Identification of Genomic Regions Implicated in Susceptibility to Schistosoma mansoni Infection in a Murine Backcross Genetic Model
Abstract
1. Introduction
2. Results
2.1. There Was a Different Susceptibility to Schistosomiasis in Mice Related to the Genetic Background but Not to Mouse Sex
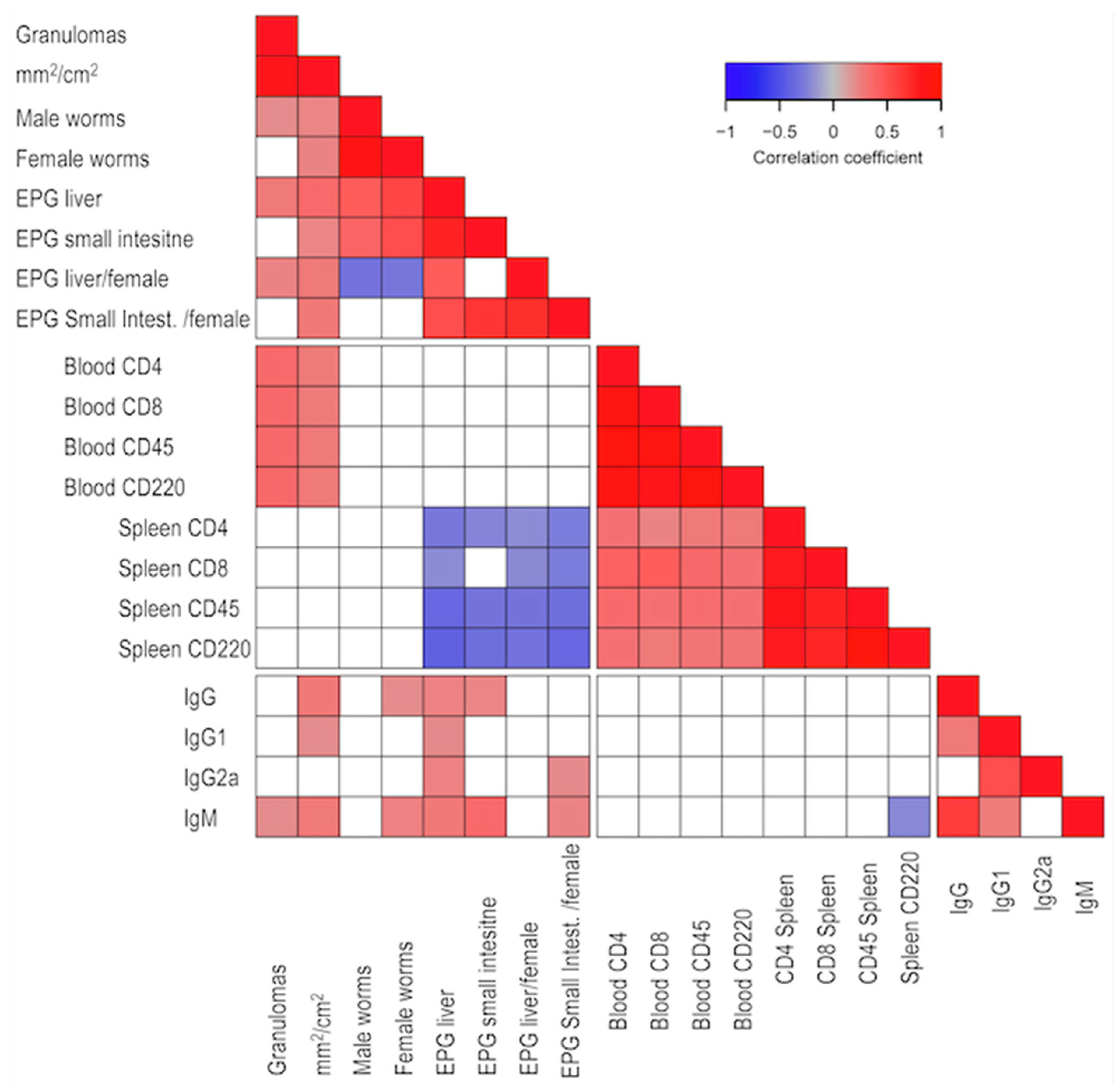
2.2. Parasitological, Pathological, and White Blood Cell Populations Correlated in the BX Cohort
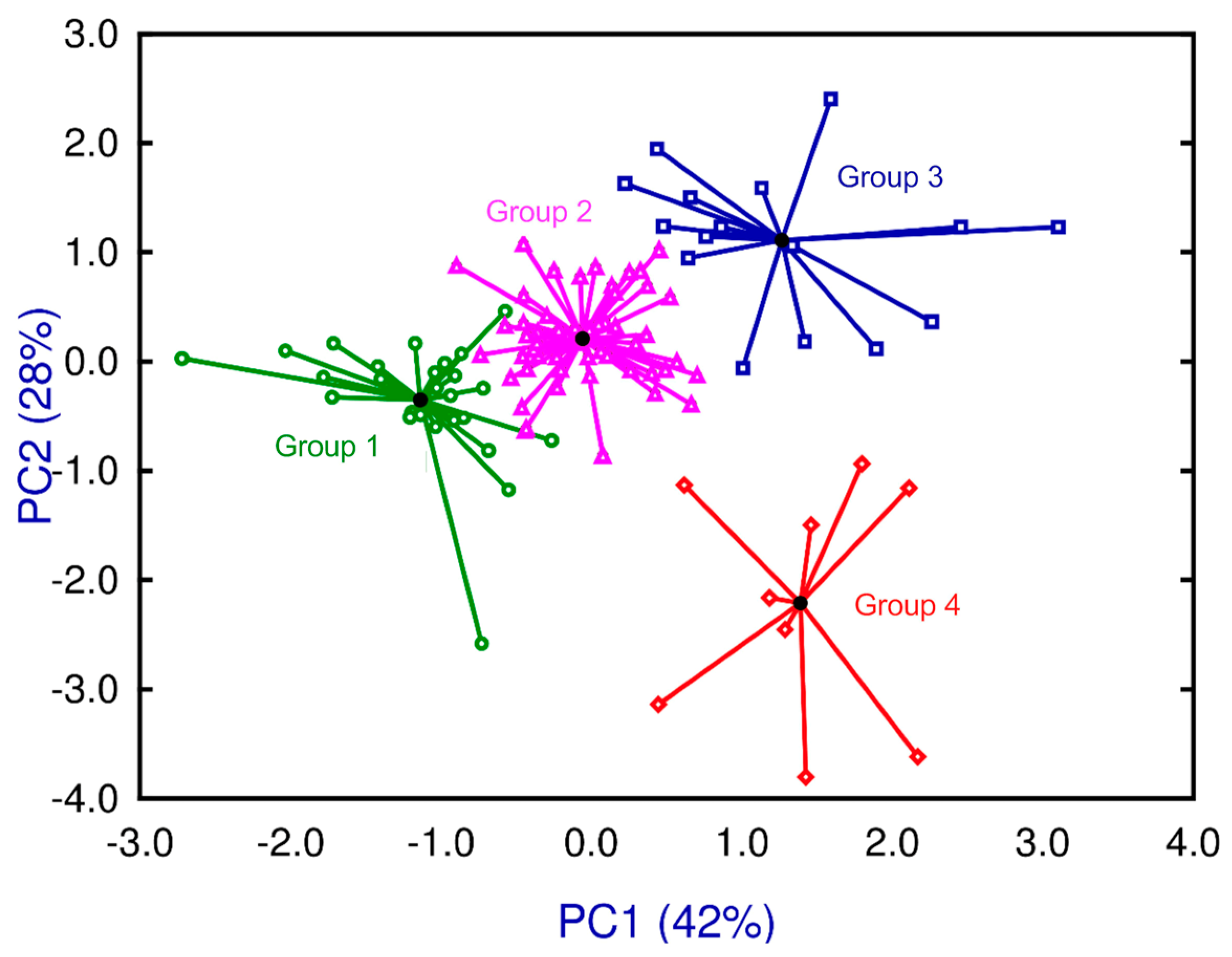
2.3. Identification of Different Degrees of Schistosomiasis Disease in the BX Mouse Cohort
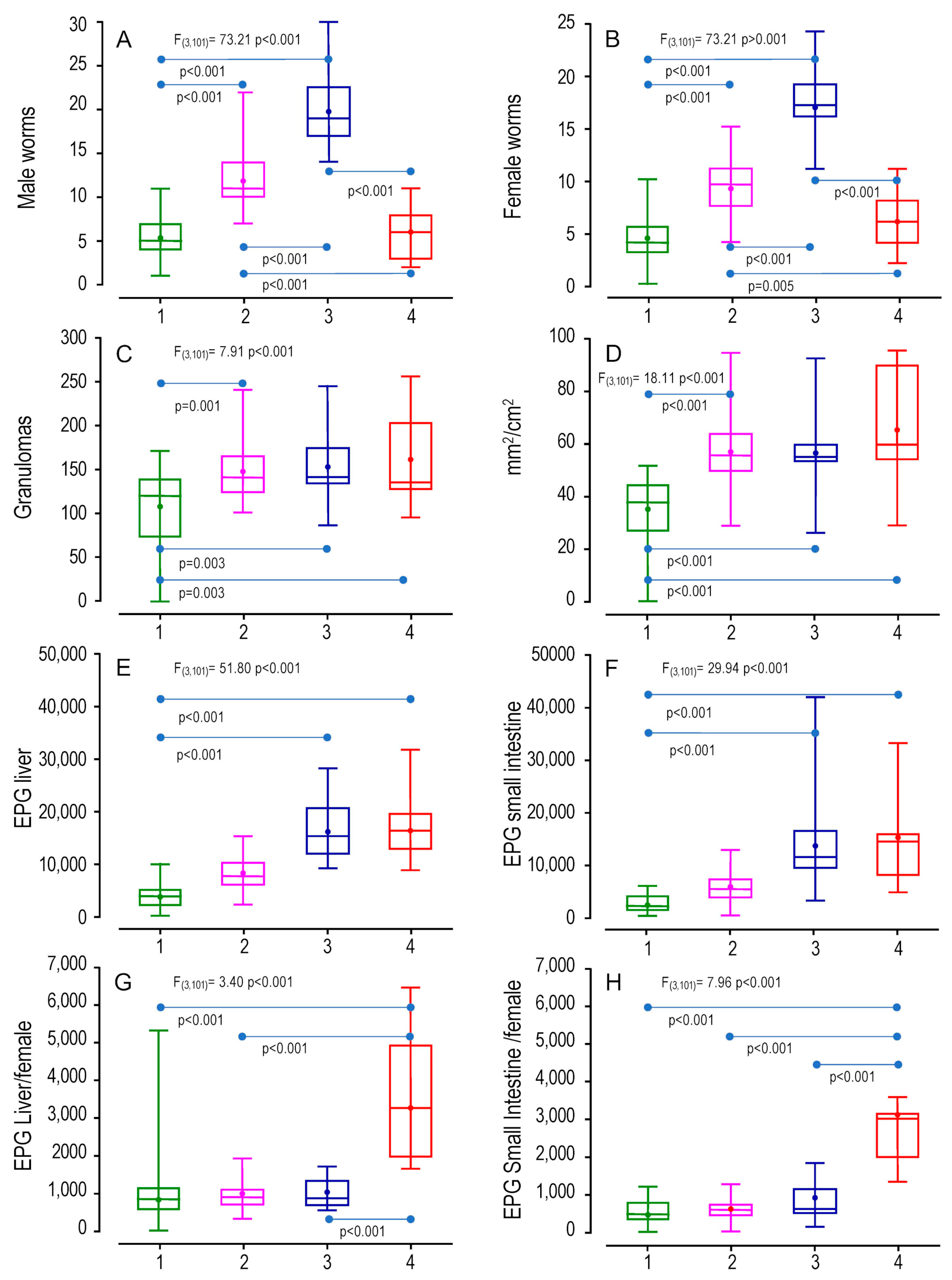
2.4. Pathophenotypic Variation in the BX Cohort in the Groups Identified
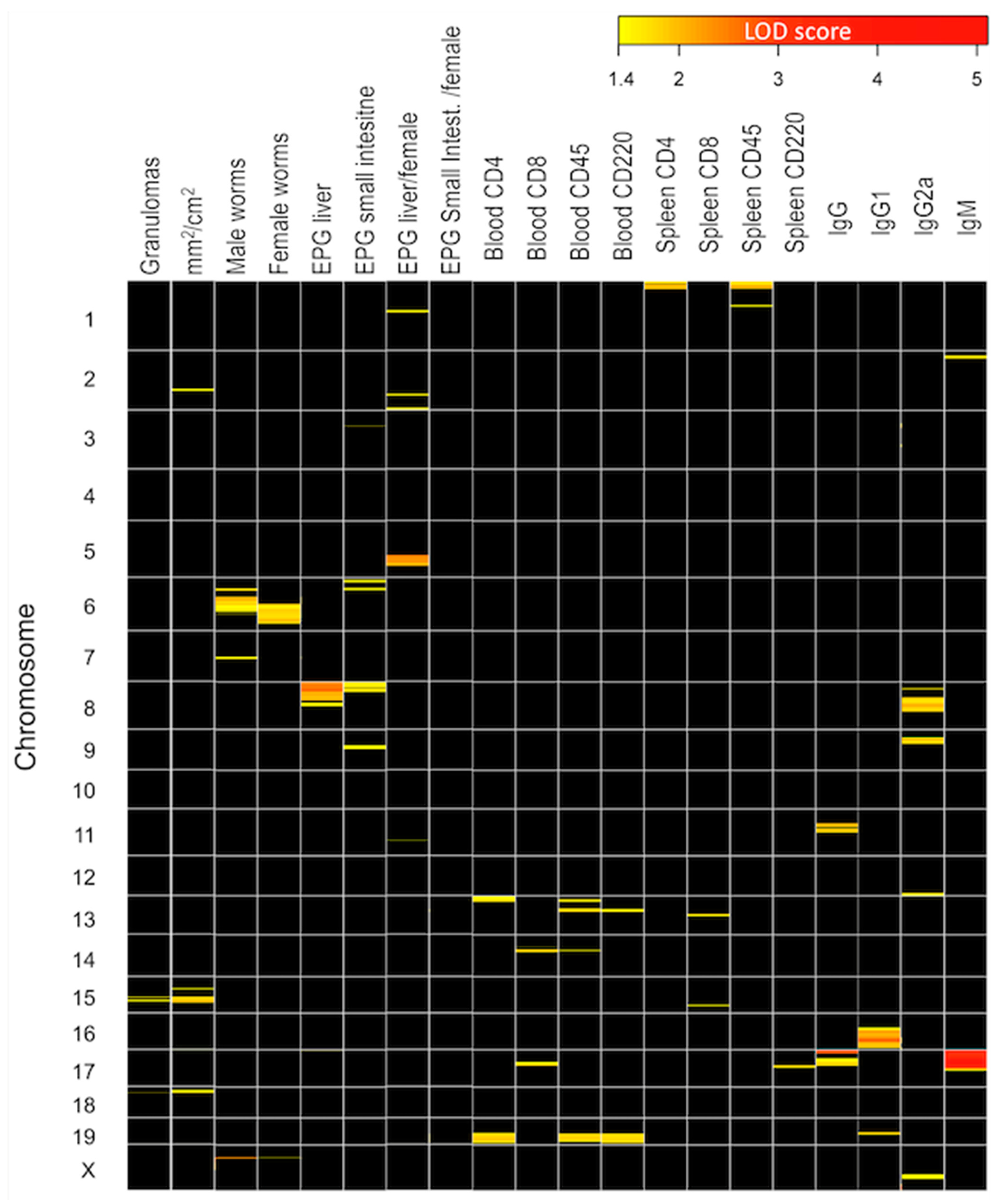
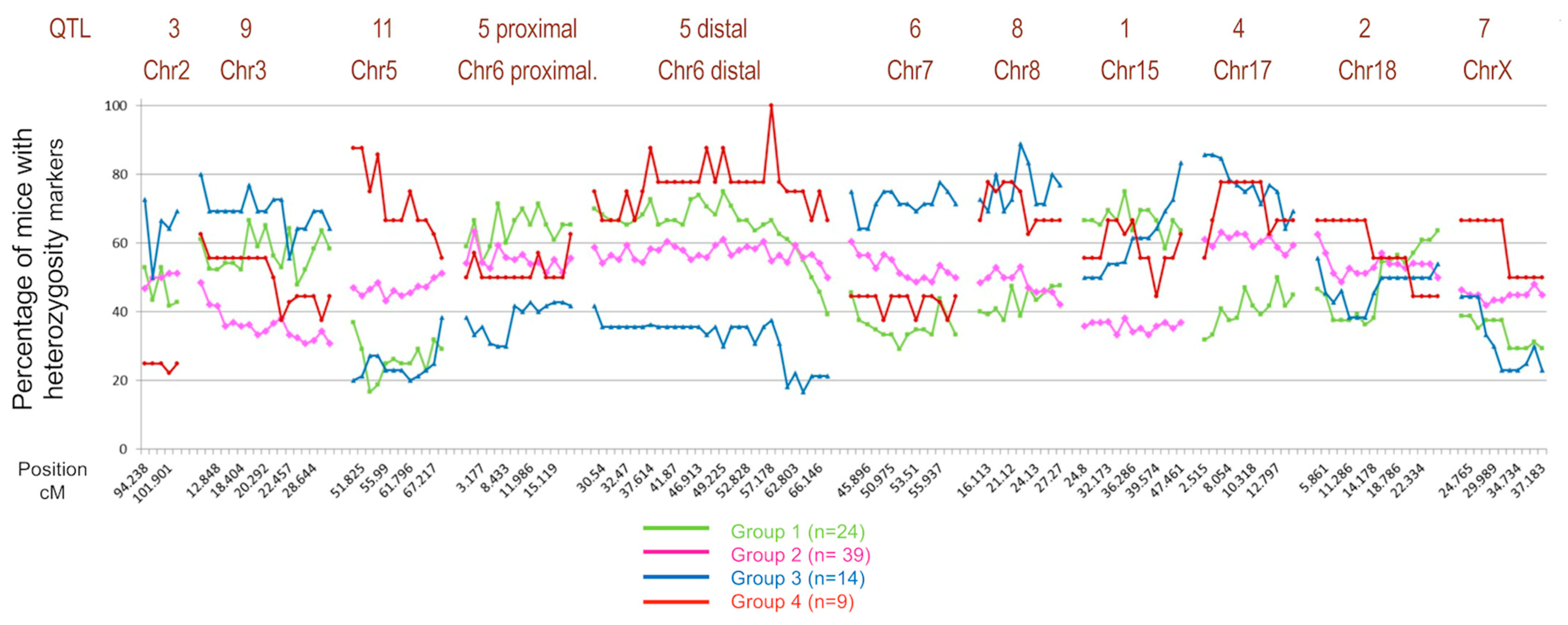
2.5. Identifying Chromosomal Regions Associated with Susceptibility
2.6. Identification of Genomic Regions Linked to Disease Susceptibility across the Four Groups Identified in the BX
3. Discussion
4. Materials and Methods
4.1. Animals and Parasites
4.2. BX Mouse Cohort Infection and Parasitological and Pathological Traits Obtained
4.3. Peripheral and Spleen White Blood Cell Subpopulations Quantified via Flow Cytometry
4.4. Detection of Anti-S. mansoni Antibodies in Mice Infected with the Parasite Using ELISA
4.5. SNP Genotyping, Quality Control, and Linkage Analysis
4.6. Data Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kyu, H.H.; Abate, D.; Abate, K.H.; Abay, S.M.; Abbafati, C.; Abbasi, N.; Abbastabar, H.; Abd-Allah, F.; Abdela, J.; Abdelalim, A.; et al. Global, Regional, and National Disability-Adjusted Life-Years (DALYs) for 359 Diseases and Injuries and Healthy Life Expectancy (HALE) for 195 Countries and Territories, 1990–2017: A Systematic Analysis for the Global Burden of Disease Study 2017. Lancet 2018, 392, 1859–1922. [Google Scholar] [CrossRef] [PubMed]
- Abath, F.G.C.; Morais, C.N.L.; Montenegro, C.E.L.; Wynn, T.A.; Montenegro, S.M.L. Immunopathogenic Mechanisms in Schistosomiasis: What Can Be Learnt from Human Studies? Trends Parasitol. 2006, 22, 85–91. [Google Scholar] [CrossRef]
- Burke, M.L.; Jones, M.K.; Gobert, G.N.; Li, Y.S.; Ellis, M.K.; McManus, D.P. Immunopathogenesis of Human Schistosomiasis. Parasite Immunol. 2009, 31, 163–176. [Google Scholar] [CrossRef] [PubMed]
- Marquet, S.; Abel, L.; Hillaire, D.; Dessein, H.; Kalil, J.; Feingold, J.; Weissenbach, J.; Dessein, A.J. Genetic Localization of a Locus Controlling the Intensity of Infection by Schistosoma mansoni on Chromosome 5q31–q33. Nat. Genet. 1996, 14, 181–184. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, V.; Piper, K.; Couissinier-Paris, P.; Bacelar, O.; Dessein, H.; Dessein, A.J. Genetic Control of Schistosome Infections by the SM1 Locus of the 5q31–q33 Region Is Linked to Differentiation of Type 2 Helper T Lymphocytes. Infect. Immun. 1999, 67, 4689–4692. [Google Scholar] [CrossRef] [PubMed]
- Chevillard, C.; Moukoko, C.E.; Elwali, N.-E.M.A.; Bream, J.H.; Kouriba, B.; Argiro, L.; Rahoud, S.; Mergani, A.; Henri, S.; Gaudart, J.; et al. IFN-γ Polymorphisms (IFN-γ +2109 and IFN-γ +3810) Are Associated with Severe Hepatic Fibrosis in Human Hepatic Schistosomiasis (Schistosoma mansoni). J. Immunol. 2003, 171, 5596–5601. [Google Scholar] [CrossRef]
- Dessein, A.; Chevillard, C.; Arnaud, V.; Hou, X.; Hamdoun, A.A.; Dessein, H.; He, H.; Abdelmaboud, S.A.; Luo, X.; Li, J.; et al. Variants of CTGF Are Associated with Hepatic Fibrosis in Chinese, Sudanese, and Brazilians Infected with Schistosomes. J. Exp. Med. 2009, 206, 2321–2328. [Google Scholar] [CrossRef]
- Cox, A.; Ackert-Bicknell, C.L.; Dumont, B.L.; Yueming, D.; Bell, J.T.; Brockmann, G.A.; Wergedal, J.E.; Bult, C.; Paigen, B.; Flint, J.; et al. A New Standard Genetic Map for the Laboratory Mouse. Genetics 2009, 182, 1335–1344. [Google Scholar] [CrossRef]
- Smith, P.M.; Shainheit, M.G.; Bazzone, L.E.; Rutitzky, L.I.; Poltorak, A.; Stadecker, M.J. Genetic Control of Severe Egg-Induced Immunopathology and IL-17 Production in Murine Schistosomiasis. J. Immunol. 2009, 183, 3317–3323. [Google Scholar] [CrossRef]
- Campino, S.; Kwiatkowski, D.; Dessein, A. Mendelian and Complex Genetics of Susceptibility and Resistance to Parasitic Infections. Semin. Immunol. 2006, 18, 411–422. [Google Scholar] [CrossRef]
- Gatlin, M.R.; Black, C.L.; Mwinzi, P.N.; Secor, W.E.; Karanja, D.M.; Colley, D.G. Association of the Gene Polymorphisms IFN-γ +874, IL-13-1055 and IL-4-590 with Patterns of Reinfection with Schistosoma mansoni. PLoS Neglected Trop. Dis. 2009, 3, e375. [Google Scholar] [CrossRef] [PubMed]
- Dessein, A.J.; Hillaire, D.; Elwali, N.E.M.A.; Marquet, S.; Mohamed-Ali, Q.; Mirghani, A.; Henri, S.; Abdelhameed, A.A.; Saeed, O.K.; Magzoub, M.M.A.; et al. Severe Hepatic Fibrosis in Schistosoma mansoni Infection Is Controlled by a Major Locus That Is Closely Linked to the Interferon-γ Receptor Gene. Am. J. Hum. Genet. 1999, 65, 709–721. [Google Scholar] [CrossRef] [PubMed]
- Blanton, R.E.; Salam, E.A.; Ehsan, A.; King, C.H.; Goddard, K.A.B. Schistosomal Hepatic Fibrosis and the Interferon Gamma Receptor: A Linkage Analysis Using Single-Nucleotide Polymorphic Markers. Eur. J. Hum. Genet. 2005, 13, 660–668. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mangano, V.D.; Modiano, D. Host Genetics and Parasitic Infections. Clin. Microbiol. Infect. 2014, 20, 1265–1275. [Google Scholar] [CrossRef] [PubMed]
- Pérez Del Villar, L.; Vicente, B.; Blanco-Gómez, A.; Castellanos, A.; Pérez-Losada, J.; Muro, A. Identifying Phenotypes Involved in Susceptibility to Schistosoma mansoni Infection in F1B6CBA Mice. Acta Parasitol. 2014, 59, 529–539. [Google Scholar] [CrossRef]
- Abbafati, C.; Machado, D.B.; Cislaghi, B.; Salman, O.M.; Karanikolos, M.; McKee, M.; Abbas, K.M.; Brady, O.J.; Larson, H.J.; Trias-Llimós, S.; et al. Global Age-Sex-Specific Fertility, Mortality, Healthy Life Expectancy (HALE), and Population Estimates in 204 Countries and Territories, 1950–2019: A Comprehensive Demographic Analysis for the Global Burden of Disease Study 2019. Lancet 2020, 396, 1160–1203. [Google Scholar] [CrossRef]
- Roth, G.A.; Abate, D.; Abate, K.H.; Abay, S.M.; Abbafati, C.; Abbasi, N.; Abbastabar, H.; Abd-Allah, F.; Abdela, J.; Abdelalim, A.; et al. Global, Regional, and National Age-Sex-Specific Mortality for 282 Causes of Death in 195 Countries and Territories, 1980–2017: A Systematic Analysis for the Global Burden of Disease Study 2017. Lancet 2018, 392, 1736–1788. [Google Scholar] [CrossRef]
- Abiola, O.; Angel, J.M.; Avner, P.; Bachmanov, A.A.; Belknap, J.K.; Bennett, B.; Blankenhorn, E.P.; Blizard, D.A.; Bolivar, V.; Brockmann, G.A.; et al. The Nature and Identification of Quantitative Trait Loci: A Community’s View. Nat. Rev. Genet. 2003, 4, 911–916. [Google Scholar]
- Kruglyak, L.; Lander, E.S. High-Resolution Genetic Mapping of Complex Traits. Am. J. Hum. Genet. 1995, 56, 1212–1223. [Google Scholar]
- Cheever, A.W.; Dunn, M.A.; Dean, D.A.; Duvall, R.H. Differences in Hepatic Fibrosis in ICR, C3H, and C57BL/6 Mice Infected with Schistosoma mansoni. Am. J. Trop. Med. Hyg. 1983, 32, 1364–1369. [Google Scholar] [CrossRef]
- Dean, D.A.; Bukowski, M.A.; Cheever, A.W. Relationship between Acquired Resistance, Portal Hypertension, and Lung Granulomas in Ten Strains of Mice Infected with Schistosoma mansoni. Am. J. Trop. Med. Hyg. 1981, 30, 806–814. [Google Scholar] [CrossRef]
- Cheever, A.W.; Duvall, R.H.; Hallack, T.A.; Minker, R.G.; Malley, J.D.; Malley, K.G. Variation of Hepatic Fibrosis and Granuloma Size among Mouse Strains Infected with Schistosoma mansoni. Am. J. Trop. Med. Hyg. 1987, 37, 85–97. [Google Scholar] [CrossRef] [PubMed]
- Colley, D.G.; Secor, W.E. Immunology of Human Schistosomiasis. Parasite Immunol. 2014, 36, 347–357. [Google Scholar] [CrossRef] [PubMed]
- Pérez del Villar, L.; Gutiérrez, M.; Vicente, B.; Castellanos, A.; Pérez-Losada, J.; Muro, A. Identifying Phenotypes Involved in Resistance to Schistosoma mansoni Infection in Inbred Mice. Trop. Med. Int. Health 2011, 16, 214–215. [Google Scholar]
- Fong, C.R.; Moron, N.A.; Kuris, A.M. Two’s a Crowd? Crowding Effect in a Parasitic Castrator Drives Differences in Reproductive Resource Allocation in Single vs Double Infections. Parasitology 2017, 144, 662–668. [Google Scholar] [CrossRef]
- Anthony, B.J.; Ramm, G.A.; McManus, D.P. Role of Resident Liver Cells in the Pathogenesis of Schistosomiasis. Trends Parasitol. 2012, 28, 572–579. [Google Scholar] [CrossRef] [PubMed]
- Pearce, E.J.; MacDonald, A.S. The Immunobiology of Schistosomiasis. Nat. Rev. Immunol. 2002, 2, 499–511. [Google Scholar] [CrossRef]
- Wilson, M.S.; Mentink-Kane, M.M.; Pesce, J.T.; Ramalingam, T.R.; Thompson, R.; Wynn, T.A. Immunopathology of Schistosomiasis. Immunol. Cell Biol. 2007, 85, 148–154. [Google Scholar] [CrossRef] [PubMed]
- Pedras-Vasconcelos, J.A.; Pearce, E.J. Type 1 CD8+ T Cell Responses during Infection with the Helminth Schistosoma mansoni. J. Immunol. 1996, 157, 3046–3053. [Google Scholar] [CrossRef]
- Stevens, T.L.; Bossie, A.; Sanders, V.M.; Fernandez-Botran, R.; Coffman, R.L.; Mosmann, T.R.; Vitetta, E.S. Regulation of Antibody Isotype Secretion by Subsets of Antigen-Specific Helper T Cells. Nature 1988, 334, 255–258. [Google Scholar] [CrossRef]
- Lawrence, R.A.; Allen, J.E.; Osborne, J.; Maizels, R.M. Adult and Microfilarial Stages of the Filarial Parasite Brugia Malayi Stimulate Contrasting Cytokine and Ig Isotype Responses in BALB/c Mice. J. Immunol. 1994, 153, 1216–1224. [Google Scholar] [CrossRef] [PubMed]
- Castellanos-Martín, A.; Castillo-Lluva, S.; Sáez-Freire, M.d.M.; Blanco-Gómez, A.; Hontecillas-Prieto, L.; Patino-Alonso, C.; Galindo-Villardon, P.; Pérez del Villar, L.; Martín-Seisdedos, C.; Isidoro-Garcia, M.; et al. Unraveling Heterogeneous Susceptibility and the Evolution of Breast Cancer Using a Systems Biology Approach. Genome Biol. 2015, 16, 40. [Google Scholar] [CrossRef]
- Blanco-Gómez, A.; Castillo-Lluva, S.; del Mar Sáez-Freire, M.; Hontecillas-Prieto, L.; Mao, J.H.; Castellanos-Martín, A.; Pérez-Losada, J. Missing Heritability of Complex Diseases: Enlightenment by Genetic Variants from Intermediate Phenotypes. BioEssays 2016, 38, 664–673. [Google Scholar] [CrossRef] [PubMed]
- Sáez-Freire, M.d.M.; Blanco-Gómez, A.; Castillo-Lluva, S.; Gómez-Vecino, A.; Galvis-Jiménez, J.M.; Martín-Seisdedos, C.; Isidoro-García, M.; Hontecillas-Prieto, L.; García-Cenador, M.B.; García-Criado, F.J.; et al. Supplementary Data for the Biological Age Linked to Oxidative Stress Modifies Breast Cancer Aggressiveness. Data Brief 2018, 18, 1172–1184. [Google Scholar] [CrossRef]
- Vicente, B.; López-Abán, J.; Rojas-Caraballo, J.; Del Olmo, E.; Fernández-Soto, P.; Muro, A. Protection against Schistosoma mansoni Infection Using a Fasciola hepatica-Derived Fatty Acid Binding Protein from Different Delivery Systems. Parasit Vectors 2016, 9, 216. [Google Scholar] [CrossRef] [PubMed]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 Years of Image Analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Turner, J.D.; Bourke, C.D.; Meurs, L.; Mbow, M.; Dièye, T.N.; Mboup, S.; Polman, K.; Mountford, A.P. Circulating CD14brightCD16+ “Intermediate” Monocytes Exhibit Enhanced Parasite Pattern Recognition in Human Helminth Infection. PLoS Neglected Trop. Dis. 2014, 8, e2817. [Google Scholar] [CrossRef]
- Debnath, S.; Nath, B.; Chakrabarti, A. Flow Cytometric Analysis of Protein Aggregates. Protein Pept. Lett. 2018, 24, 969–973. [Google Scholar] [CrossRef]
- Broman, K.W.; Wu, H.; Sen, Ś.; Churchill, G.A. R/Qtl: QTL Mapping in Experimental Crosses. Bioinformatics 2003, 19, 889–890. [Google Scholar] [CrossRef]
- Lander, E.; Kruglyak, L. Genetic Dissection of Complex Traits: Guidelines for Interpreting and Reporting Linkage Results. Nat. Genet. 1995, 11, 241–247. [Google Scholar] [CrossRef]
- Pérez-López, C. Métodos Estadísticos Avanzados Con SPSS.; Ediciones Paraninfo S.A.: Madrid, Spain, 2005. [Google Scholar]
- Armitage, P.; Berry, G.; Matthews, J.N.S. Statistical Methods in Medical Research; Blackwell Science: Oxford, UK, 2002. [Google Scholar]
- Bradsley, W. Simfit: Simulation, Statistical Analysis, Curve Fitting and Graph Plotting; Manchester University: Manchester, UK, 2017. [Google Scholar]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hernández-Goenaga, J.; López-Abán, J.; Blanco-Gómez, A.; Vicente, B.; Burguillo, F.J.; Pérez-Losada, J.; Muro, A. Identification of Genomic Regions Implicated in Susceptibility to Schistosoma mansoni Infection in a Murine Backcross Genetic Model. Int. J. Mol. Sci. 2023, 24, 14768. https://doi.org/10.3390/ijms241914768
Hernández-Goenaga J, López-Abán J, Blanco-Gómez A, Vicente B, Burguillo FJ, Pérez-Losada J, Muro A. Identification of Genomic Regions Implicated in Susceptibility to Schistosoma mansoni Infection in a Murine Backcross Genetic Model. International Journal of Molecular Sciences. 2023; 24(19):14768. https://doi.org/10.3390/ijms241914768
Chicago/Turabian StyleHernández-Goenaga, Juan, Julio López-Abán, Adrián Blanco-Gómez, Belén Vicente, Francisco Javier Burguillo, Jesús Pérez-Losada, and Antonio Muro. 2023. "Identification of Genomic Regions Implicated in Susceptibility to Schistosoma mansoni Infection in a Murine Backcross Genetic Model" International Journal of Molecular Sciences 24, no. 19: 14768. https://doi.org/10.3390/ijms241914768
APA StyleHernández-Goenaga, J., López-Abán, J., Blanco-Gómez, A., Vicente, B., Burguillo, F. J., Pérez-Losada, J., & Muro, A. (2023). Identification of Genomic Regions Implicated in Susceptibility to Schistosoma mansoni Infection in a Murine Backcross Genetic Model. International Journal of Molecular Sciences, 24(19), 14768. https://doi.org/10.3390/ijms241914768





