Interactions between Quantum Dots and G-Actin
Abstract
:1. Introduction
2. Results
2.1. Identification of QD Binding Protein
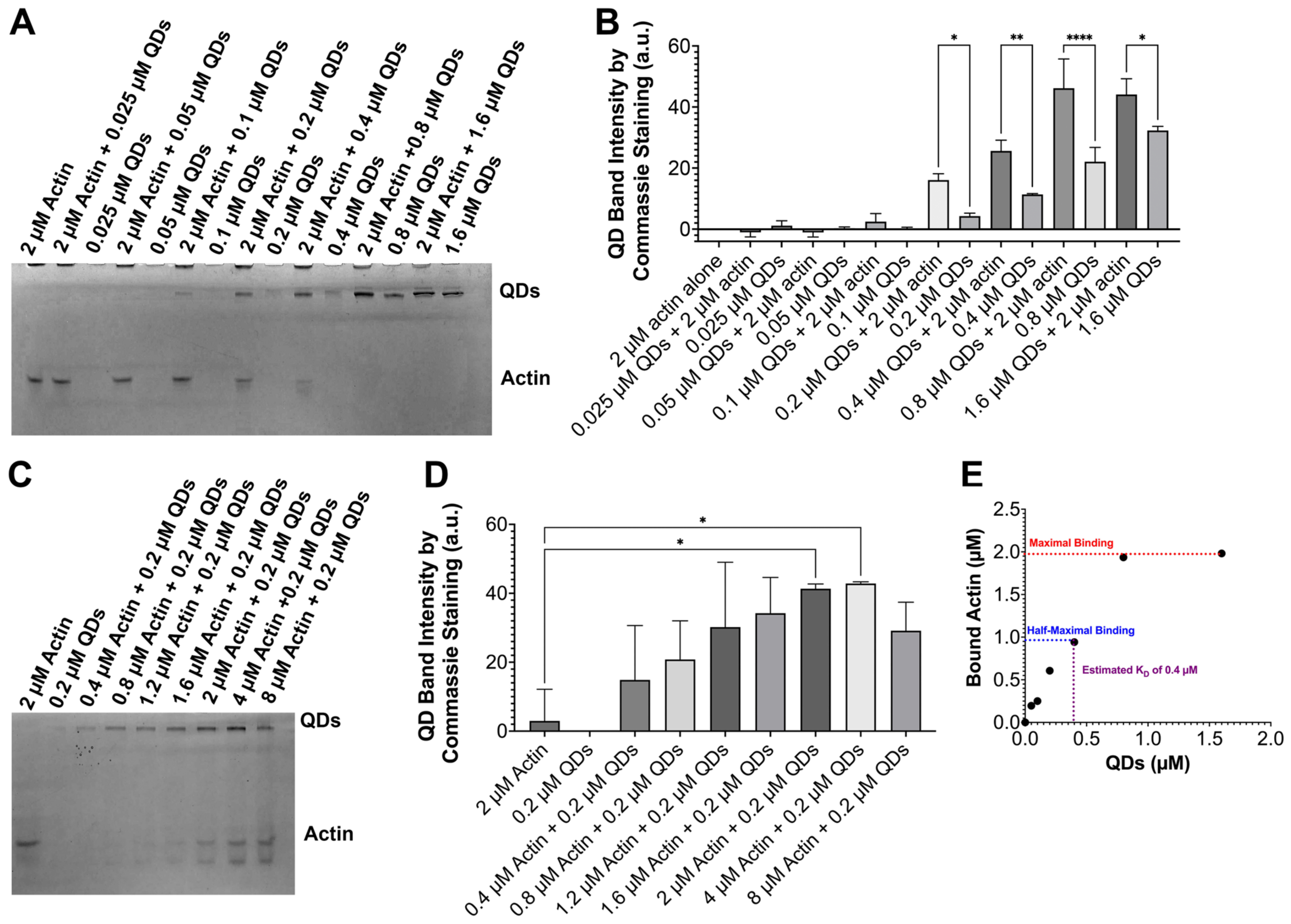
2.2. Validation of QD–Actin Interaction via a Native Gel Analysis
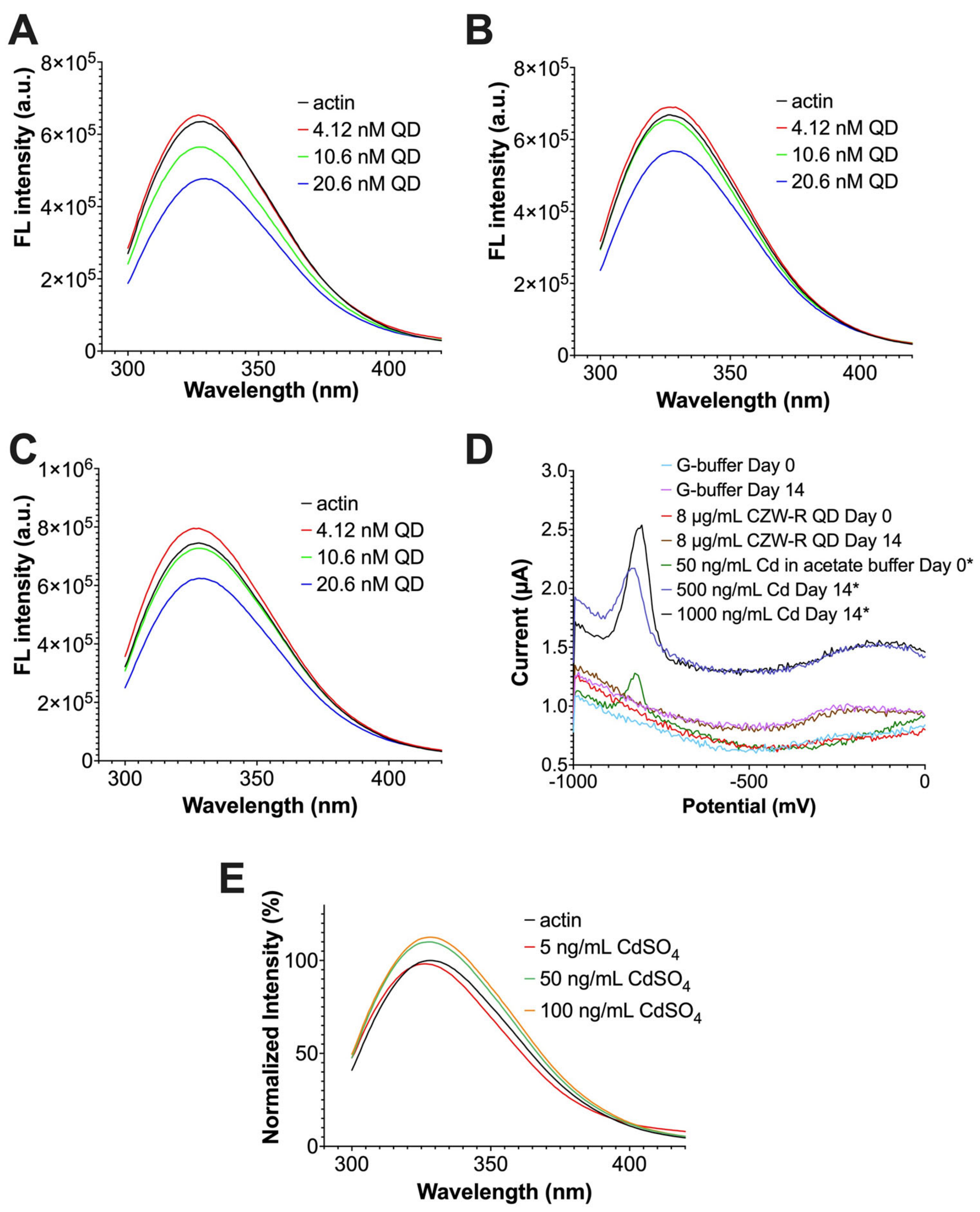
2.3. The Quenching of G-Actin’s Intrinsic Fluorescence by QDs
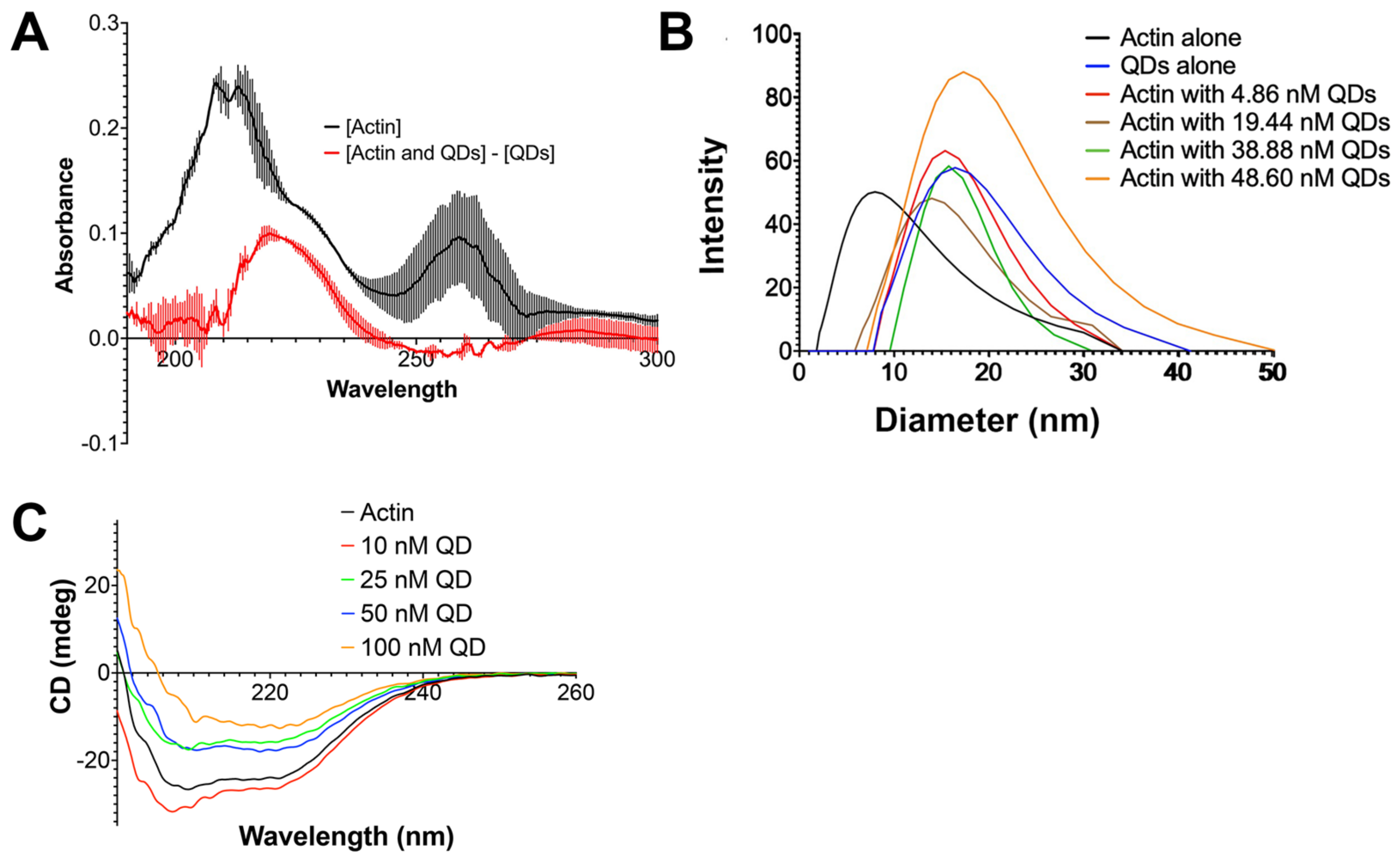
2.4. Complex Formation Assessment Using UV–Vis Absorption Spectroscopy
2.5. G-Actin Hydrodynamic Diameter Increased in the Presence of CdSe/ZnS QDs
2.6. The Alteration of Actin’s Secondary Structure by CdSe/ZnS QDs
3. Discussion
4. Materials and Methods
4.1. CdSe/ZnS QD Characteristics
4.2. Yeast Lysate Preparation
4.3. Proteomic Shotgun Analysis
4.4. Actin Preparation
4.5. Native Gel Electrophoresis
4.6. Fluorometer-Based Actin Fluorescence Quenching
4.7. Cd2+ Ion Leakage Detection
4.8. Ultraviolet–Visible (UV–Vis) Absorption Spectroscopy
4.9. Dynamic Light Scattering (DLS)
4.10. Circular Dichroism
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Herrera-Ochoa, D.; Pacheco-Liñán, P.J.; Bravo, I.; Garzón-Ruiz, A. A Novel Quantum Dot-Based PH Probe for Long-Term Fluorescence Lifetime Imaging Microscopy Experiments in Living Cells. ACS Appl. Mater. Interfaces 2022, 14, 2578–2586. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Li, W.; Yin, L.; Liu, Y.; Guo, H.; Lai, J.; Han, Y.; Li, G.; Li, M.; Zhang, J.; et al. Full-Color Fluorescent Carbon Quantum Dots. Sci. Adv. 2020, 6, eabb6772. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Tang, M. The Cytotoxicity of Core-Shell or Non-Shell Structure Quantum Dots and Reflection on Environmental Friendly: A Review. Environ. Res. 2021, 194, 110593. [Google Scholar] [CrossRef] [PubMed]
- Vasudevan, D.; Gaddam, R.R.; Trinchi, A.; Cole, I. Core-Shell Quantum Dots: Properties and Applications. J. Alloys Compd. 2015, 636, 395–404. [Google Scholar] [CrossRef]
- Abdul Ghani, S.F.; Wright, M.; Paramo, J.G.; Bottrill, M.; Green, M.; Long, N.; Thanou, M. Three Bisphosphonate Ligands Improve the Water Solubility of Quantum Dots. Faraday Discuss. 2014, 175, 153–169. [Google Scholar] [CrossRef] [PubMed]
- Khan, K.U.; Minhas, M.U.; Badshah, S.F.; Suhail, M.; Ahmad, A.; Ijaz, S. Overview of Nanoparticulate Strategies for Solubility Enhancement of Poorly Soluble Drugs. Life Sci. 2022, 291, 120301. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Lin, S.; Li, Q.; Jiang, S.; Wang, P. Recent Advances in Techniques for Enhancing the Solubility of Hydrophobic Drugs. Pak. J. Pharm. Sci. 2022, 35, 95–112. [Google Scholar]
- Göke, K.; Bunjes, H. Drug Solubility in Lipid Nanocarriers: Influence of Lipid Matrix and Available Interfacial Area. Int. J. Pharm. 2017, 529, 617–628. [Google Scholar] [CrossRef] [PubMed]
- Azam, N.; Najabat Ali, M.; Javaid Khan, T. Carbon Quantum Dots for Biomedical Applications: Review and Analysis. Front. Mater. 2021, 8, 272. [Google Scholar] [CrossRef]
- Ding, L.; Wang, X.; Li, J.; Huang, J.; Li, Z. Synthesis of Fluorescent Carbon Quantum Dots and Their Application in the Plant Cell Imaging. J. Wuhan. Univ. Technol. Mater. Sci. Ed. 2018, 33, 1546–1550. [Google Scholar] [CrossRef]
- Zhao, C.; Song, X.; Liu, Y.; Fu, Y.; Ye, L.; Wang, N.; Wang, F.; Li, L.; Mohammadniaei, M.; Zhang, M.; et al. Synthesis of Graphene Quantum Dots and Their Applications in Drug Delivery. J. Nanobiotech. 2020, 18, 142. [Google Scholar] [CrossRef] [PubMed]
- Qiu, J.; Zhang, R.; Li, J.; Sang, Y.; Tang, W.; Gil, P.R.; Liu, H. Fluorescent Graphene Quantum Dots as Traceable, PH-Sensitive Drug Delivery Systems. Int. J. Nanomed. 2015, 10, 6709–6724. [Google Scholar] [CrossRef]
- Mansuriya, B.D.; Altintas, Z. Applications of Graphene Quantum Dots in Biomedical Sensors. Sensors 2020, 20, 1072. [Google Scholar] [CrossRef] [PubMed]
- Thakur, S.; Sharma, S. Biomedical Applications of Single-Particle Based Material: Quantum Dots. Int. J. Radiol. Radiat. Ther. 2022, 9, 121–127. [Google Scholar] [CrossRef]
- Panja, A.; Patra, P. A Review on Quantum Dots (QDs) and Their Biomedical Applications. 4open 2023, 6, 1. [Google Scholar] [CrossRef]
- Abdellatif, A.A.H.; Tawfeek, H.M.; Younis, M.A.; Alsharidah, M.; Al Rugaie, O. Biomedical Applications of Quantum Dots: Overview, Challenges, and Clinical Potential. Int. J. Nanomed. 2022, 17, 1951–1970. [Google Scholar] [CrossRef]
- Le, N.; Kim, K. Current Advances in the Biomedical Applications of Quantum Dots: Promises and Challenges. Int. J. Mol. Sci. 2023, 24, 12682. [Google Scholar] [CrossRef]
- Truskewycz, A.; Yin, H.; Halberg, N.; Lai, D.T.H.; Ball, A.S.; Truong, V.K.; Rybicka, A.M.; Cole, I. Carbon Dot Therapeutic Platforms: Administration, Distribution, Metabolism, Excretion, Toxicity, and Therapeutic Potential. Small 2022, 18, 2106342. [Google Scholar] [CrossRef]
- Bottrill, M.; Green, M. Some Aspects of Quantum Dot Toxicity. Chem. Commun. 2011, 47, 7039–7050. [Google Scholar] [CrossRef]
- Valizadeh, A.; Mikaeili, H.; Samiei, M.; Farkhani, S.M.; Zarghami, N.; Kouhi, M.; Akbarzadeh, A.; Davaran, S. Quantum Dots: Synthesis, Bioapplications, and Toxicity. Nanoscale Res. Lett. 2012, 7, 480. [Google Scholar] [CrossRef]
- Hauck, T.S.; Anderson, R.E.; Fischer, H.C.; Newbigging, S.; Chan, W.C.W. In Vivo Quantum-Dot Toxicity Assessment. Small 2010, 6, 138–144. [Google Scholar] [CrossRef] [PubMed]
- Wong, B.S.E.; Hu, Q.; Baeg, G.H. Epigenetic Modulations in Nanoparticle-Mediated Toxicity. Food Chem. Toxicol. 2017, 109, 746–752. [Google Scholar] [CrossRef]
- Han, X.; Lai, L.; Tian, F.; Jiang, F.L.; Xiao, Q.; Li, Y.; Yu, Q.; Li, D.; Wang, J.; Zhang, Q.; et al. Toxicity of CdTe Quantum Dots on Yeast Saccharomyces Cerevisiae. Small 2012, 8, 2680–2689. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Zhang, T.; Tang, M. Toxicity of Quantum Dots on Target Organs and Immune System. J. Appl. Toxicol. 2022, 42, 17–40. [Google Scholar] [CrossRef] [PubMed]
- Zhong, L.; Zhang, L.; Li, Y.; Liang, X.; Kong, L.; Shen, X.; Wu, T. Assessment of the Toxicity of Quantum Dots through Biliometric Analysis. Int. J. Environ. Res. Public Health 2021, 18, 5768. [Google Scholar] [CrossRef] [PubMed]
- Duan, J.; Yu, Y.; Li, Y.; Yu, Y.; Li, Y.; Huang, P.; Zhou, X.; Peng, S.; Sun, Z. Developmental Toxicity of CdTe QDs in Zebrafish Embryos and Larvae. J. Nanoparticle Res. 2013, 15, 1700. [Google Scholar] [CrossRef]
- Xu, Z.Q.; Lai, L.; Li, D.W.; Li, R.; Xiang, C.; Jiang, F.L.; Sun, S.F.; Liu, Y. Toxicity of CdTe QDs with Different Sizes Targeted to HSA Investigated by Two Electrochemical Methods. Mol. Biol. Rep. 2013, 40, 1009–1019. [Google Scholar] [CrossRef]
- Liu, Z.; Li, F.; Luo, Y.; Li, M.; Hu, G.; Pu, X.; Tang, T.; Wen, J.; Li, X.; Li, W. Size Effect of Graphene Quantum Dots on Photoluminescence. Molecules 2021, 26, 3922. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Lu, Y.; Li, J.; Liu, R.; Zhu, X. Effect of Graphene Quantum Dot Size on Plant Growth. Nanoscale 2020, 12, 15045–15049. [Google Scholar] [CrossRef]
- Mo, D.; Hu, L.; Zeng, G.; Chen, G.; Wan, J.; Yu, Z.; Huang, Z.; He, K.; Zhang, C.; Cheng, M. Cadmium-Containing Quantum Dots: Properties, Applications, and Toxicity. Appl. Microbiol. Biotechnol. 2017, 101, 2713–2733. [Google Scholar] [CrossRef]
- Zhang, B.; Wang, Y.; Hu, R.; Roy, I.; Yong, K.T. Cadmium-Free Quantum Dots for Biophotonic Imaging and Sensing. In Handbook of Photonics for Biomedical Engineering; Springer: Berlin/Heidelberg, Germany, 2014. [Google Scholar]
- Tekle, C.; Van Deurs, B.; Sandvig, K.; Iversen, T.G. Cellular Trafficking of Quantum Dot-Ligand Bioconjugates and Their Induction of Changes in Normal Routing of Unconjugated Ligands. Nano Lett. 2008, 8, 1858–1865. [Google Scholar] [CrossRef] [PubMed]
- Tan, S.J.; Jana, N.R.; Gao, S.; Patra, P.K.; Ying, J.Y. Surface-Ligand-Dependent Cellular Interaction, Subcellular Localization, and Cytotoxicity of Polymer-Coated Quantum Dots. Chem. Mater. 2010, 22, 2239–2247. [Google Scholar] [CrossRef]
- Yu, Y.Q.; Chen, W.Q.; Li, X.H.; Liu, M.; He, X.H.; Liu, Y.; Jiang, F.L. Quantum Dots Meet Enzymes: Hydrophobicity of Surface Ligands and Size Do Matter. Langmuir 2022, 39, 3967–3978. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, K.C.; Willmore, W.G.; Tayabali, A.F. Cadmium Telluride Quantum Dots Cause Oxidative Stress Leading to Extrinsic and Intrinsic Apoptosis in Hepatocellular Carcinoma HepG2 Cells. Toxicology 2013, 306, 114–123. [Google Scholar] [CrossRef]
- Harris, S.; Kim, K. Apoptotic Pathway Protein Expression Variance in Metal Oxide and Quantum Dot Treated HeLa Cells. MicroPubl. Biol. 2023, 2023. [Google Scholar] [CrossRef]
- Liu, Y.Y.; Chang, Q.; Sun, Z.X.; Liu, J.; Deng, X.; Liu, Y.; Cao, A.; Wang, H. Fate of CdSe/ZnS Quantum Dots in Cells: Endocytosis, Translocation and Exocytosis. Colloids Surf. B Biointerfaces 2021, 208, 112140. [Google Scholar] [CrossRef]
- Zhang, M.Q.; Wang, Z.G.; Fu, D.D.; Zhang, J.M.; Liu, H.Y.; Liu, S.L.; Pang, D.W. Quantum Dots Tracking Endocytosis and Transport of Proteins Displayed by Mammalian Cells. Anal. Chem. 2022, 94, 7567–7575. [Google Scholar] [CrossRef]
- Le, N.; Routh, J.; Kirk, C.; Wu, Q.; Patel, R.; Keyes, C.; Kim, K. Red CdSe/ZnS QDs’ Intracellular Trafficking and Its Impact on Yeast Polarization and Actin Filament. Cells 2023, 12, 484. [Google Scholar] [CrossRef]
- Matea, C.T.; Mocan, T.; Tabaran, F.; Pop, T.; Mosteanu, O.; Puia, C.; Iancu, C.; Mocan, L. Quantum Dots in Imaging, Drug Delivery and Sensor Applications. Int. J. Nanomed. 2017, 12, 5421–5431. [Google Scholar] [CrossRef]
- Nair, A.; Haponiuk, J.T.; Thomas, S.; Gopi, S. Natural Carbon-Based Quantum Dots and Their Applications in Drug Delivery: A Review. Biomed. Pharmacother. 2020, 132, 110834. [Google Scholar] [CrossRef]
- Bagalkot, V.; Zhang, L.; Levy-Nissenbaum, E.; Jon, S.; Kantoff, P.W.; Langery, R.; Farokhzad, O.C. Quantum Dot-Aptamer Conjugates for Synchronous Cancer Imaging, Therapy, and Sensing of Drug Delivery Based on Bi-Fluorescence Resonance Energy Transfer. Nano Lett. 2007, 7, 3065–3070. [Google Scholar] [CrossRef] [PubMed]
- Chakravarthy, K.V.; Davidson, B.A.; Helinski, J.D.; Ding, H.; Law, W.-C.; Yong, K.-T.; Prasad, P.N.; Knight, P.R. Doxorubicin Conjugated Quantum Dots to Target Alveolar Macrophages/Inflammation. Nanomedicine 2010, 7, 88. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Liu, H.; Liu, J.; Haley, K.N.; Treadway, J.A.; Larson, J.P.; Ge, N.; Peale, F.; Bruchez, M.P. Immunofluorescent Labeling of Cancer Marker Her2 and Other Cellular Targets with Semiconductor Quantum Dots. Nat. Biotechnol. 2003, 21, 41–46. [Google Scholar] [CrossRef]
- Zheng, M.; Ruan, S.; Liu, S.; Sun, T.; Qu, D.; Zhao, H.; Xie, Z.; Gao, H.; Jing, X.; Sun, Z. Self-Targeting Fluorescent Carbon Dots for Diagnosis of Brain Cancer Cells. ACS Nano 2015, 9, 11455–11461. [Google Scholar] [CrossRef] [PubMed]
- Pilch, J.; Kowalik, P.; Kowalczyk, A.; Bujak, P.; Kasprzak, A.; Paluszkiewicz, E.; Augustin, E.; Nowicka, A.M. Foliate-Targeting Quantum Dots-β-Cyclodextrin Nanocarrier for Efficient Delivery of Unsymmetrical Bisacridines to Lung and Prostate Cancer Cells. Int. J. Mol. Sci. 2022, 23, 1261. [Google Scholar] [CrossRef] [PubMed]
- Preeyanka, N.; Akhuli, A.; Dey, H.; Chakraborty, D.; Rahaman, A.; Sarkar, M. Realization of a Model-Free Pathway for Quantum Dot-Protein Interaction Beyond Classical Protein Corona or Protein Complex. Langmuir 2022, 38, 10704–10715. [Google Scholar] [CrossRef]
- Akhuli, A.; Chakraborty, D.; Agrawal, A.K.; Sarkar, M. Probing the Interaction of Bovine Serum Albumin with Copper Nanoclusters: Realization of Binding Pathway Different from Protein Corona. Langmuir 2021, 37, 1823–1837. [Google Scholar] [CrossRef]
- Qu, S.; Qiao, Z.; Zhong, W.; Liang, K.; Jiang, X.; Shang, L. Chirality-Dependent Dynamic Evolution of the Protein Corona on the Surface of Quantum Dots. ACS Appl. Mater. Interfaces 2022, 14, 44147–44157. [Google Scholar] [CrossRef]
- Littlechild, J.A. Protein Structure and Function. In Introduction to Biological and Small Molecule Drug Research and Development: Theory and Case Studies; Academic Press: Cambridge, MA, USA, 2013; pp. 57–79. [Google Scholar] [CrossRef]
- Dominguez, R.; Holmes, K.C. Actin Structure and Function. Annu. Rev. Biophys. 2011, 40, 169. [Google Scholar] [CrossRef]
- Goode, B.L.; Eskin, J.A.; Wendland, B. Actin and Endocytosis in Budding Yeast. Genetics 2014, 199, 315–358. [Google Scholar] [CrossRef]
- Galletta, B.J.; Mooren, O.L.; Cooper, J.A. Actin Dynamics and Endocytosis in Yeast and Mammals. Curr. Opin. Biotechnol. 2010, 21, 604–610. [Google Scholar] [CrossRef] [PubMed]
- Nickaeen, M.; Berro, J.; Pollard, T.D.; Slepchenko, B.M. Actin Assembly Produces Sufficient Forces for Endocytosis in Yeast. Mol. Biol. Cell 2019, 30, 2014–2024. [Google Scholar] [CrossRef] [PubMed]
- Moseley, J.B.; Goode, B.L. The Yeast Actin Cytoskeleton: From Cellular Function to Biochemical Mechanism. Microbiol. Mol. Biol. Rev. 2006, 70, 605–645. [Google Scholar] [CrossRef] [PubMed]
- Chambers, J.R.; Sauer, K. Detection of Cyclic Di-GMP Binding Proteins Utilizing a Biotinylated Cyclic Di-GMP Pulldown Assay. Methods Mol. Biol. 2017, 1657, 317–329. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Q.; Huang, S.; Qi, Z.D.; Zhou, B.; He, Z.K.; Liu, Y. Conformation, Thermodynamics and Stoichiometry of HSA Adsorbed to Colloidal CdSe/ZnS Quantum Dots. Biochim. Biophys. Acta 2008, 1784, 1020–1027. [Google Scholar] [CrossRef]
- Lichota, A.; Szabelski, M.; Krokosz, A. Quenching of Protein Fluorescence by Fullerenol C60(OH)36 Nanoparticles. Int. J. Mol. Sci. 2022, 23, 12382. [Google Scholar] [CrossRef]
- Feldman, I.; Young, D.; McGuire, R. Static and Dynamic Quenching of Protein Fluorescence. I. Bovine Serum Albumin. Biopolymers 1975, 14, 335–351. [Google Scholar] [CrossRef]
- Raghuraman, H.; Chatterjee, S.; Das, A. Site-Directed Fluorescence Approaches for Dynamic Structural Biology of Membrane Peptides and Proteins. Front. Mol. Biosci. 2019, 6, 96. [Google Scholar] [CrossRef]
- Hashempour, S.; Shahabadi, N.; Adewoye, A.; Murphy, B.; Rouse, C.; Salvatore, B.A.; Stratton, C.; Mahdavian, E. Binding Studies of AICAR and Human Serum Albumin by Spectroscopic, Theoretical, and Computational Methodologies. Molecules 2020, 25, 5410. [Google Scholar] [CrossRef]
- Deepa, H.R.; Thipperudrappa, J.; Kumar, H.S. Effect of Temperature on Fluorescence Quenching and Emission Characteristics of Laser Dyes. J. Phys. Conf. Ser. 2020, 1473, 012046. [Google Scholar] [CrossRef]
- Fraiji, L.K.; Hayes, D.M.; Werner1, T.C. Static and Dynamic Fluorescence Quenching Experiments for the Physical Chemistry Laboratory. J. Chem. Educ. 1992, 69, 424. [Google Scholar] [CrossRef]
- Duah-Williams, L.; Hawkridge, F.M. The Temperature Dependence of the Kinetics of Cyanide Dissociation from the Cyanide Complex of Myoglobin Studied by Cyclic Voltammetry. J. Electroanal. Chem. 1999, 466, 177–186. [Google Scholar] [CrossRef]
- Doyle, T.C.; Hansen, J.E.; Reisler, E. Tryptophan Fluorescence of Yeast Actin Resolved via Conserved Mutations. Biophys. J. 2001, 80, 427–434. [Google Scholar] [CrossRef]
- Mei, J.; Yang, L.Y.; Lai, L.; Xu, Z.Q.; Wang, C.; Zhao, J.; Jin, J.C.; Jiang, F.L.; Liu, Y. The Interactions between CdSe Quantum Dots and Yeast Saccharomyces Cerevisiae: Adhesion of Quantum Dots to the Cell Surface and the Protection Effect of ZnS Shell. Chemosphere 2014, 112, 92–99. [Google Scholar] [CrossRef]
- Le, N.; Zhang, M.; Kim, K. Quantum Dots and Their Interaction with Biological Systems. Int. J. Mol. Sci. 2022, 23, 10763. [Google Scholar] [CrossRef]
- Yan, R.; Yu, B.Q.; Yin, M.M.; Zhou, Z.Q.; Xiang, X.; Han, X.L.; Liu, Y.; Jiang, F.L. The Interactions of CdTe Quantum Dots with Serum Albumin and Subsequent Cytotoxicity: The Influence of Homologous Ligands. Toxicol. Res. 2018, 7, 147. [Google Scholar] [CrossRef]
- Saptarshi, S.R.; Duschl, A.; Lopata, A.L. Interaction of Nanoparticles with Proteins: Relation to Bio-Reactivity of the Nanoparticle. J. Nanobiotechnol. 2013, 11, 26. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Li, H.; Meng, P.; Li, K.; Xiong, Y.; Zhang, S.; Yang, Y.; Yin, A.; Huang, P. Interactions between CdTe Quantum Dots and Plasma Proteins: Kinetics, Thermodynamics and Molecular Structure Changes. Colloids Surf. B Biointerfaces 2020, 189, 110881. [Google Scholar] [CrossRef] [PubMed]
- Rottner, K.; Faix, J.; Bogdan, S.; Linder, S.; Kerkhoff, E. Actin Assembly Mechanisms at a Glance. J. Cell Sci. 2017, 130, 3427–3435. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Nakamura, F. Actin-Associated Proteins and Small Molecules Targeting the Actin Cytoskeleton. Int. J. Mol. Sci. 2022, 23, 2118. [Google Scholar] [CrossRef] [PubMed]
- Qin, C.; Li, W.; Li, Q.; Yin, W.; Zhang, X.; Zhang, Z.; Zhang, X.E.; Cui, Z. Real-Time Dissection of Dynamic Uncoating of Individual Influenza Viruses. Proc. Natl. Acad. Sci. USA 2019, 116, 2577–2582. [Google Scholar] [CrossRef]
- Liu, H.; Liu, Y.; Liu, S.; Pang, D.-W.; Xiao, G. Clathrin-Mediated Endocytosis in Living Host Cells Visualized through Quantum Dot Labeling of Infectious Hematopoietic Necrosis Virus. J. Virol. 2011, 85, 6252–6262. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Su, W.; Wu, H.; Yuan, T.; Yuan, C.; Liu, J.; Deng, G.; Gao, X.; Chen, Z.; Bao, Y.; et al. Targeted Tumour Theranostics in Mice via Carbon Quantum Dots Structurally Mimicking Large Amino Acids|Enhanced Reader. Nat. Biomed. Eng. 2020, 4, 704–716. [Google Scholar] [CrossRef] [PubMed]
- Ryu, I.; Ryu, J.Y.; Choe, G.; Kwon, H.; Park, H.; Cho, Y.S.; Du, R.; Yim, S. In Vivo Plain X-Ray Imaging of Cancer Using Perovskite Quantum Dot Scintillators. Adv. Funct. Mater. 2021, 31, 2102334. [Google Scholar] [CrossRef]
- Zhang, M.; Vojtech, L.; Ye, Z.; Hladik, F.; Nance, E. Quantum Dot Labeling and Visualization of Extracellular Vesicles. ACS Appl. Nano Mater. 2020, 3, 7211–7222. [Google Scholar] [CrossRef]
- Voura, E.B.; Jaiswal, J.K.; Mattoussi, H.; Simon, S.M. Tracking Metastatic Tumor Cell Extravasation with Quantum Dot Nanocrystals and Fluorescence Emission-Scanning Microscopy. Nat. Med. 2004, 10, 993–998. [Google Scholar] [CrossRef]
- Díaz-García, V.M.; Guerrero, S.; Díaz-Valdivia, N.; Lobos-González, L.; Kogan, M.; Pérez-Donoso, J.M.; Quest, A.F.G. Biomimetic Quantum Dot-Labeled B16F10 Murine Melanoma Cells as a Tool to Monitor Early Steps of Lung Metastasis by in Vivo Imaging. Int. J. Nanomed. 2018, 13, 6391. [Google Scholar] [CrossRef]
- Wang, H.; Nienhaus, K.; Shang, L.; Nienhaus, G.U. Highly Luminescent Positively Charged Quantum Dots Interacting with Proteins and Cells. Chin. J. Chem. 2022, 40, 2685. [Google Scholar] [CrossRef]
- Luo, H.; Li, B.; Liu, J.; Liu, Y.; Xiao, Q.; Huang, S. Investigation on Conformational Variation and Fibrillation of Human Serum Albumin Affected by Molybdenum Disulfide Quantum Dots. Int. J. Biol. Macromol. 2021, 190, 999–1006. [Google Scholar] [CrossRef]
- Wang, Q.; Chen, W.Q.; Liu, X.Y.; Liu, Y.; Jiang, F.L. Thermodynamic Implications and Time Evolution of the Interactions of Near-Infrared PbS Quantum Dots with Human Serum Albumin. ACS Omega 2021, 6, 5569–5581. [Google Scholar] [CrossRef]
- Horstmann, C.; Kim, D.S.; Campbell, C.; Kim, K. Transcriptome Profile Alteration with Cadmium Selenide/Zinc Sulfide Quantum Dots in Saccharomyces Cerevisiae. Biomolecules 2019, 9, 653. [Google Scholar] [CrossRef]
- Zhang, M.; Kim, D.S.; Patel, R.; Wu, Q.; Kim, K. Intracellular Trafficking and Distribution of Cd and InP Quantum Dots in HeLa and ML-1 Thyroid Cancer Cells. Nanomaterials 2022, 12, 1517. [Google Scholar] [CrossRef] [PubMed]
- Horstmann, C.; Kim, K. Comparing Transcriptome Profiles of Saccharomyces Cerevisiae Cells Exposed to Cadmium Selenide/Zinc Sulfide and Indium Phosphide/Zinc Sulfide. Genes 2021, 12, 428. [Google Scholar] [CrossRef]
- Horstmann, C.; Davenport, V.; Zhang, M.; Peters, A.; Kim, K. Transcriptome Profile Alterations with Carbon Nanotubes, Quantum Dots, and Silver Nanoparticles: A Review. Genes 2021, 12, 794. [Google Scholar] [CrossRef] [PubMed]
- Hens, B.; Smothers, J.; Rizvanovic, H.; Patel, R.; Wu, Q.; Kim, K. The Future of Anticancer Drugs: A Cytotoxicity Assessment Study of CdSe/ZnS Quantum Dots. J. Nanotheranostics 2020, 1, 19–38. [Google Scholar] [CrossRef]
- Davenport, V.; Horstmann, C.; Patel, R.; Wu, Q.; Kim, K. An Assessment of InP/ZnS as Potential Anti-cancer Therapy: Quantum Dot Treatment Induces Stress on HeLa Cells. J. Nanotheranostics 2021, 2, 16–32. [Google Scholar] [CrossRef]
- Xu, Y.M.; Tan, H.W.; Zheng, W.; Liang, Z.L.; Yu, F.Y.; Wu, D.D.; Yao, Y.; Zhong, Q.H.; Yan, R.; Lau, A.T.Y. Cadmium Telluride Quantum Dot-Exposed Human Bronchial Epithelial Cells: A Further Study of the Cellular Response by Proteomics. Toxicol. Res. 2019, 8, 994–1001. [Google Scholar] [CrossRef]
- Zhang, Y.; Schnoes, A.M.; Clapp, A.R. Dithiocarbamates as Capping Ligands for Water-Soluble Quantum Dots. ACS Appl. Mater. Interfaces 2010, 2, 3384–3395. [Google Scholar] [CrossRef]
- Gao, Y.; Aerts, M.; Sandeep, C.S.S.; Talgorn, E.; Savenije, T.J.; Kinge, S.; Siebbeles, L.D.A.; Houtepen, A.J. Photoconductivity of PbSe Quantum-Dot Solids: Dependence on Ligand Anchor Group and Length. ACS Nano 2012, 6, 9606–9614. [Google Scholar] [CrossRef]
- Tan, Y.; Jin, S.; Hamers, R.J. Photostability of Cdse Quantum Dots Functionalized with Aromatic Dithiocarbamate Ligands. ACS Appl. Mater. Interfaces 2013, 5, 12975–12983. [Google Scholar] [CrossRef]
- Pilch, J.; Matysiak-Brynda, E.; Kowalczyk, A.; Bujak, P.; Mazerska, Z.; Nowicka, A.M.; Augustin, E. New Unsymmetrical Bisacridine Derivatives Noncovalently Attached to Quaternary Quantum Dots Improve Cancer Therapy by Enhancing Cytotoxicity toward Cancer Cells and Protecting Normal Cells. ACS Appl. Mater. Interfaces 2020, 12, 17276–17289. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Shi, W.; Chen, W.; Li, X.; Ma, H. Fluorescent Carbon Nanodots Conjugated with Folic Acid for Distinguishing Folate-Receptor-Positive Cancer Cells from Normal Cells. J. Mater. Chem. 2012, 22, 12568–12573. [Google Scholar] [CrossRef]
- Campbell, E.; Hasan, M.T.; Gonzalez Rodriguez, R.; Akkaraju, G.R.; Naumov, A.V. Doped Graphene Quantum Dots for Intracellular Multicolor Imaging and Cancer Detection. ACS Biomater. Sci. Eng. 2019, 5, 4671–4682. [Google Scholar] [CrossRef] [PubMed]
- Lin, G.; Ouyang, Q.; Hu, R.; Ding, Z.; Tian, J.; Yin, F.; Xu, G.; Chen, Q.; Wang, X.; Yong, K.T. In Vivo Toxicity Assessment of Non-Cadmium Quantum Dots in BALB/c Mice. Nanomedicine 2015, 11, 341–350. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Chen, Y.; Xu, G.; Liu, D.; Yang, Z.; Chen, T.; Wang, X.; Jiang, W.; Xue, D.; Lin, G. In Vivo Comparison of the Biodistribution and Toxicity of InP/ZnS Quantum Dots with Different Surface Modifications. Int. J. Nanomed. 2020, 15, 1951–1965. [Google Scholar] [CrossRef]
- Yaghini, E.; Turner, H.; Pilling, A.; Naasani, I.; MacRobert, A.J. In Vivo Biodistribution and Toxicology Studies of Cadmium-Free Indium-Based Quantum Dot Nanoparticles in a Rat Model. Nanomedicine 2018, 14, 2644–2655. [Google Scholar] [CrossRef]
- Song, Y.; Wu, Y.; Wang, H.; Liu, S.; Song, L.; Li, S.; Tan, M. Carbon Quantum Dots from Roasted Atlantic Salmon (Salmo salar L.): Formation, Biodistribution and Cytotoxicity. Food Chem. 2019, 293, 387–395. [Google Scholar] [CrossRef]
- Xu, L.; Dai, Y.; Wang, Z.; Zhao, J.; Li, F.; White, J.C.; Xing, B. Graphene Quantum Dots in Alveolar Macrophage: Uptake-Exocytosis, Accumulation in Nuclei, Nuclear Responses and DNA Cleavage. Part. Fibre Toxicol. 2018, 15, 45. [Google Scholar] [CrossRef]

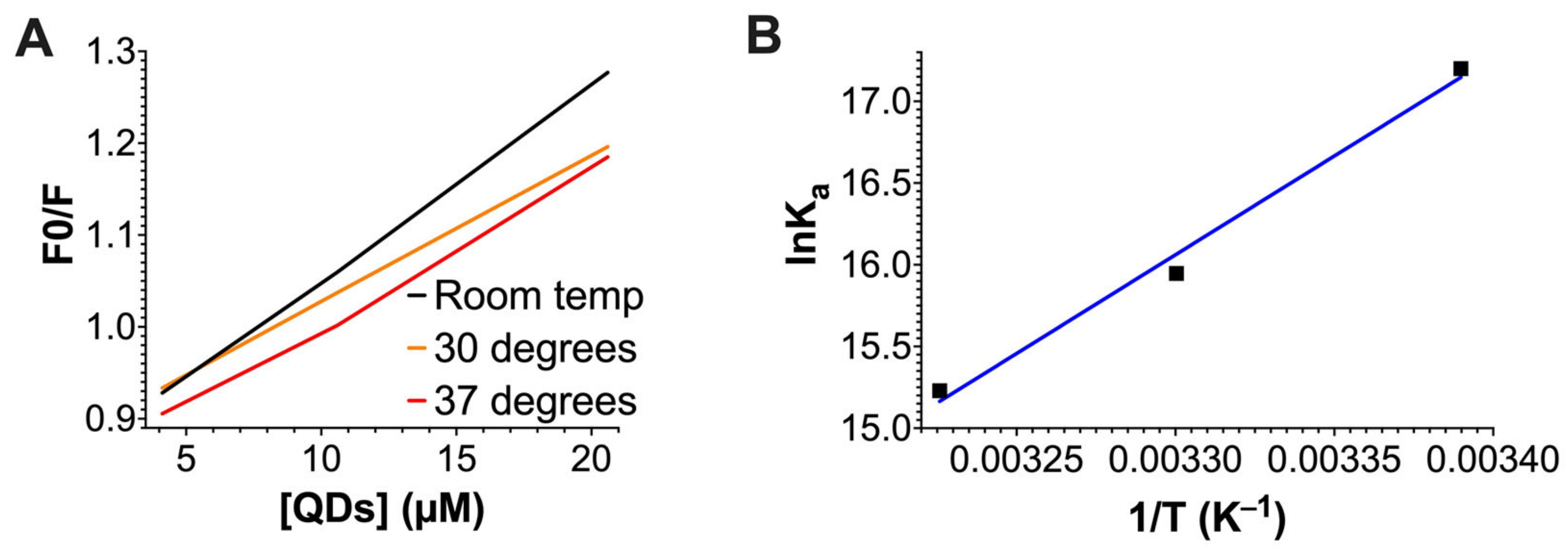
| T (K) | Ksv (L mol−1) | R2 | Ka (L/mol−1) | R2 | ∆G (kJ mol−1) | ∆H (kJ mol−1) | ∆S (J mol−1 K−1) |
|---|---|---|---|---|---|---|---|
| 310 | 8.98 × 106 | 0.969 | 4.11 × 106 ± 3.91× 105 | 0.826 | −39.25 ± 0.22 | −18.69 ± 4.5 | 65.99 ± 14.5 |
| 303 | 9.52 × 106 | 0.986 | 4.33 × 106 ± 1.14 × 106 | 0.872 | −38.49 ± 0.46 | ||
| 295 | 1.34 × 107 | 0.980 | 5.90 × 106 ± 1.85 × 106 | 0.808 | −38.24 ± 0.88 | ||
| Percent Change | ||||
|---|---|---|---|---|
| Wavelength | 10 nM | 25 nM | 50 nM | 100 nM |
| 222 nm | 34.1 ± 30.8 | 56.1 ± 17.2 | 47.7 ± 19.9 | 69.4 ± 1.5 |
| 208 nm | 30.4 ± 32.3 | 53.8 ± 13.8 | 54.3 ± 15.2 | 88.8 ± 2.0 |
| 218 nm | 32.9 ± 30.9 | 55.0 ± 17.7 | 47.3 ± 19.9 | 71.3 ± 1.9 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Le, N.; Chand, A.; Braun, E.; Keyes, C.; Wu, Q.; Kim, K. Interactions between Quantum Dots and G-Actin. Int. J. Mol. Sci. 2023, 24, 14760. https://doi.org/10.3390/ijms241914760
Le N, Chand A, Braun E, Keyes C, Wu Q, Kim K. Interactions between Quantum Dots and G-Actin. International Journal of Molecular Sciences. 2023; 24(19):14760. https://doi.org/10.3390/ijms241914760
Chicago/Turabian StyleLe, Nhi, Abhishu Chand, Emma Braun, Chloe Keyes, Qihua Wu, and Kyoungtae Kim. 2023. "Interactions between Quantum Dots and G-Actin" International Journal of Molecular Sciences 24, no. 19: 14760. https://doi.org/10.3390/ijms241914760
APA StyleLe, N., Chand, A., Braun, E., Keyes, C., Wu, Q., & Kim, K. (2023). Interactions between Quantum Dots and G-Actin. International Journal of Molecular Sciences, 24(19), 14760. https://doi.org/10.3390/ijms241914760






