Derivation of Arbas Cashmere Goat Induced Pluripotent Stem Cells in LCDM with Trophectoderm Lineage Differentiation and Interspecies Chimeric Abilities
Abstract
:1. Introduction
2. Results
2.1. LCDM Supports the Generation of giPSCs through Somatic Cell Reprogramming
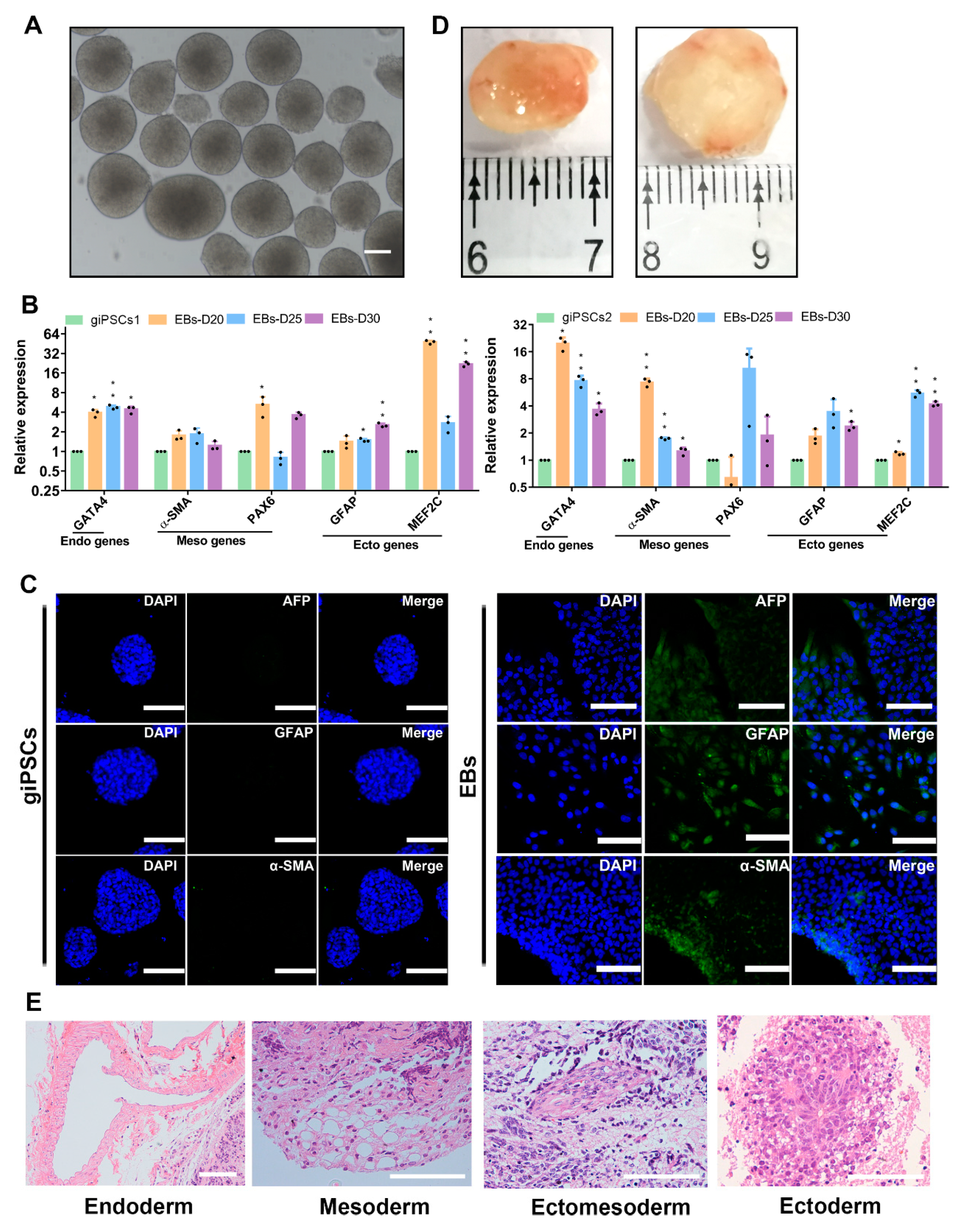
2.2. giPSCs Have the Potential to Differentiate into the TE Lineages
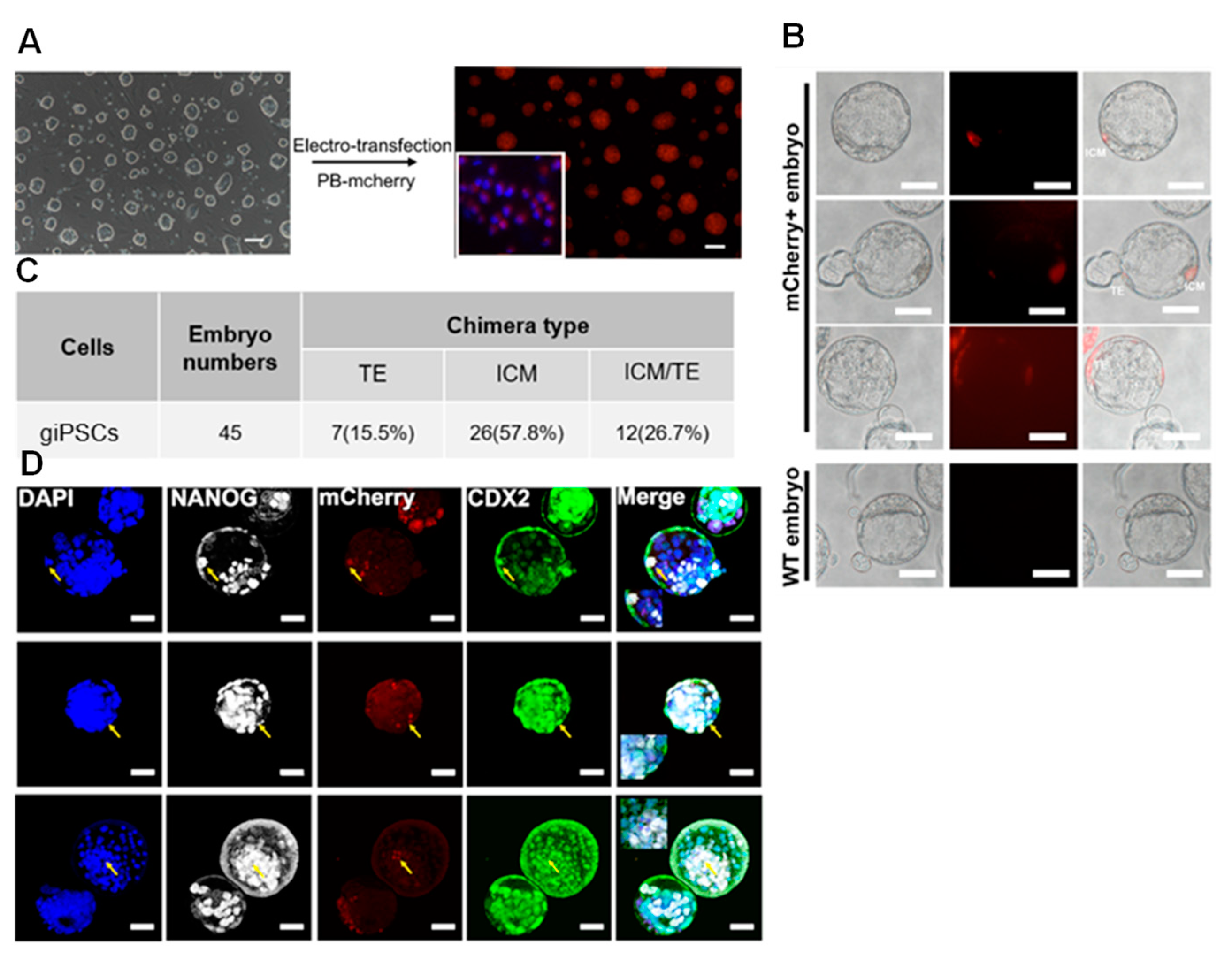
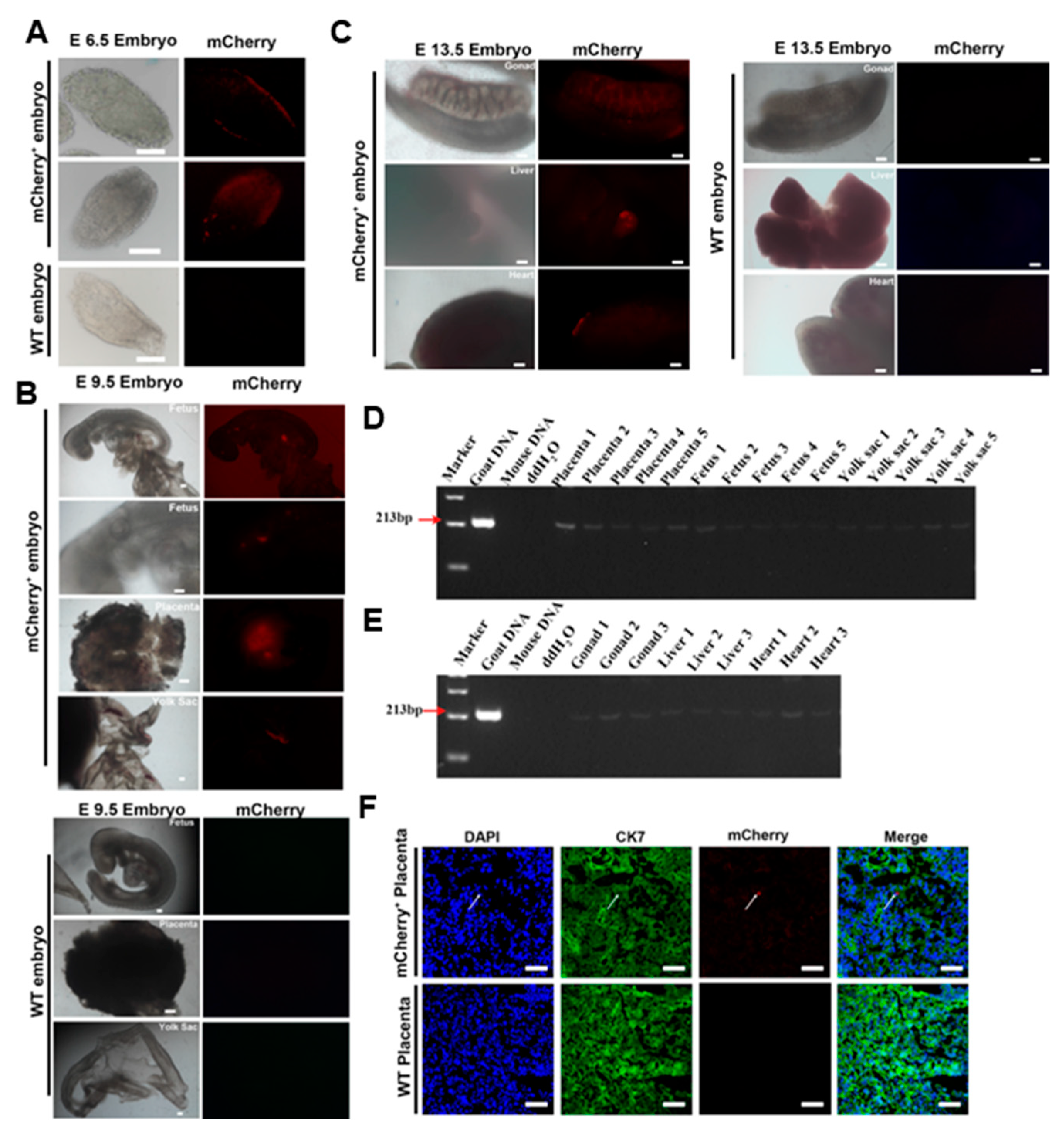
2.3. giPSCs Contribute to Embryonic and Extraembryonic Tissue in Chimeric Embryos
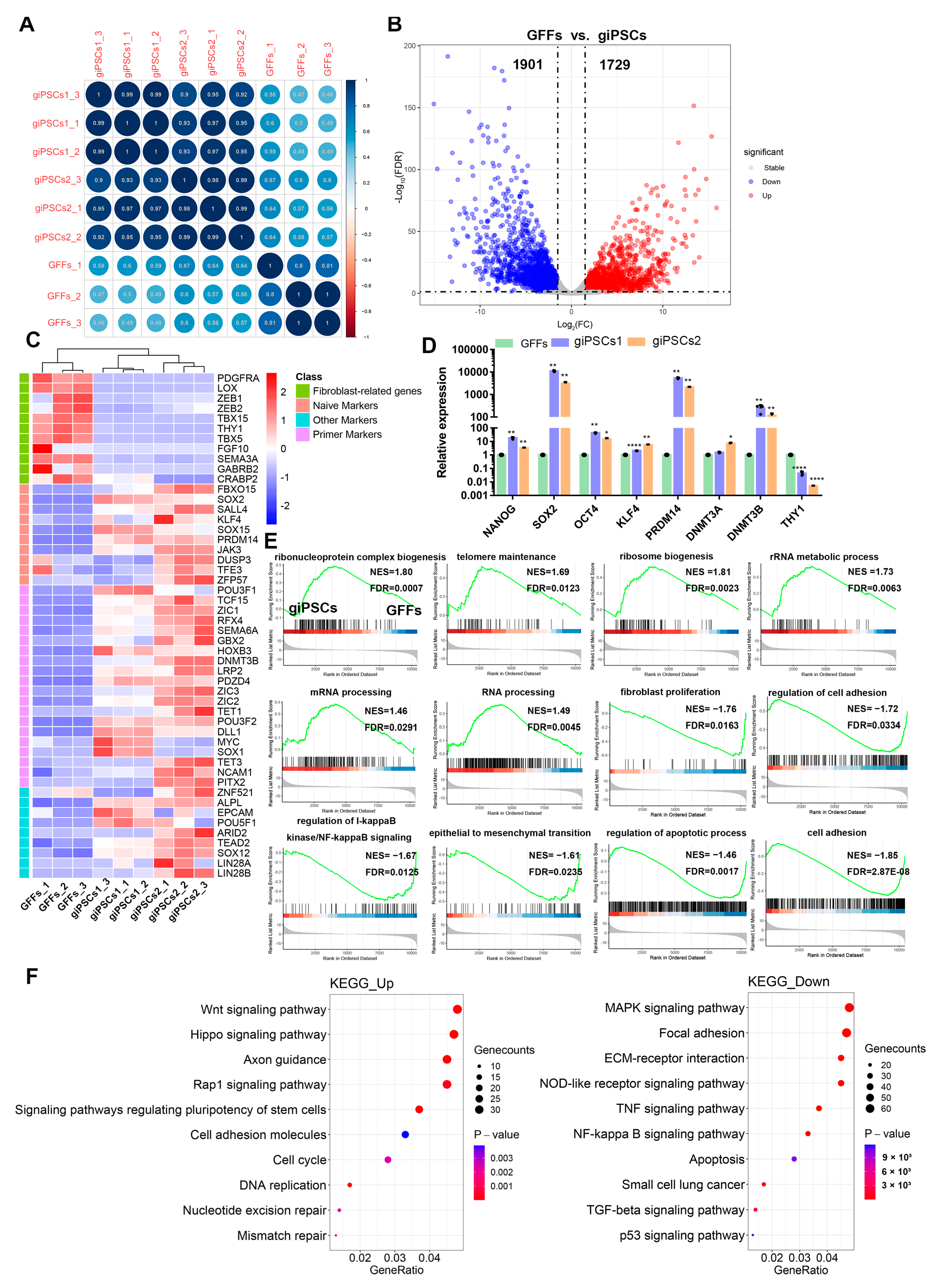
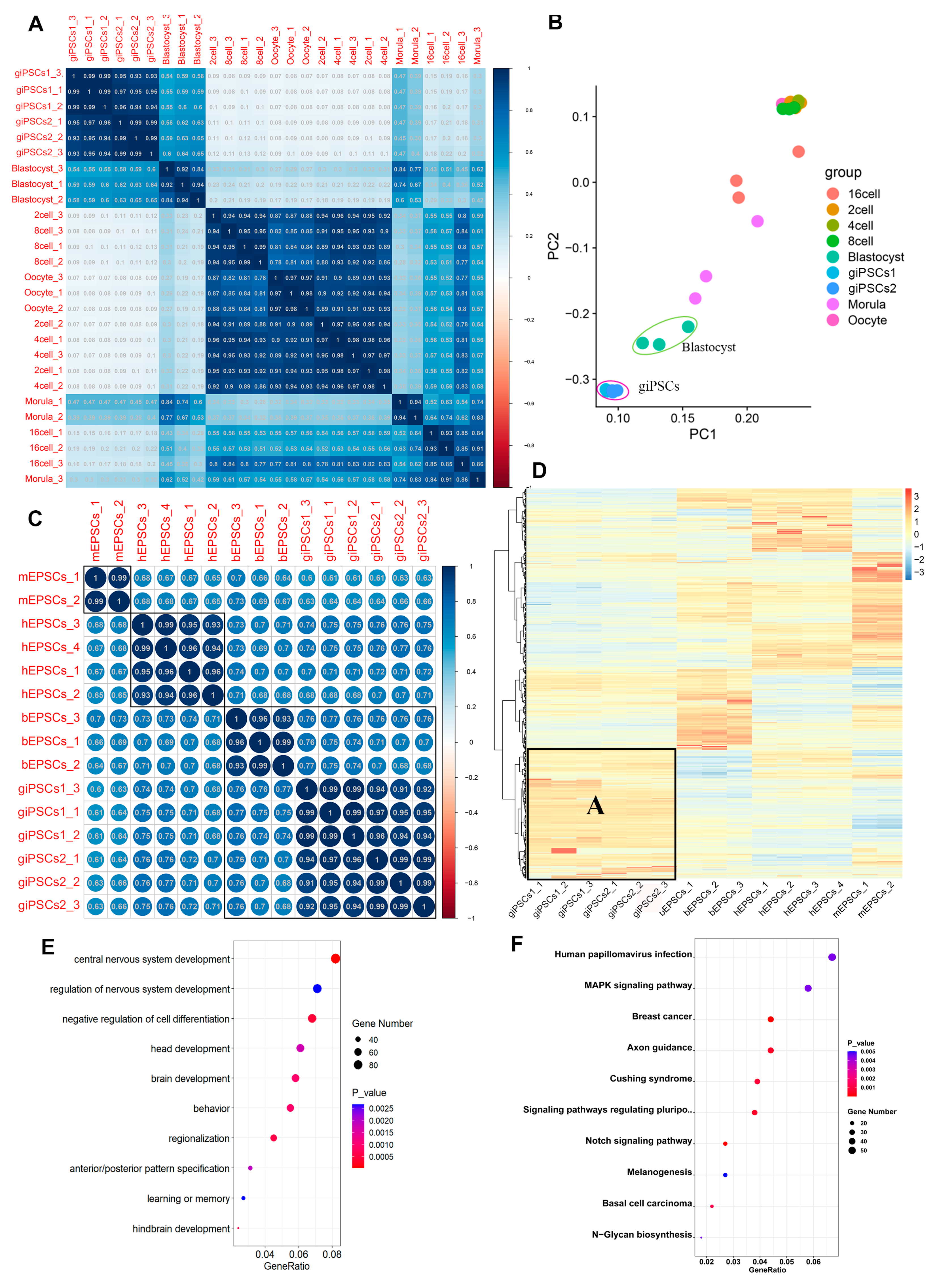
2.4. giPSCs Resemble Goat Blastocysts and Differ from GFFs and EPSCs of Other Species
3. Discussion
4. Materials and Methods
4.1. Animal Experiments
4.2. Generation of giPSCs through the Reprogramming of Somatic Cells
4.3. AP Staining
4.4. Karyotype Analysis
4.5. EBs Formation and In Vitro Differentiation
4.6. Immunofluorescence Staining
4.7. RT-PCR
4.8. Detecting Copy Number of Transgenes in giPSCs by Absolute Real-Time Quantitative PCR
4.9. Teratoma Formation
4.10. Differentiation of giPSCs into the TE Lineage
4.11. Western Blotting
4.12. Collection and In Vitro Culture of Mouse Embryos
4.13. Microinjection of giPSCs into 4- to 8-Cell Embryos and Early Blastocysts and Detection of the Developmental Fate of giPSCs
4.14. Flow Cytometry
4.15. Cell Fusion
4.16. RNA-seq and Analysis
4.17. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Proffen, B.L.; McElfresh, M.; Fleming, B.C.; Murray, M.M. A comparative anatomical study of the human knee and six animal species. Knee 2012, 19, 493–499. [Google Scholar] [CrossRef] [PubMed]
- Zhou, K.; Wu, G.; Li, Y.; Zhao, L.; Zhou, R.; Zhu, Q.; Huang, X.; Mu, D.; Hua, Y. Protective effects of indomethacin and dexamethasone in a goat model with intrauterine balloon aortic valvuloplasty. J. Biomed. Sci. 2012, 19, 74. [Google Scholar] [CrossRef] [PubMed]
- Martin, G.R. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc. Natl. Acad. Sci. USA 1981, 78, 7634–7638. [Google Scholar] [CrossRef] [PubMed]
- Thomson, J.A.; Itskovitz-Eldor, J.; Shapiro, S.S.; Waknitz, M.A.; Swiergiel, J.J.; Marshall, V.S.; Jones, J.M. Embryonic stem cell lines derived from human blastocysts. Science 1998, 282, 1145–1147. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef] [PubMed]
- Yue, L.; Pei, Y.; Zhong, L.; Yang, H.; Wang, Y.; Zhang, W.; Chen, N.; Zhu, Q.; Gao, J.; Zhi, M.; et al. Mthfd2 Modulates Mitochondrial Function and DNA Repair to Maintain the Pluripotency of Mouse Stem Cells. Stem Cell Rep. 2020, 15, 529–545. [Google Scholar] [CrossRef] [PubMed]
- Hao, F.; Yan, W.; Li, X.; Wang, H.; Wang, Y.; Hu, X.; Liu, X.; Liang, H.; Liu, D. Generation of Cashmere Goats Carrying an EDAR Gene Mutant Using CRISPR-Cas9-Mediated Genome Editing. Int. J. Biol. Sci. 2018, 14, 427–436. [Google Scholar] [CrossRef] [PubMed]
- Skrzyszowska, M.; Samiec, M. Generating Cloned Goats by Somatic Cell Nuclear Transfer-Molecular Determinants and Application to Transgenics and Biomedicine. Int. J. Mol. Sci. 2021, 22, 7490. [Google Scholar] [CrossRef]
- Kumar De, A.; Malakar, D.; Akshey, Y.S.; Jena, M.K.; Dutta, R. Isolation and characterization of embryonic stem cell-like cells from in vitro produced goat (Capra hircus) embryos. Anim. Biotechnol. 2011, 22, 181–196. [Google Scholar] [CrossRef]
- Ren, J.; Pak, Y.; He, L.; Qian, L.; Gu, Y.; Li, H.; Rao, L.; Liao, J.; Cui, C.; Xu, X.; et al. Generation of hircine-induced pluripotent stem cells by somatic cell reprogramming. Cell Res. 2011, 21, 849–853. [Google Scholar] [CrossRef] [PubMed]
- Wei, Q.; Xi, Q.; Liu, X.; Meng, K.; Zhao, X.; Ma, B. Characterization of goat inner cell mass derived cells in double kinase inhibition condition. Biochem. Biophys. Res. Commun. 2017, 483, 325–331. [Google Scholar] [CrossRef] [PubMed]
- Tai, D.; Liu, P.; Gao, J.; Jin, M.; Xu, T.; Zuo, Y.; Liang, H.; Liu, D. Generation of Arbas Cashmere Goat Induced Pluripotent Stem Cells Through Fibroblast Reprogramming. Cell. Reprogramming 2015, 17, 297–305. [Google Scholar] [CrossRef] [PubMed]
- Beddington, R.S.; Robertson, E.J. An assessment of the developmental potential of embryonic stem cells in the midgestation mouse embryo. Development 1989, 105, 733–737. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Ryan, D.J.; Wang, W.; Tsang, J.C.; Lan, G.; Masaki, H.; Gao, X.; Antunes, L.; Yu, Y.; Zhu, Z.; et al. Establishment of mouse expanded potential stem cells. Nature 2017, 550, 393–397. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Nowak-Imialek, M.; Chen, X.; Chen, D.; Herrmann, D.; Ruan, D.; Chen, A.C.H.; Eckersley-Maslin, M.A.; Ahmad, S.; Lee, Y.L.; et al. Establishment of porcine and human expanded potential stem cells. Nat. Cell Biol. 2019, 21, 687–699. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Liu, B.; Xu, J.; Wang, J.; Wu, J.; Shi, C.; Xu, Y.; Dong, J.; Wang, C.; Lai, W.; et al. Derivation of Pluripotent Stem Cells with In Vivo Embryonic and Extraembryonic Potency. Cell 2017, 169, 243–257.e25. [Google Scholar] [CrossRef] [PubMed]
- Bao, S.; Tang, W.W.; Wu, B.; Kim, S.; Li, J.; Li, L.; Kobayashi, T.; Lee, C.; Chen, Y.; Wei, M.; et al. Derivation of hypermethylated pluripotent embryonic stem cells with high potency. Cell Res. 2018, 28, 22–34. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Zhou, S.; Liu, M.; Chen, L.; Zhang, X.; Yang, X.; Cui, H.; Jiang, C.; Lv, Y.; Yan, T.; et al. Porcine Pluripotent Stem Cells Established from LCDM Medium with Characteristics Differ from Human and Mouse Extended Pluripotent Stem Cells. Stem Cells 2022, 40, 751–762. [Google Scholar] [CrossRef] [PubMed]
- Xiang, J.; Wang, H.; Zhang, Y.; Wang, J.; Liu, F.; Han, X.; Lu, Z.; Li, C.; Li, Z.; Gao, Y.; et al. LCDM medium supports the derivation of bovine extended pluripotent stem cells with embryonic and extraembryonic potency in bovine-mouse chimeras from iPSCs and bovine fetal fibroblasts. FEBS J. 2021, 288, 4394–4411. [Google Scholar] [CrossRef]
- Niwa, H.; Toyooka, Y.; Shimosato, D.; Strumpf, D.; Takahashi, K.; Yagi, R.; Rossant, J. Interaction between Oct3/4 and Cdx2 determines trophectoderm differentiation. Cell 2005, 123, 917–929. [Google Scholar] [CrossRef] [PubMed]
- Todd, C.D.; Deniz, Ö.; Taylor, D.; Branco, M.R. Functional evaluation of transposable elements as enhancers in mouse embryonic and trophoblast stem cells. eLife 2019, 8, e44344. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Sun, J.; Ling, Y.; Ming, H.; Chen, Z.; Fang, F.; Liu, Y.; Cao, H.; Ding, J.; Cao, Z.; et al. Transcription profiles of oocytes during maturation and embryos during preimplantation development in vivo in the goat. Reprod. Fertil. Dev. 2020, 32, 714–725. [Google Scholar] [CrossRef]
- Zhao, L.; Gao, X.; Zheng, Y.; Wang, Z.; Zhao, G.; Ren, J.; Zhang, J.; Wu, J.; Wu, B.; Chen, Y.; et al. Establishment of bovine expanded potential stem cells. Proc. Natl. Acad. Sci. USA 2021, 118, e2018505118. [Google Scholar] [CrossRef] [PubMed]
- Boyer, L.A.; Lee, T.I.; Cole, M.F.; Johnstone, S.E.; Levine, S.S.; Zucker, J.R.; Guenther, M.G.; Kumar, R.M.; Murray, H.L.; Jenner, R.G.; et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 2005, 122, 947–956. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Dey, S.K. Roadmap to embryo implantation: Clues from mouse models. Nat. Rev. Genet. 2006, 7, 185–199. [Google Scholar] [CrossRef]
- Lee, M.T.; Bonneau, A.R.; Giraldez, A.J. Zygotic genome activation during the maternal-to-zygotic transition. Annu. Rev. Cell Dev. Biol. 2014, 30, 581–613. [Google Scholar] [CrossRef]
- Kues, W.A.; Sudheer, S.; Herrmann, D.; Carnwath, J.W.; Havlicek, V.; Besenfelder, U.; Lehrach, H.; Adjaye, J.; Niemann, H. Genome-wide expression profiling reveals distinct clusters of transcriptional regulation during bovine preimplantation development in vivo. Proc. Natl. Acad. Sci. USA 2008, 105, 19768–19773. [Google Scholar] [CrossRef] [PubMed]
- Kuckenberg, P.; Buhl, S.; Woynecki, T.; van Fürden, B.; Tolkunova, E.; Seiffe, F.; Moser, M.; Tomilin, A.; Winterhager, E.; Schorle, H. The transcription factor TCFAP2C/AP-2gamma cooperates with CDX2 to maintain trophectoderm formation. Mol. Cell Biol. 2010, 30, 3310–3320. [Google Scholar] [CrossRef]
- Benchetrit, H.; Jaber, M.; Zayat, V.; Sebban, S.; Pushett, A.; Makedonski, K.; Zakheim, Z.; Radwan, A.; Maoz, N.; Lasry, R.; et al. Direct Induction of the Three Pre-implantation Blastocyst Cell Types from Fibroblasts. Cell Stem Cell 2019, 24, 983–994.e7. [Google Scholar] [CrossRef]
- De Los Angeles, A.; Ferrari, F.; Xi, R.; Fujiwara, Y.; Benvenisty, N.; Deng, H.; Hochedlinger, K.; Jaenisch, R.; Lee, S.; Leitch, H.G.; et al. Hallmarks of pluripotency. Nature 2015, 525, 469–478. [Google Scholar] [CrossRef]
- Zheng, C.; Hu, Y.; Sakurai, M.; Pinzon-Arteaga, C.A.; Li, J.; Wei, Y.; Okamura, D.; Ravaux, B.; Barlow, H.R.; Yu, L.; et al. Cell competition constitutes a barrier for interspecies chimerism. Nature 2021, 592, 272–276. [Google Scholar] [CrossRef]
- Wu, J.; Platero-Luengo, A.; Sakurai, M.; Sugawara, A.; Gil, M.A.; Yamauchi, T.; Suzuki, K.; Bogliotti, Y.S.; Cuello, C.; Morales Valencia, M.; et al. Interspecies Chimerism with Mammalian Pluripotent Stem Cells. Cell 2017, 168, 473–486.e15. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Greely, H.T.; Jaenisch, R.; Nakauchi, H.; Rossant, J.; Belmonte, J.C. Stem cells and interspecies chimaeras. Nature 2016, 540, 51–59. [Google Scholar] [CrossRef] [PubMed]
- De Los Angeles, A.; Wu, J. New concepts for generating interspecies chimeras using human pluripotent stem cells. Protein Cell 2022, 13, 234–238. [Google Scholar] [CrossRef] [PubMed]
- Shen, H.; Yang, M.; Li, S.; Zhang, J.; Peng, B.; Wang, C.; Chang, Z.; Ong, J.; Du, P. Mouse totipotent stem cells captured and maintained through spliceosomal repression. Cell 2021, 184, 2843–2859.e20. [Google Scholar] [CrossRef]
- Posfai, E.; Schell, J.P.; Janiszewski, A.; Rovic, I.; Murray, A.; Bradshaw, B.; Yamakawa, T.; Pardon, T.; El Bakkali, M.; Talon, I.; et al. Evaluating totipotency using criteria of increasing stringency. Nat. Cell Biol. 2021, 23, 49–60. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Chen, S.; Xu, Y.; Lyu, Y.; Wang, J.; Du, Y.; Sun, Y.; Liu, H.; Zhou, H.; Lai, W.; et al. Chemically defined and xeno-free culture condition for human extended pluripotent stem cells. Nat. Commun. 2021, 12, 3017. [Google Scholar] [CrossRef] [PubMed]
- Hanna, M.; Sahito, R.G.A.; Rateb, M.; Kachiwal, A.B.; Seddiek, H.A.; Bhutto, B.; Hescheler, J. Generation of transgene-free induced pluripotent stem cells from cardiac fibroblasts of goat embryos. J. Stem Cells Regen. Med. 2020, 16, 34–43. [Google Scholar]
- Zhao, L.; Wang, Z.; Zhang, J.; Yang, J.; Gao, X.; Wu, B.; Zhao, G.; Bao, S.; Hu, S.; Liu, P.; et al. Characterization of the single-cell derived bovine induced pluripotent stem cells. Tissue Cell 2017, 49, 521–527. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Lin, J.; Yu, Q.H.; Hu, W.W.; Yang, Q. Copy number and integration sites in growth hormone transgenic goats. Genet. Mol. Res. 2015, 14, 2006–2014. [Google Scholar] [CrossRef] [PubMed]
- Bou, G.; Guo, J.; Fang, Y.; Li, X.; Wei, R.; Li, Y.; Liu, Z. Interspecies cell fusion between mouse embryonic stem cell and porcine pluripotent cell. Reprod. Domest. Anim. 2021, 56, 1095–1103. [Google Scholar] [CrossRef]
- Prado, A.; Partearroyo, M.A.; Mencía, M.; Goñi, F.M.; Barberá-Guillem, E. Surfactant enhancement of polyethyleneglycol-induced cell fusion. FEBS Lett. 1989, 259, 149–152. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, W.; Chang, L.; Han, Y.; Sun, L.; Gong, X.; Tang, H.; Liu, Z.; Deng, H.; Ye, Y.; et al. Vitamin C alleviates aging defects in a stem cell model for Werner syndrome. Protein Cell 2016, 7, 478–488. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. Omics A J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Suomi, T.; Seyednasrollah, F.; Jaakkola, M.K.; Faux, T.; Elo, L.L. ROTS: An R package for reproducibility-optimized statistical testing. PLoS Comput. Biol. 2017, 13, e1005562. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, F.; Wang, J.; Yue, Y.; Li, C.; Zhang, X.; Xiang, J.; Wang, H.; Li, X. Derivation of Arbas Cashmere Goat Induced Pluripotent Stem Cells in LCDM with Trophectoderm Lineage Differentiation and Interspecies Chimeric Abilities. Int. J. Mol. Sci. 2023, 24, 14728. https://doi.org/10.3390/ijms241914728
Liu F, Wang J, Yue Y, Li C, Zhang X, Xiang J, Wang H, Li X. Derivation of Arbas Cashmere Goat Induced Pluripotent Stem Cells in LCDM with Trophectoderm Lineage Differentiation and Interspecies Chimeric Abilities. International Journal of Molecular Sciences. 2023; 24(19):14728. https://doi.org/10.3390/ijms241914728
Chicago/Turabian StyleLiu, Fang, Jing Wang, Yongli Yue, Chen Li, Xuemin Zhang, Jinzhu Xiang, Hanning Wang, and Xueling Li. 2023. "Derivation of Arbas Cashmere Goat Induced Pluripotent Stem Cells in LCDM with Trophectoderm Lineage Differentiation and Interspecies Chimeric Abilities" International Journal of Molecular Sciences 24, no. 19: 14728. https://doi.org/10.3390/ijms241914728
APA StyleLiu, F., Wang, J., Yue, Y., Li, C., Zhang, X., Xiang, J., Wang, H., & Li, X. (2023). Derivation of Arbas Cashmere Goat Induced Pluripotent Stem Cells in LCDM with Trophectoderm Lineage Differentiation and Interspecies Chimeric Abilities. International Journal of Molecular Sciences, 24(19), 14728. https://doi.org/10.3390/ijms241914728






