | 3.14 | −2.2 | ||
| 3 | −3.9845 | Arg 80 (A) | H-donor | 3.27 | −1.0 |
| Val 92 (A) | H-donor | 3.05 | −1.2 | ||
| Phe 91 (A) | Pi-H | 4.03 | −0.6 | ||
| 4 | −3.8762 | Asn 50 (B) | H-donor | 3.37 | −1.8 |
| Asn 81 (A) | H-acceptor | 3.52 | −0.8 | ||
| 5 | −3.8744 | Phe 91 (A) | H-donor | 3.07 | −1.6 |
| Phe 91 (A) | H-acceptor | 3.15 | −1.2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Su, H.-H.; Huang, Y.-H.; Lien, Y.; Yang, P.-C.; Huang, C.-Y. Crystal Structure of DNA Replication Protein SsbA Complexed with the Anticancer Drug 5-Fluorouracil. Int. J. Mol. Sci. 2023, 24, 14899. https://doi.org/10.3390/ijms241914899
Su H-H, Huang Y-H, Lien Y, Yang P-C, Huang C-Y. Crystal Structure of DNA Replication Protein SsbA Complexed with the Anticancer Drug 5-Fluorouracil. International Journal of Molecular Sciences. 2023; 24(19):14899. https://doi.org/10.3390/ijms241914899
Chicago/Turabian StyleSu, Hsin-Hui, Yen-Hua Huang, Yi Lien, Po-Chun Yang, and Cheng-Yang Huang. 2023. "Crystal Structure of DNA Replication Protein SsbA Complexed with the Anticancer Drug 5-Fluorouracil" International Journal of Molecular Sciences 24, no. 19: 14899. https://doi.org/10.3390/ijms241914899
APA StyleSu, H.-H., Huang, Y.-H., Lien, Y., Yang, P.-C., & Huang, C.-Y. (2023). Crystal Structure of DNA Replication Protein SsbA Complexed with the Anticancer Drug 5-Fluorouracil. International Journal of Molecular Sciences, 24(19), 14899. https://doi.org/10.3390/ijms241914899







