N6-Methyladenine Progressively Accumulates in Mitochondrial DNA during Aging
Abstract
:1. Introduction
2. Materials and Methods
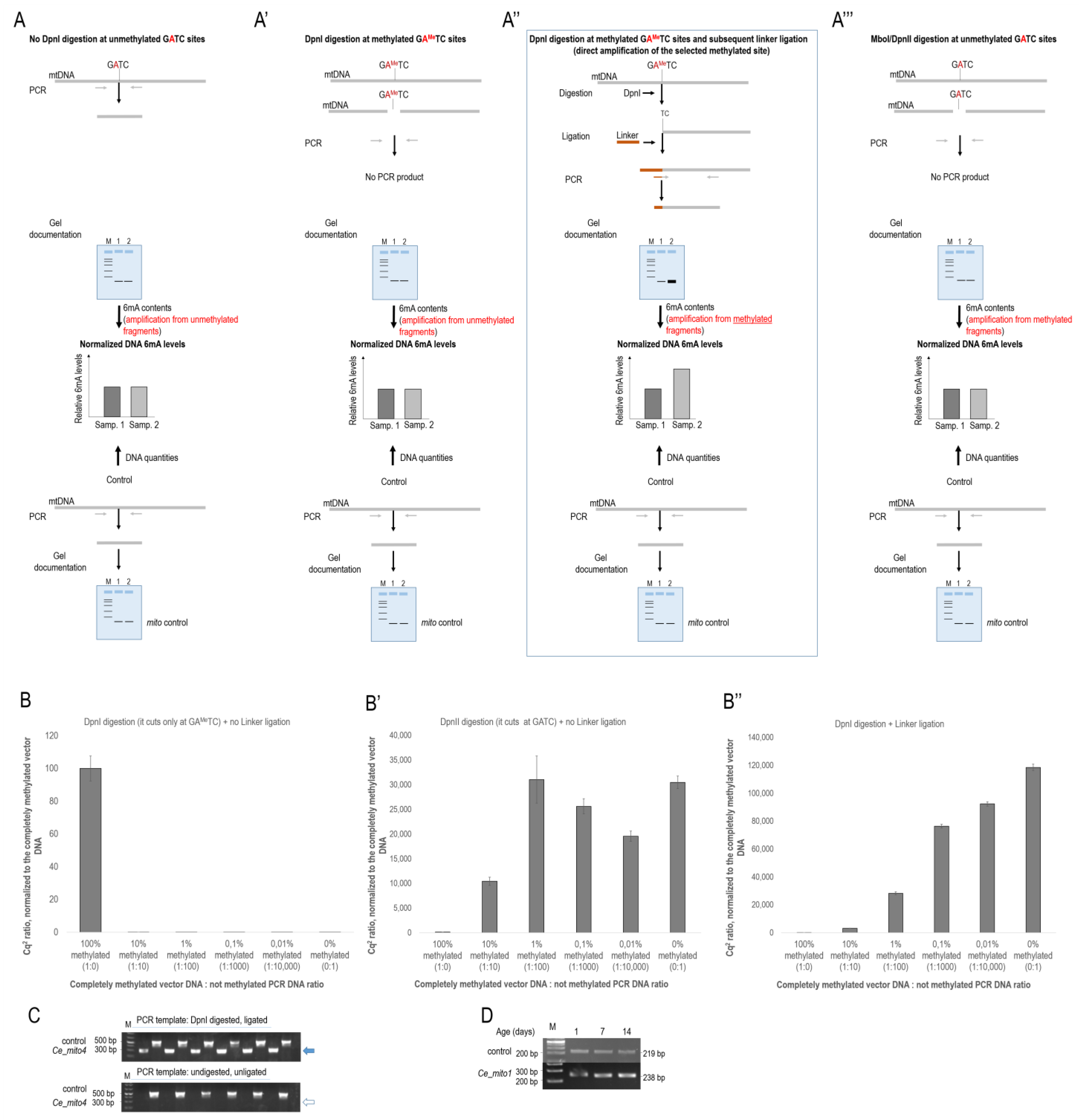
2.1. In Vitro Comparison of Different Methods in Determining Relative N6-Methyladenine Levels
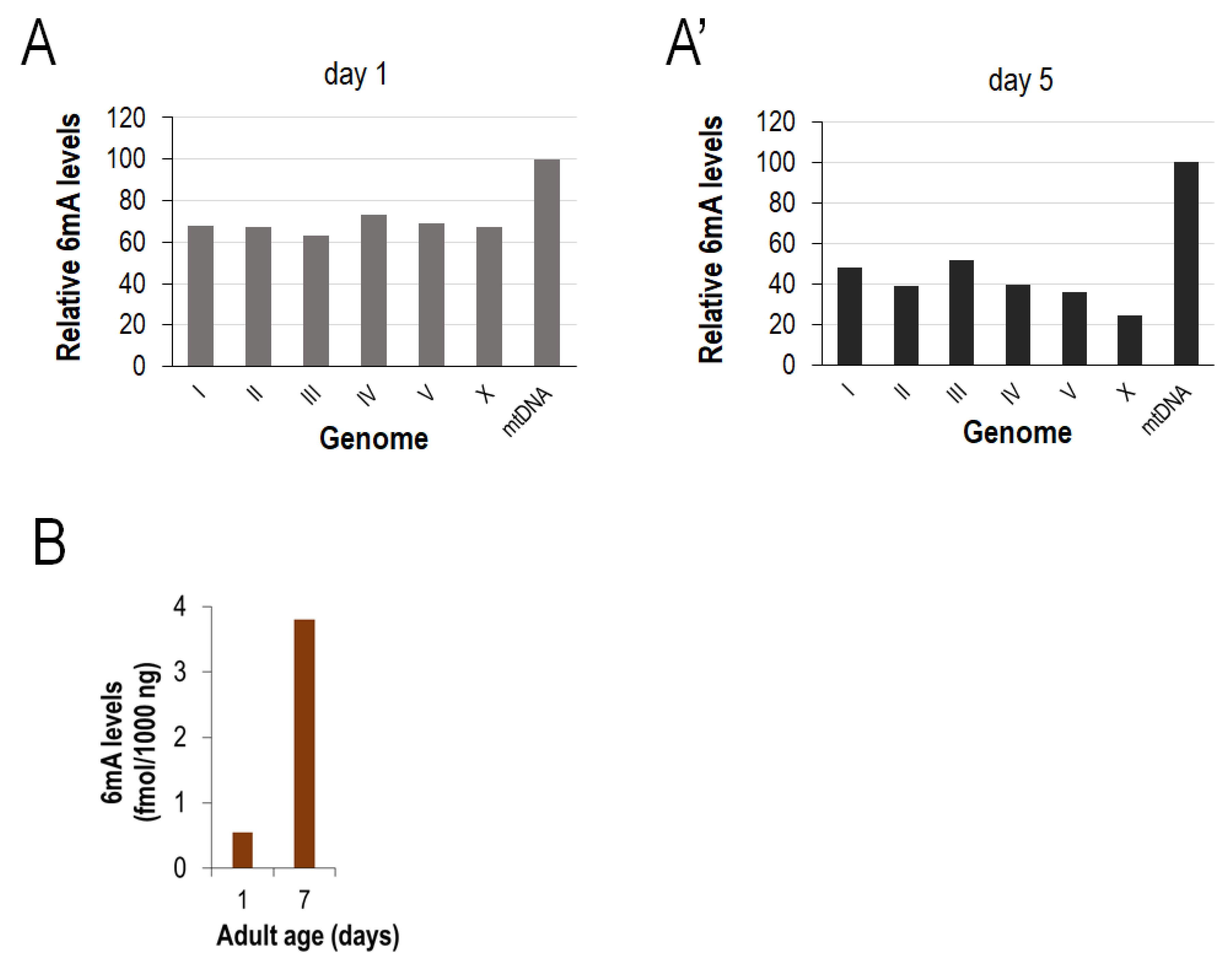
2.2. Determination of the Relative N6-Methyladenine Levels at Different mtDNA Sites
2.3. qPCR Quantification of the Relative mtDNA 6mA Levels in Canine Frontal Cortex
2.4. SMRT Sequencing and Data Analysis
2.5. LC-MS/MS Analysis
2.6. Generation of a Translational Fusion NMAD-1::GFP Reporter
2.7. TMRE Staining
2.8. Isolation of the Mitochondria
2.9. Western Blotting
2.10. Downregulation of Drosophila Tet and Mt2
2.11. Quantifying N6-Methyladenine Levels in Canine mtDNA by qRT-PCR
2.12. Statistical Analysis
3. Results
3.1. Reliable PCR-Based Identification of Relative 6mA Levels at Different mtDNA Sites
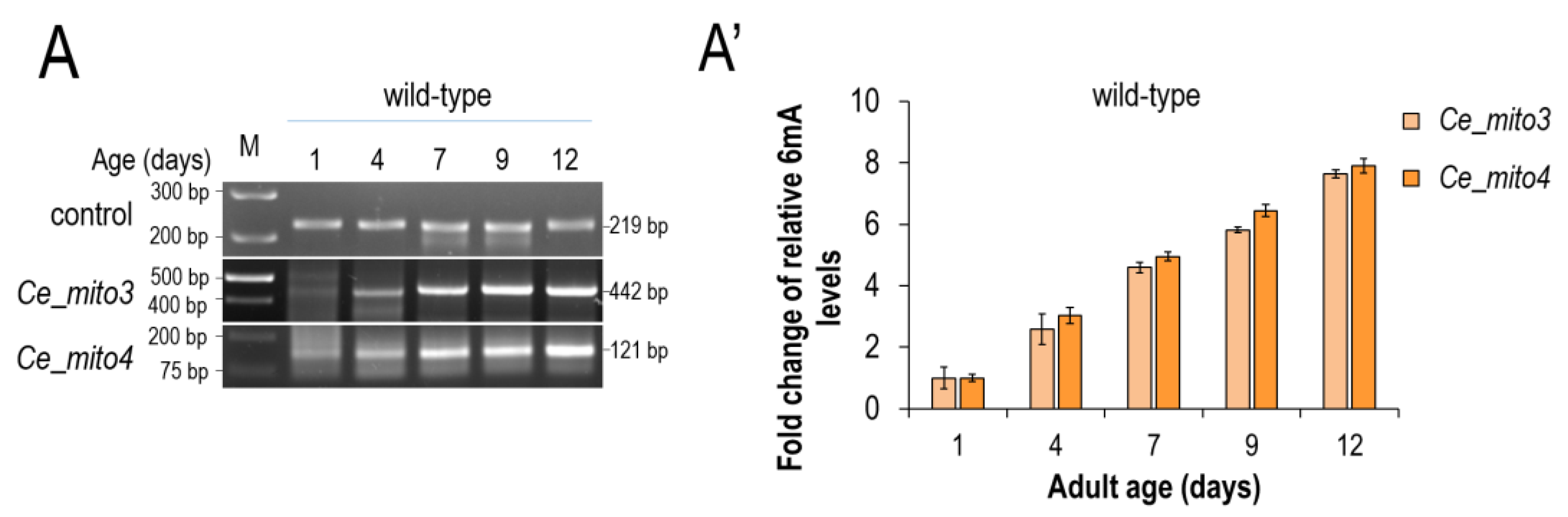
3.2. N6-Adenine Methylation Increases Progressively with Age in the mtDNA of C. elegans
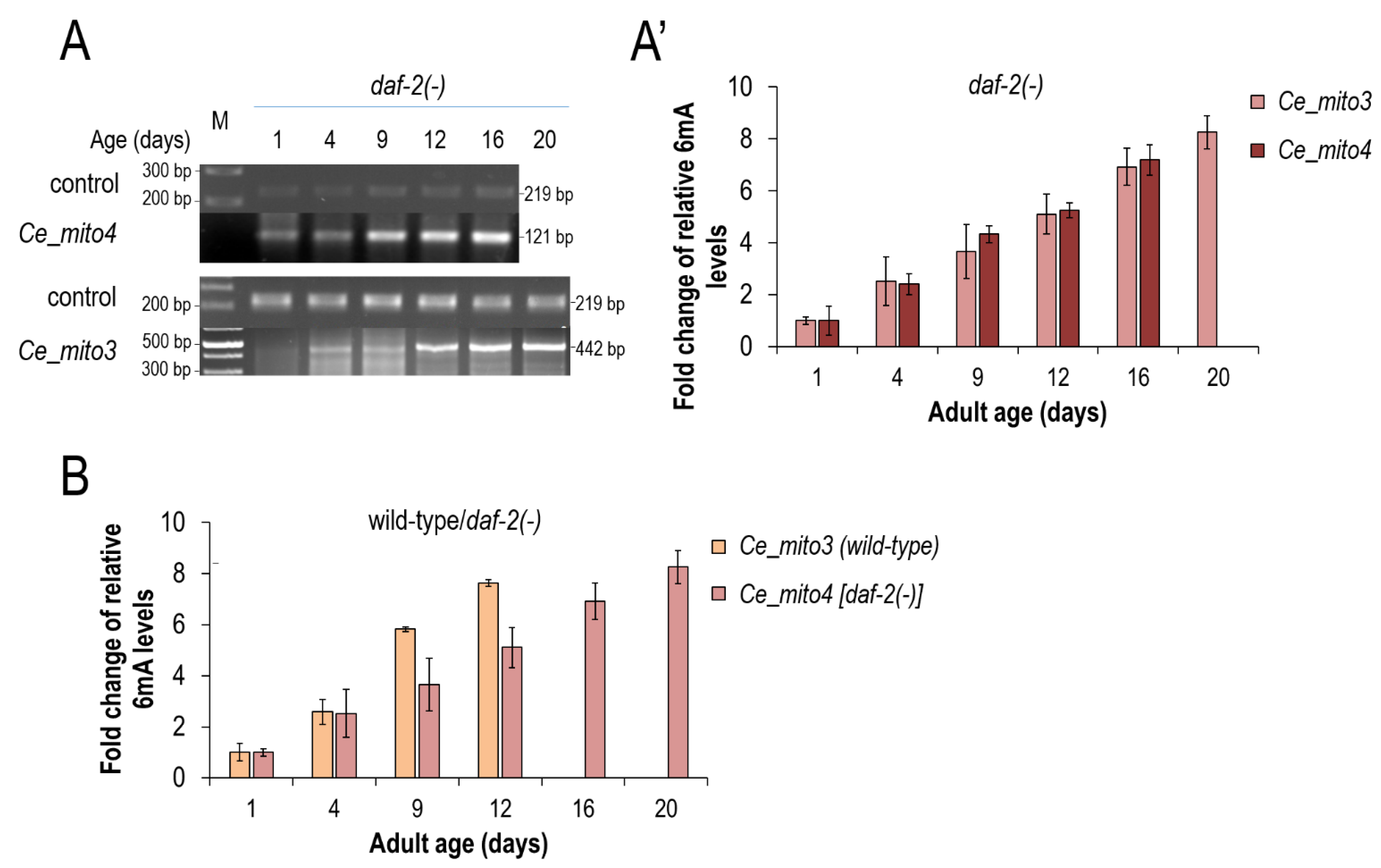
3.3. mtDNA N6-Adenine Methylation Takes Place at a Lower Rate in Long-Lived Mutant Nematodes
3.4. DAMT-1 DNA N6-Adenine Methyltransferase and NMAD-1 N6-Methyladenine Demethylase Influence mtDNA 6mA Levels
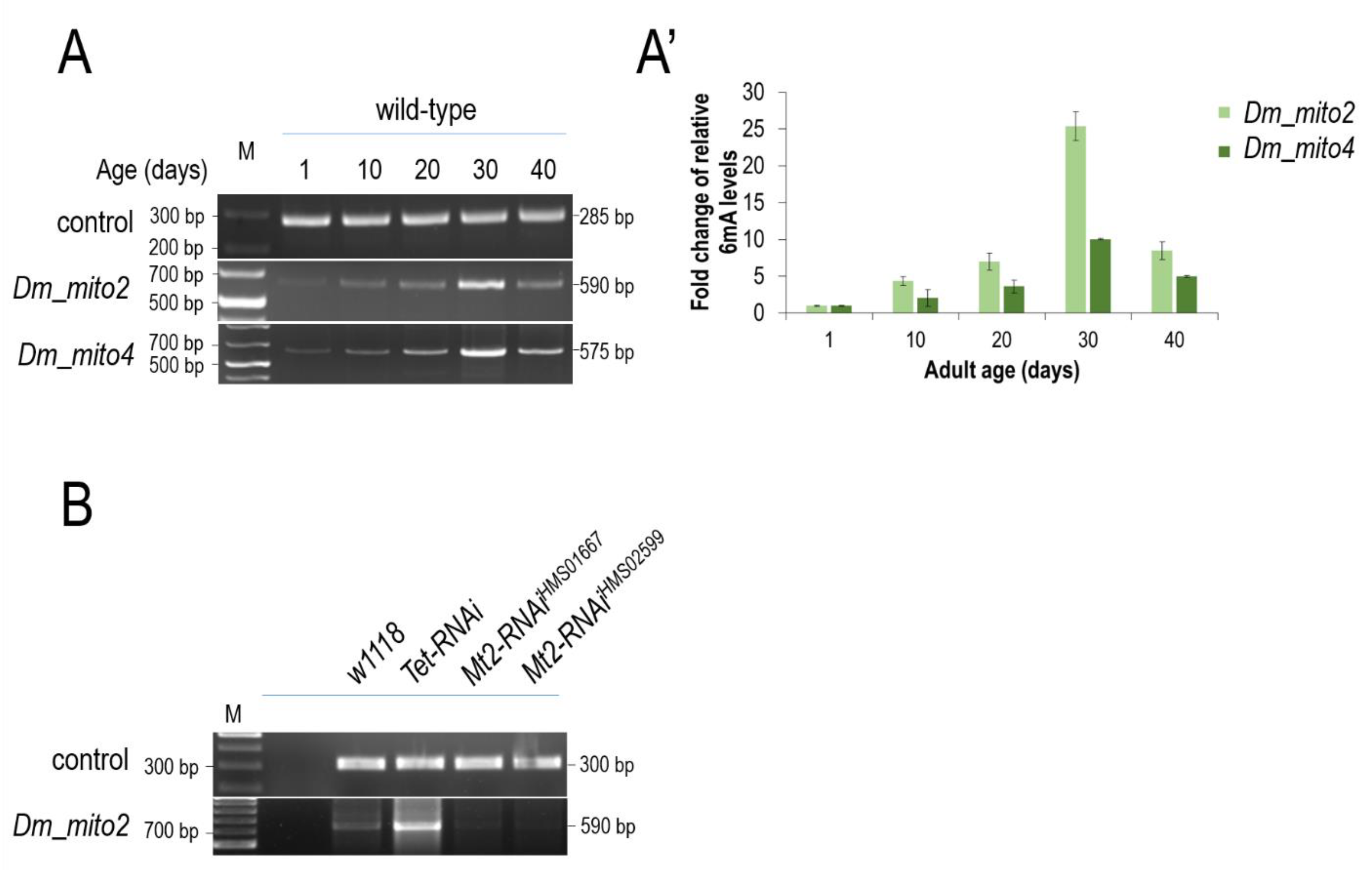
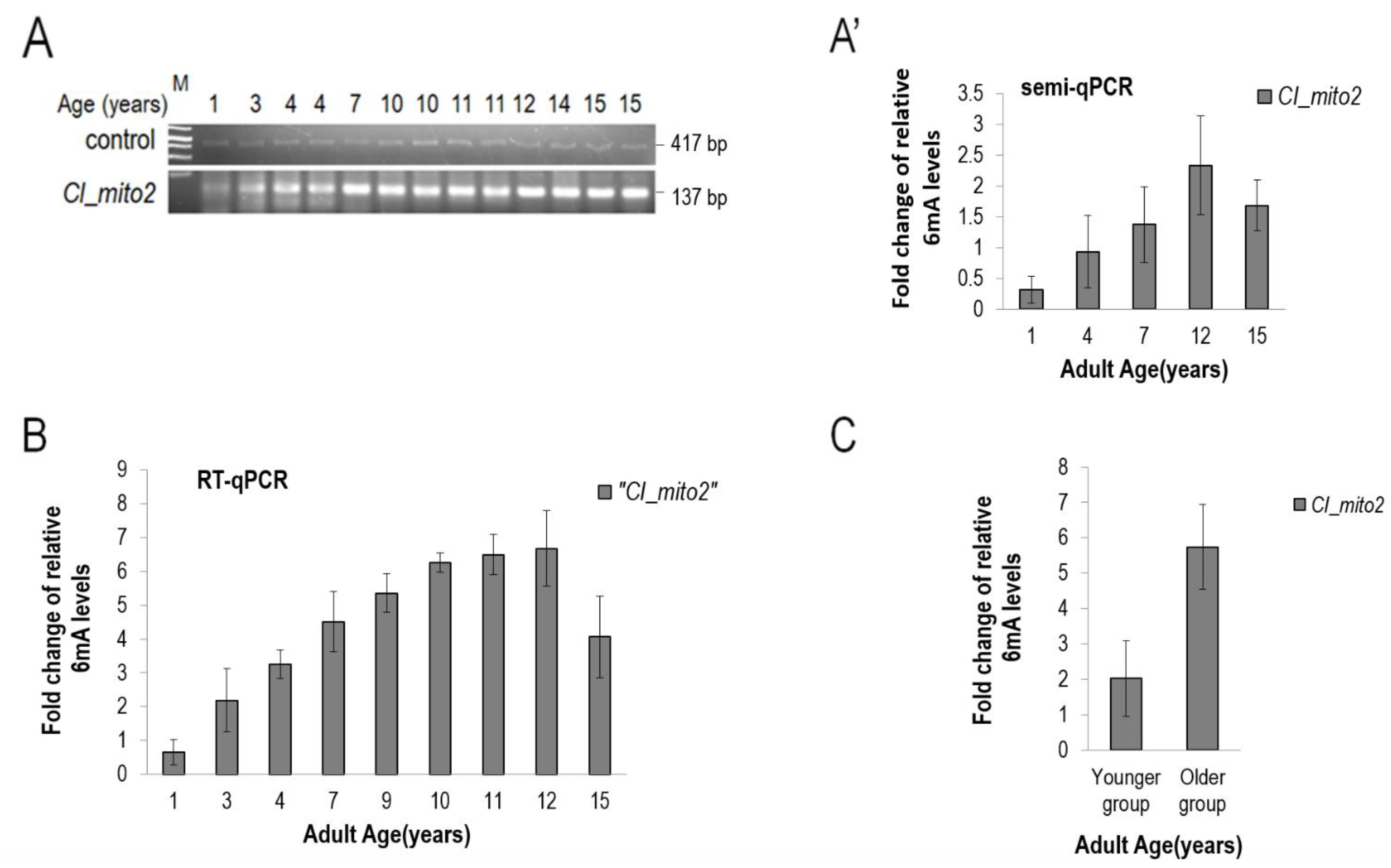
3.5. The Presence of 6mA Is also Detectable in the mtDNA of Drosophila and Dog, and Its Levels Increase with Age
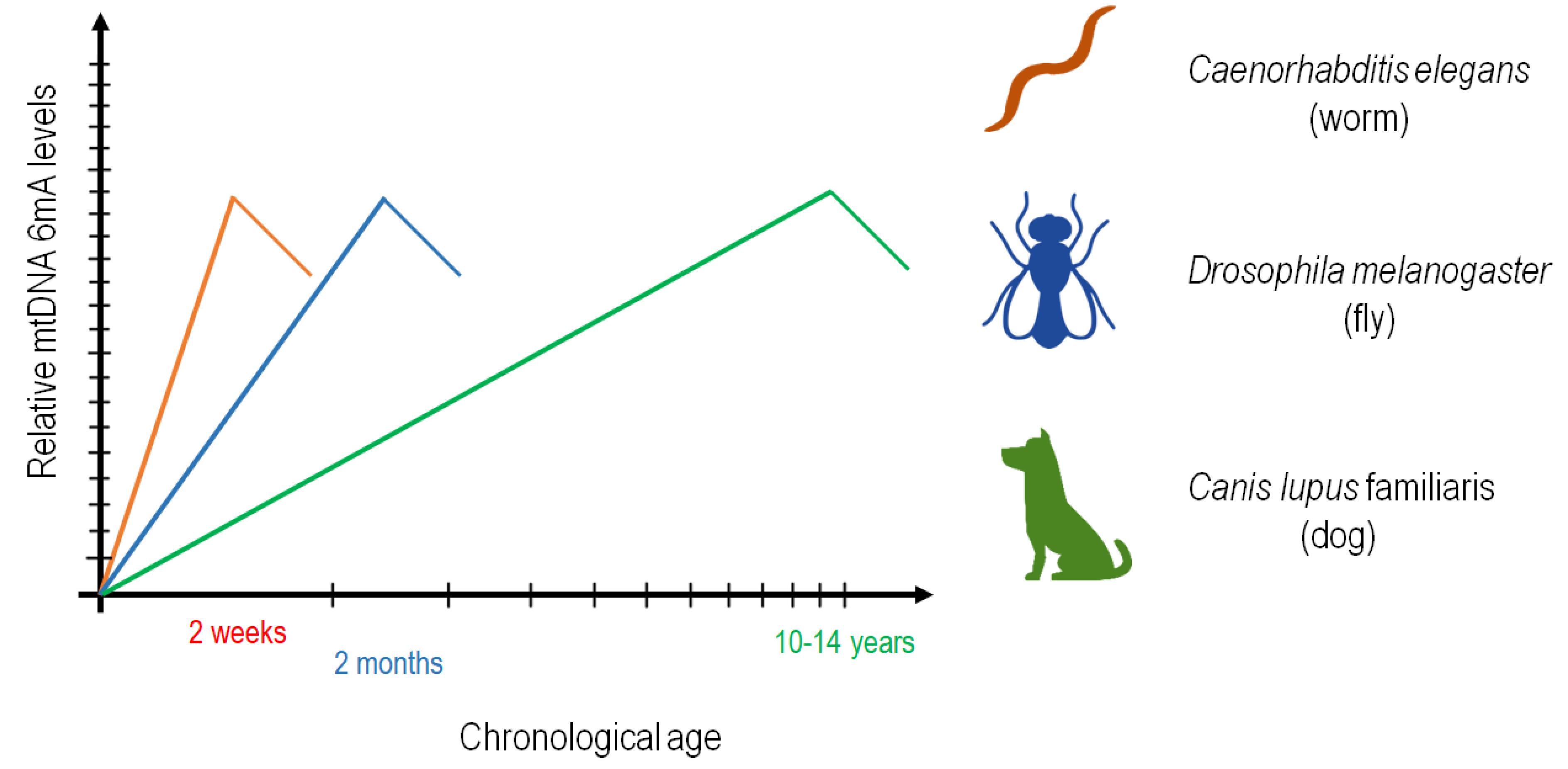
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sturm, A.; Vellai, T. How does maternal age affect genomic stability in the offspring? Aging Cell 2022, 21, e13612. [Google Scholar] [CrossRef]
- Jin, K.; Simpkins, J.W.; Ji, X.; Leis, M.; Stambler, I. The Critical Need to Promote Research of Aging and Aging-related Diseases to Improve Health and Longevity of the Elderly Population. Aging Dis. 2015, 6, 1–5. [Google Scholar] [CrossRef]
- Benayoun, B.A.; Pollina, E.A.; Brunet, A. Epigenetic regulation of ageing: Linking environmental inputs to genomic stability. Nat. Rev. Mol. Cell Biol. 2015, 16, 593–610. [Google Scholar] [CrossRef]
- Greer, E.L.; Maures, T.J.; Ucar, D.; Hauswirth, A.G.; Mancini, E.; Lim, J.P.; Benayoun, B.A.; Shi, Y.; Brunet, A. Transgenerational epigenetic inheritance of longevity in Caenorhabditis elegans. Nature 2011, 479, 365–371. [Google Scholar] [CrossRef]
- Yang, J.H.; Hayano, M.; Griffin, P.T.; Amorim, J.A.; Bonkowski, M.S.; Apostolides, J.K.; Salfati, E.L.; Blanchette, M.; Munding, E.M.; Bhakta, M.; et al. Loss of epigenetic information as a cause of mammalian aging. Cell 2023, 186, 305–326.e27. [Google Scholar] [CrossRef]
- Sigmond, T.; Vellai, T. Lysosomal alteration links food limitation to longevity. Nat. Aging 2023, 3, 1048–1050. [Google Scholar] [CrossRef] [PubMed]
- Skvortsova, K.; Iovino, N.; Bogdanovic, O. Functions and mechanisms of epigenetic inheritance in animals. Nat. Rev. Mol. Cell Biol. 2018, 19, 774–790. [Google Scholar] [CrossRef] [PubMed]
- Horvath, S.; Raj, K. DNA methylation-based biomarkers and the epigenetic clock theory of ageing. Nat. Rev. Genet. 2018, 19, 371–384. [Google Scholar] [CrossRef]
- Oliveira, P.H.; Fang, G. Conserved DNA Methyltransferases: A Window into Fundamental Mechanisms of Epigenetic Regulation in Bacteria. Trends Microbiol. 2021, 29, 28–40. [Google Scholar] [CrossRef]
- Vanyushin, B.F. DNA methylation in plants. Curr. Top. Microbiol. Immunol. 2006, 301, 67–122. [Google Scholar] [PubMed]
- Greer, E.L.; Blanco, M.A.; Gu, L.; Sendinc, E.; Liu, J.; Aristizabal-Corrales, D.; Hsu, C.H.; Aravind, L.; He, C.; Shi, Y. DNA Methylation on N6-Adenine in C. elegans. Cell 2015, 161, 868–878. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Huang, H.; Liu, D.; Cheng, Y.; Liu, X.; Zhang, W.; Yin, R.; Zhang, D.; Zhang, P.; Liu, J.; et al. N6-methyladenine DNA modification in Drosophila. Cell 2015, 161, 893–906. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.P.; Wang, T.; Seetin, M.G.; Lai, Y.; Zhu, S.; Lin, K.; Liu, Y.; Byrum, S.D.; Mackintosh, S.G.; Zhong, M.; et al. DNA methylation on N(6)-adenine in mammalian embryonic stem cells. Nature 2016, 532, 329–333. [Google Scholar] [CrossRef] [PubMed]
- Parashar, N.C.; Parashar, G.; Nayyar, H.; Sandhir, R. N(6)-adenine DNA methylation demystified in eukaryotic genome: From biology to pathology. Biochimie 2018, 144, 56–62. [Google Scholar] [CrossRef]
- Wamucho, A.; Unrine, J.; May, J.; Tsyusko, O. Global DNA Adenine Methylation in Caenorhabditis elegans after Multigenerational Exposure to Silver Nanoparticles and Silver Nitrate. Int. J. Mol. Sci. 2023, 24, 6168. [Google Scholar] [CrossRef]
- Hao, Z.; Wu, T.; Cui, X.; Zhu, P.; Tan, C.; Dou, X.; Hsu, K.W.; Lin, Y.T.; Peng, P.H.; Zhang, L.S.; et al. N(6)-Deoxyadenosine Methylation in Mammalian Mitochondrial DNA. Mol. Cell 2020, 78, 382–395.e8. [Google Scholar] [CrossRef]
- O′Brown, Z.K.; Boulias, K.; Wang, J.; Wang, S.Y.; O’Brown, N.M.; Hao, Z.; Shibuya, H.; Fady, P.E.; Shi, Y.; He, C.; et al. Sources of artifact in measurements of 6mA and 4mC abundance in eukaryotic genomic DNA. BMC Genom. 2019, 20, 445. [Google Scholar] [CrossRef]
- Schiffers, S.; Ebert, C.; Rahimoff, R.; Kosmatchev, O.; Steinbacher, J.; Bohne, A.V.; Spada, F.; Michalakis, S.; Nickelsen, J.; Muller, M.; et al. Quantitative LC-MS Provides No Evidence for m(6) dA or m(4) dC in the Genome of Mouse Embryonic Stem Cells and Tissues. Angew. Chem. 2017, 56, 11268–11271. [Google Scholar] [CrossRef]
- Douvlataniotis, K.; Bensberg, M.; Lentini, A.; Gylemo, B.; Nestor, C.E. No evidence for DNA N (6)-methyladenine in mammals. Sci. Adv. 2020, 6, eaay3335. [Google Scholar] [CrossRef]
- Kong, Y.; Cao, L.; Deikus, G.; Fan, Y.; Mead, E.A.; Lai, W.; Zhang, Y.; Yong, R.; Sebra, R.; Wang, H.; et al. Critical assessment of DNA adenine methylation in eukaryotes using quantitative deconvolution. Science 2022, 375, 515–522. [Google Scholar] [CrossRef]
- Kenyon, C.J. The genetics of ageing. Nature 2010, 464, 504–512. [Google Scholar] [CrossRef] [PubMed]
- Dillin, A.; Hsu, A.L.; Arantes-Oliveira, N.; Lehrer-Graiwer, J.; Hsin, H.; Fraser, A.G.; Kamath, R.S.; Ahringer, J.; Kenyon, C. Rates of behavior and aging specified by mitochondrial function during development. Science 2002, 298, 2398–2401. [Google Scholar] [CrossRef]
- Cho, J.; Hur, J.H.; Walker, D.W. The role of mitochondria in Drosophila aging. Exp. Gerontol. 2011, 46, 331–334. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Yu, Z.; Hou, Y.; Zhang, L.; Fu, A. Improvement of cognitive and motor performance with mitotherapy in aged mice. Int. J. Biol. Sci. 2020, 16, 849–858. [Google Scholar] [CrossRef] [PubMed]
- Vellai, T. How the amino acid leucine activates the key cell-growth regulator mTOR. Nature 2021, 596, 192–194. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Jiang, Y.; Paxman, J.; O’Laughlin, R.; Klepin, S.; Zhu, Y.; Pillus, L.; Tsimring, L.S.; Hasty, J.; Hao, N. A programmable fate decision landscape underlies single-cell aging in yeast. Science 2020, 369, 325–329. [Google Scholar] [CrossRef]
- Sturm, A.; Saskoi, E.; Hotzi, B.; Tarnoci, A.; Barna, J.; Bodnar, F.; Sharma, H.; Kovacs, T.; Ari, E.; Weinhardt, N.; et al. Downregulation of transposable elements extends lifespan in Caenorhabditis elegans. Nat. Commun. 2023, 14, 5278. [Google Scholar] [CrossRef]
- Sun, N.; Youle, R.J.; Finkel, T. The Mitochondrial Basis of Aging. Mol. Cell 2016, 61, 654–666. [Google Scholar] [CrossRef]
- Timmons, L.; Fire, A. Specific interference by ingested dsRNA. Nature 1998, 395, 854. [Google Scholar] [CrossRef]
- Sturm, A.; Saskoi, E.; Tibor, K.; Weinhardt, N.; Vellai, T. Highly efficient RNAi and Cas9-based auto-cloning systems for C. elegans research. Nucleic Acids Res 2018, 46, e105. [Google Scholar] [CrossRef]
- Palikaras, K.; Lionaki, E.; Tavernarakis, N. Coordination of mitophagy and mitochondrial biogenesis during ageing in C. elegans. Nature 2015, 521, 525–528. [Google Scholar] [CrossRef] [PubMed]
- Kenyon, C.; Chang, J.; Gensch, E.; Rudner, A.; Tabtiang, R. A C. elegans mutant that lives twice as long as wild type. Nature 1993, 366, 461–464. [Google Scholar] [CrossRef] [PubMed]
- Kimura, K.D.; Tissenbaum, H.A.; Liu, Y.; Ruvkun, G. daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science 1997, 277, 942–946. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Niu, R.; Huang, T.; Shao, L.W.; Peng, Y.; Ding, W.; Wang, Y.; Jia, G.; He, C.; Li, C.Y.; et al. N6-methyldeoxyadenine is a transgenerational epigenetic signal for mitochondrial stress adaptation. Nat. Cell Biol. 2019, 21, 319–327. [Google Scholar] [CrossRef] [PubMed]
- Crowley, L.C.; Christensen, M.E.; Waterhouse, N.J. Measuring Mitochondrial Transmembrane Potential by TMRE Staining. Cold Spring Harb. Protoc. 2016, 2016. [Google Scholar] [CrossRef]
- Jang, J.Y.; Blum, A.; Liu, J.; Finkel, T. The role of mitochondria in aging. J. Clin. Investig. 2018, 128, 3662–3670. [Google Scholar] [CrossRef]
- Haas, R.H. Mitochondrial Dysfunction in Aging and Diseases of Aging. Biology 2019, 8, 48. [Google Scholar] [CrossRef]
- Guitton, R.; Nido, G.S.; Tzoulis, C. No evidence of extensive non-CpG methylation in mtDNA. Nucleic Acids Res. 2022, 50, 9190–9194. [Google Scholar] [CrossRef]
- Meyer, D.H.; Schumacher, B. BiT age: A transcriptome-based aging clock near the theoretical limit of accuracy. Aging Cell 2021, 20, e13320. [Google Scholar] [CrossRef]
- Sayed, N.; Huang, Y.; Nguyen, K.; Krejciova-Rajaniemi, Z.; Grawe, A.P.; Gao, T.; Tibshirani, R.; Hastie, T.; Alpert, A.; Cui, L.; et al. An inflammatory aging clock (iAge) based on deep learning tracks multimorbidity, immunosenescence, frailty and cardiovascular aging. Nat. Aging 2021, 1, 598–615. [Google Scholar] [CrossRef]
- Hwangbo, N.; Zhang, X.; Raftery, D.; Gu, H.; Hu, S.C.; Montine, T.J.; Quinn, J.F.; Chung, K.A.; Hiller, A.L.; Wang, D.; et al. A metabolomic aging clock using human CSF. J. Gerontol. Ser. A Biol. Sci. Med. Sci. 2021. [Google Scholar]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sturm, Á.; Sharma, H.; Bodnár, F.; Aslam, M.; Kovács, T.; Németh, Á.; Hotzi, B.; Billes, V.; Sigmond, T.; Tátrai, K.; et al. N6-Methyladenine Progressively Accumulates in Mitochondrial DNA during Aging. Int. J. Mol. Sci. 2023, 24, 14858. https://doi.org/10.3390/ijms241914858
Sturm Á, Sharma H, Bodnár F, Aslam M, Kovács T, Németh Á, Hotzi B, Billes V, Sigmond T, Tátrai K, et al. N6-Methyladenine Progressively Accumulates in Mitochondrial DNA during Aging. International Journal of Molecular Sciences. 2023; 24(19):14858. https://doi.org/10.3390/ijms241914858
Chicago/Turabian StyleSturm, Ádám, Himani Sharma, Ferenc Bodnár, Maryam Aslam, Tibor Kovács, Ákos Németh, Bernadette Hotzi, Viktor Billes, Tímea Sigmond, Kitti Tátrai, and et al. 2023. "N6-Methyladenine Progressively Accumulates in Mitochondrial DNA during Aging" International Journal of Molecular Sciences 24, no. 19: 14858. https://doi.org/10.3390/ijms241914858
APA StyleSturm, Á., Sharma, H., Bodnár, F., Aslam, M., Kovács, T., Németh, Á., Hotzi, B., Billes, V., Sigmond, T., Tátrai, K., Egyed, B., Téglás-Huszár, B., Schlosser, G., Charmpilas, N., Ploumi, C., Perczel, A., Tavernarakis, N., & Vellai, T. (2023). N6-Methyladenine Progressively Accumulates in Mitochondrial DNA during Aging. International Journal of Molecular Sciences, 24(19), 14858. https://doi.org/10.3390/ijms241914858






