Identification and Characterization of the BBX Gene Family in Bambusa pervariabilis × Dendrocalamopsis grandis and Their Potential Role under Adverse Environmental Stresses
Abstract
:1. Introduction
2. Results
2.1. Identification and Analysis of the BBX Gene Family
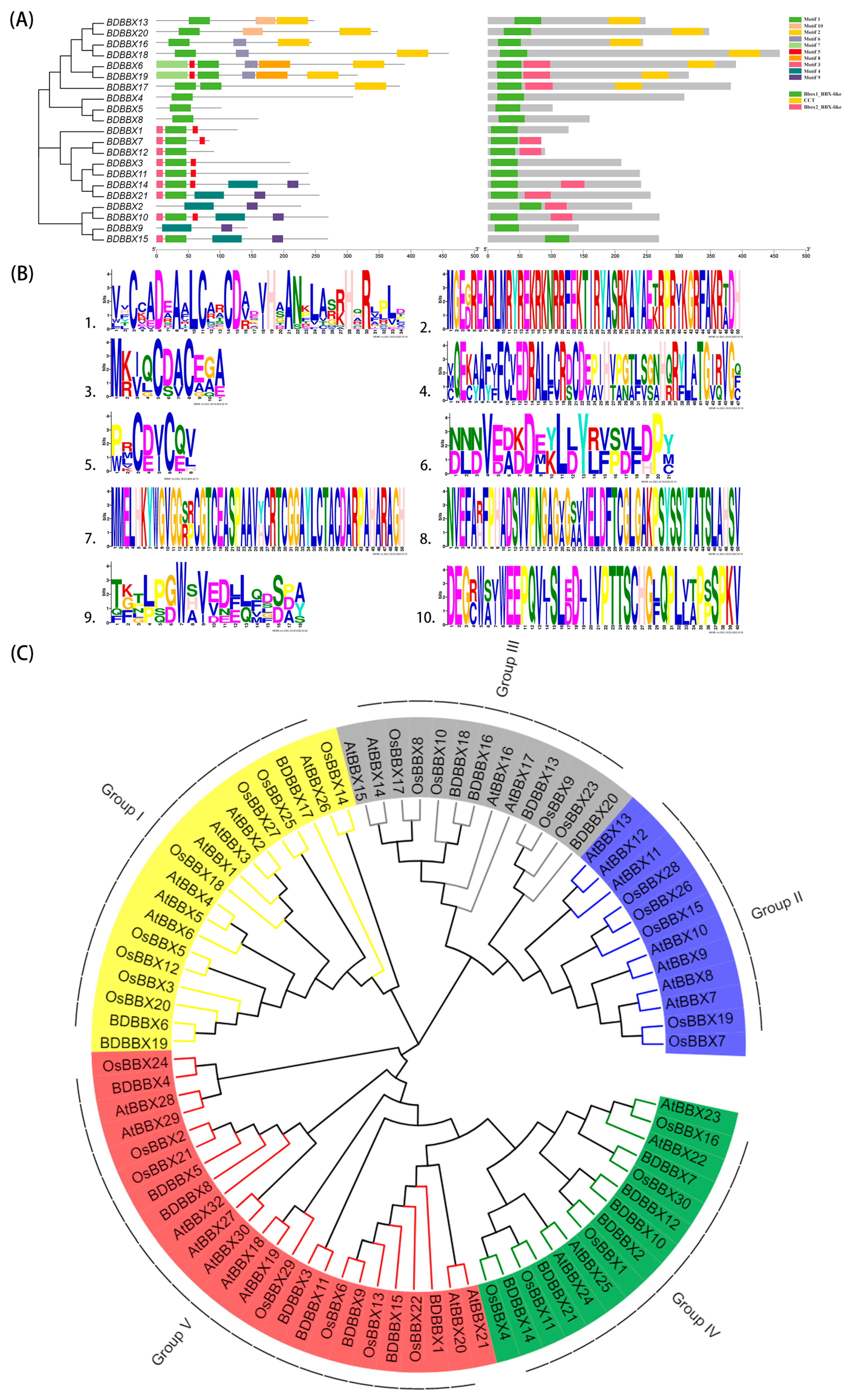
2.2. Conserved Motif Analysis of the BBX Gene Family
2.3. Phylogenetic Analysis of the BBX Gene Family
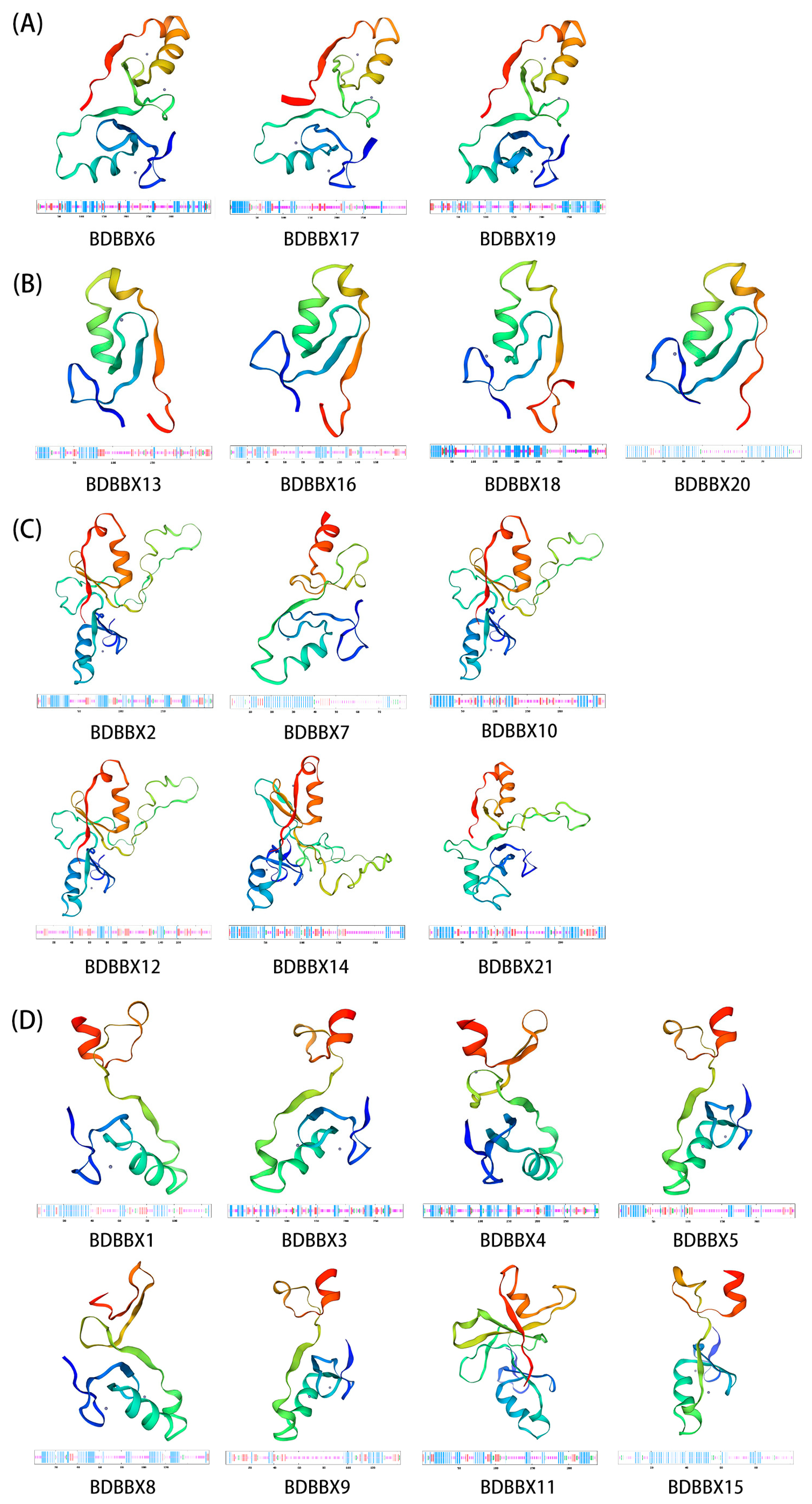
2.4. Structural Information Analysis of the BBX Gene Family
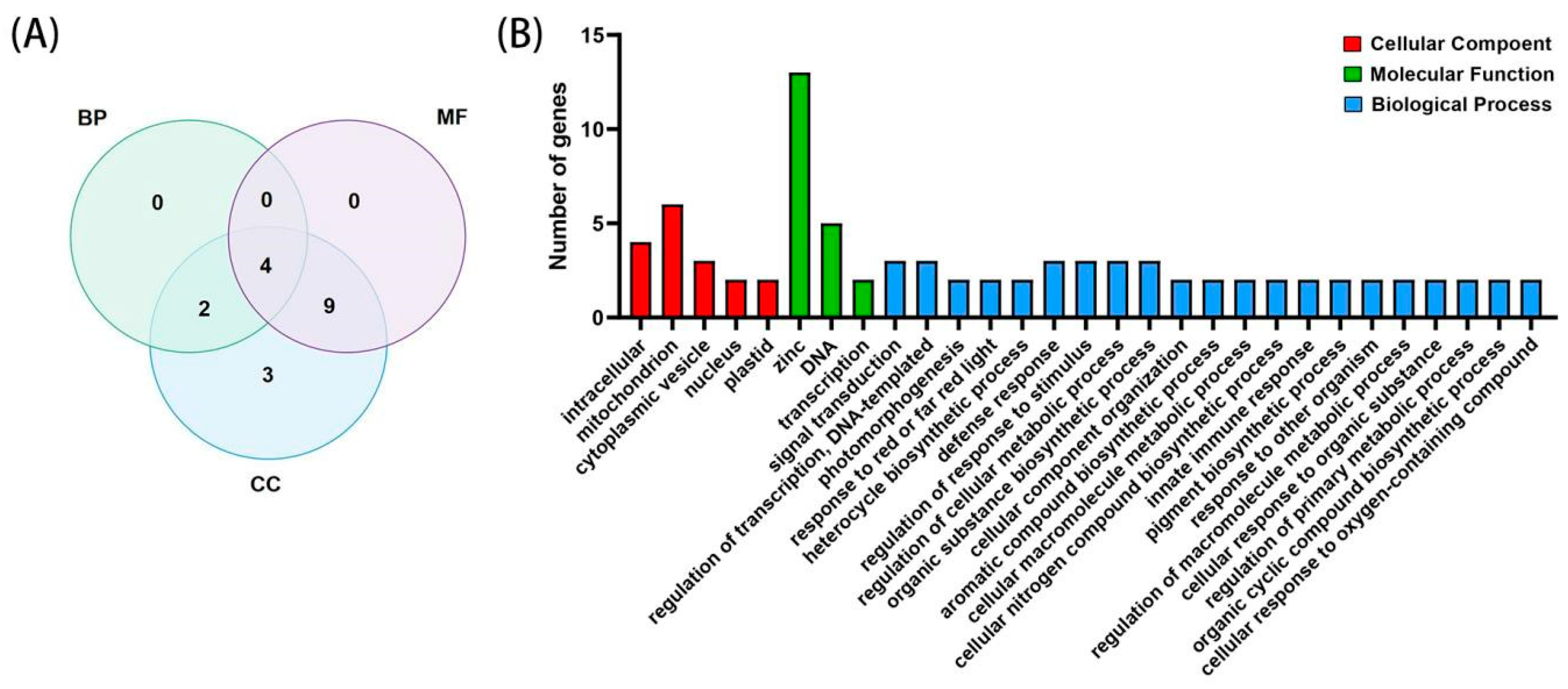
2.5. GO Analysis of the BBX Gene Family
2.6. Potential Phosphorylation Sites and Glycosylation Analysis of the BBX Gene Family
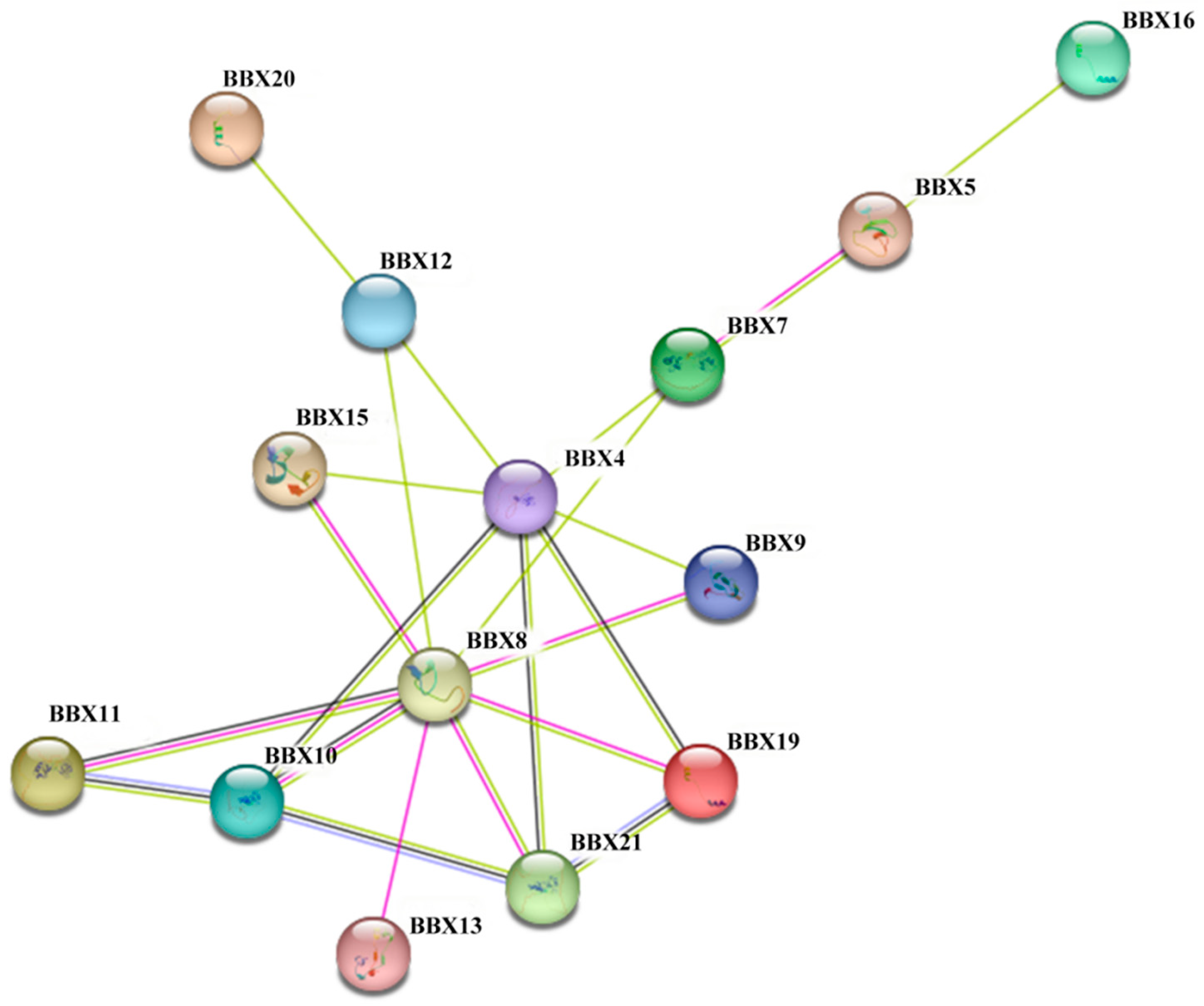
2.7. Protein–Protein Interaction Networks
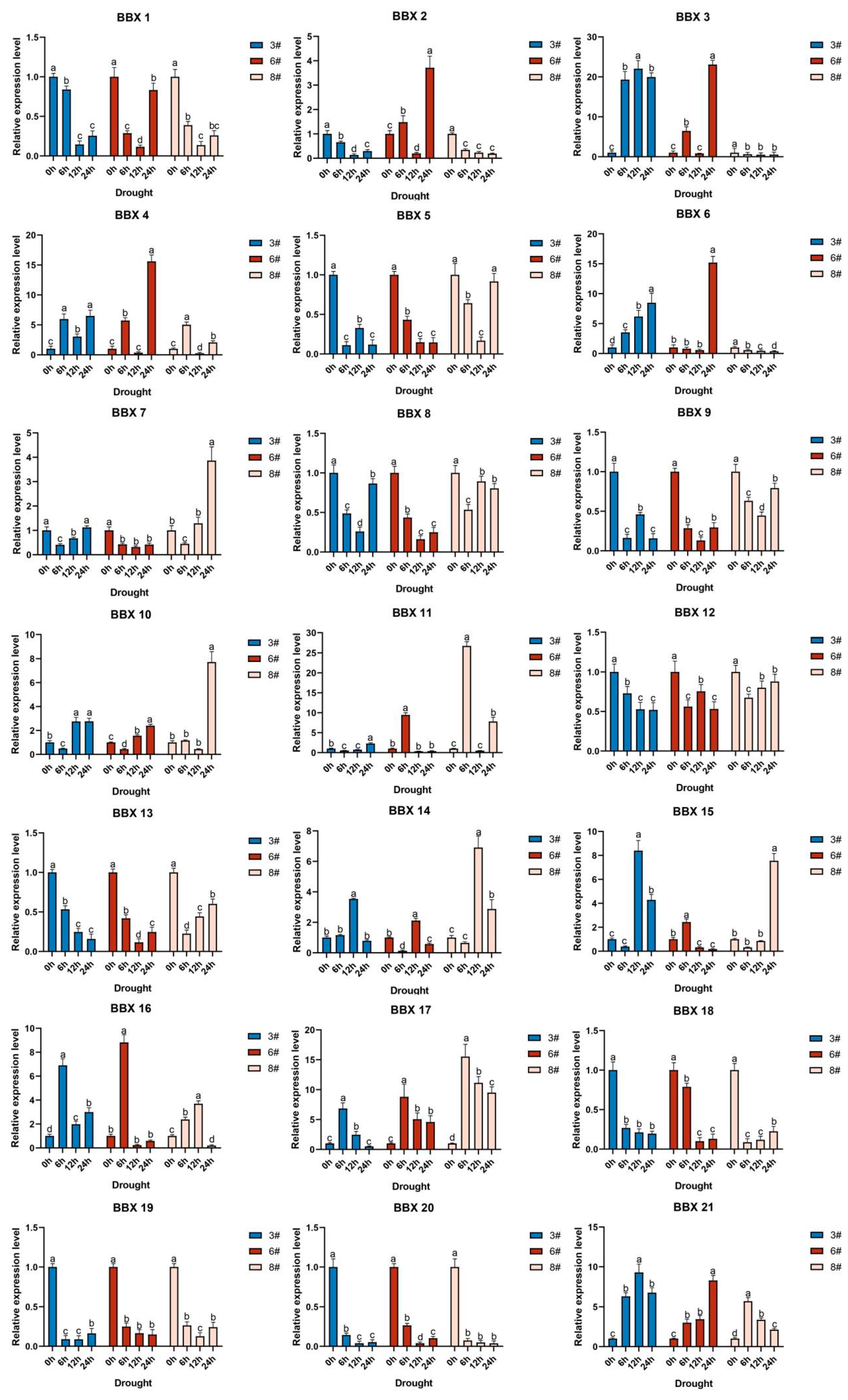
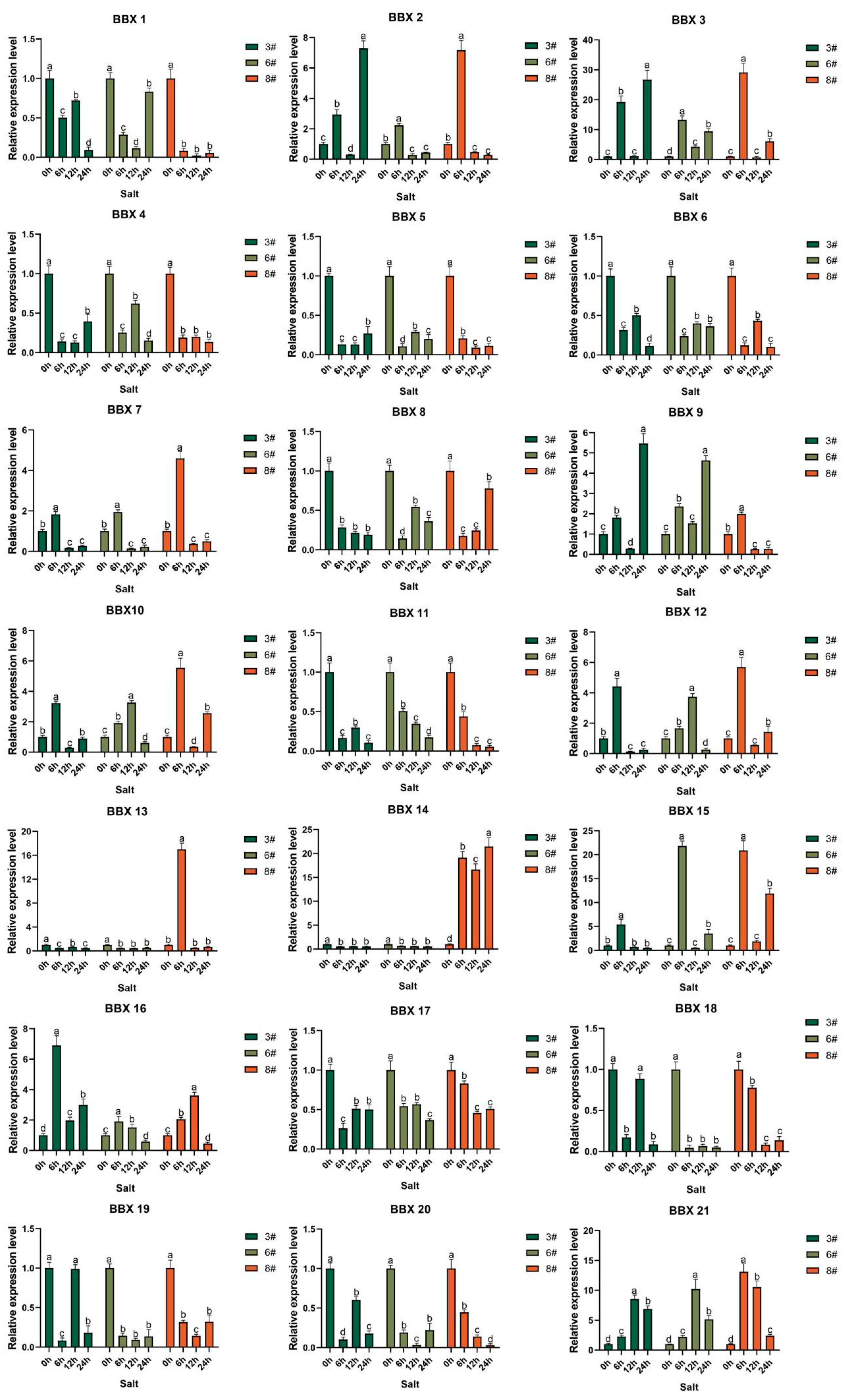
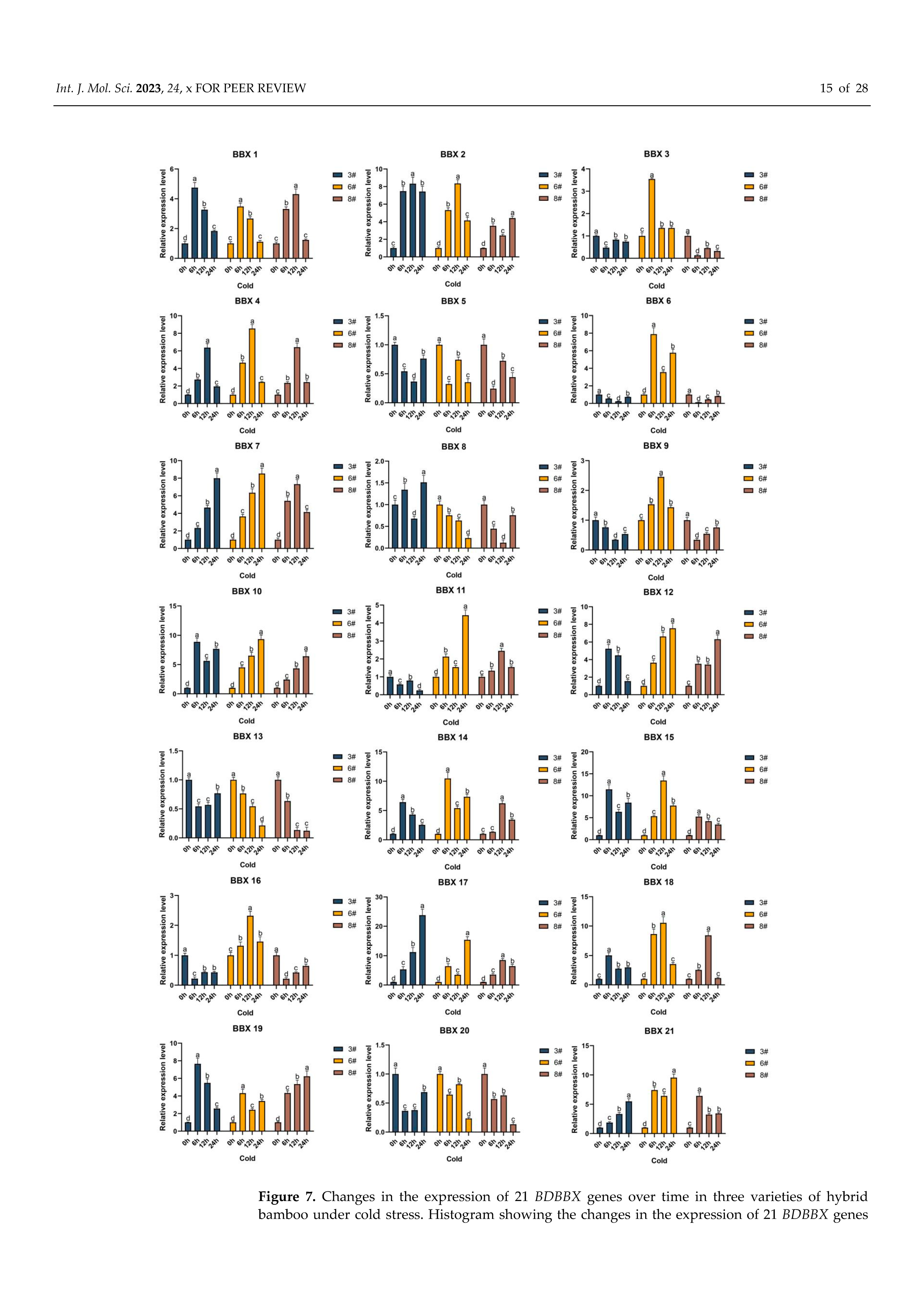
2.8. Inducible Expression Analysis of the BBX Gene in Bambusa pervariabilis × Dendrocalamopsis grandis under Abiotic Stress
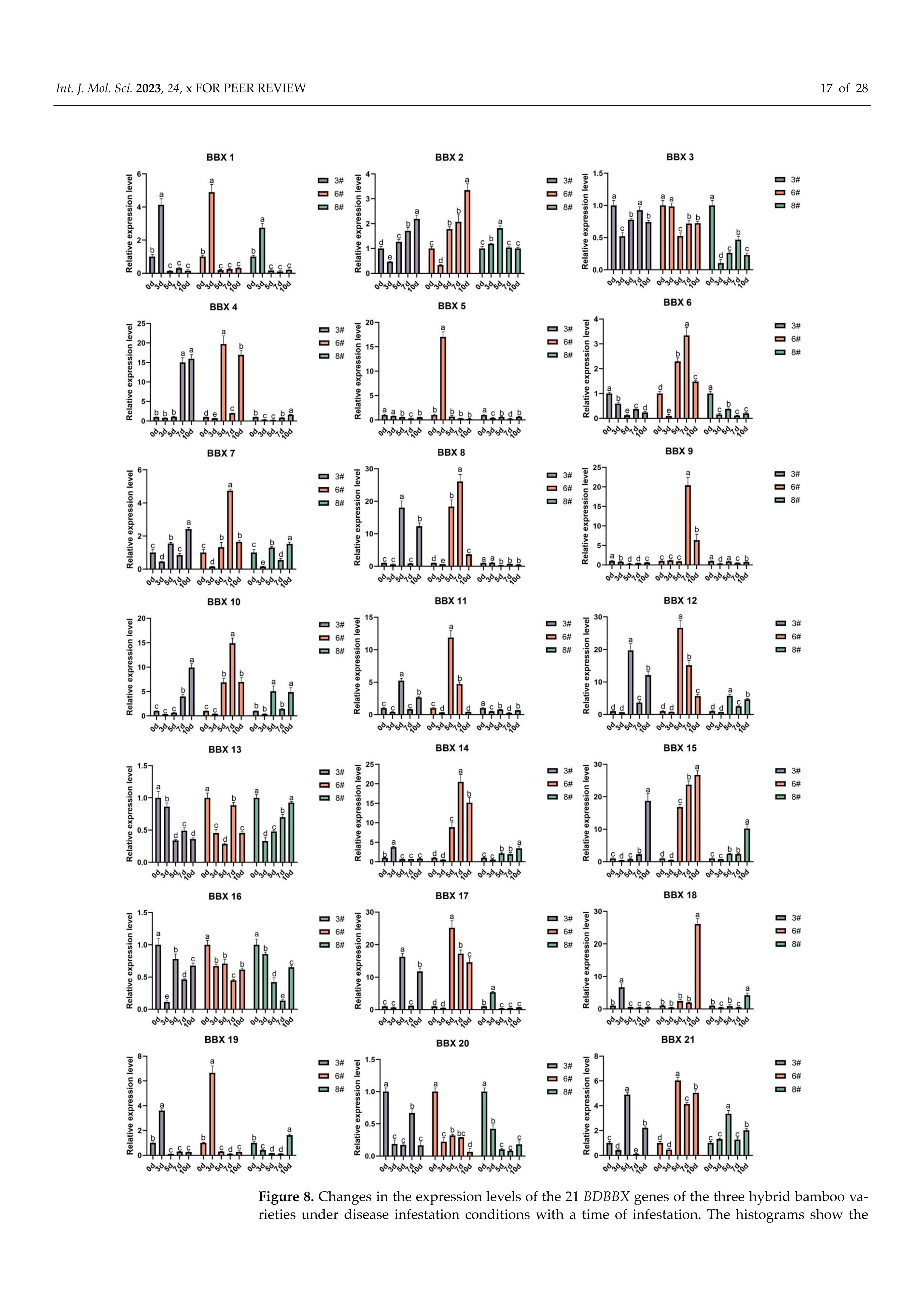
2.9. Induced Expression Analysis of the BBX Gene in Bambusa pervariabilis × Dendrocalamopsis grandis under Disease Infestation
2.10. Comparison of Amino Acid Sequences of BBX Proteins of Different Species
3. Discussion
4. Materials and Methods
4.1. Test Materials
4.2. BBX Gene Family Identification and Analysis
4.3. Analysis of the Basic Physicochemical Properties and Conserved Motifs of the BBX Gene Family
4.4. Phylogenetic Relationship Analysis of the BBX Gene Family
4.5. Structural Information Analysis of the BBX Gene Family
4.6. GO Analysis of the BBX Gene Family
4.7. Potential Phosphorylation Sites and Glycosylation Analysis of BBX Gene Family
4.8. BDBBX Protein Interaction Network Prediction and Functional Annotation
4.9. Treatment of Biotic and Abiotic Stress Conditions
disease rating)/(total plants × most serious disease rating)] × 100
4.10. Quantitative PCR Analysis
4.11. Comparison of Amino Acid Sequences of BBX Genes from Different Species
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Li, S.J.; Zhu, T.H.; Zhu, H.M.Y.; Liang, M.; Qiao, T.M.; Han, S.; Che, G.N. Purification of protein AP-toxin from Arthrinium phaeospermum causing blight in Bambusa pervariabilis × Dendrocalamopsis grandis and its metabolic effects on four bamboo varieties. Phytopathology 2013, 103, 135–145. [Google Scholar] [CrossRef] [PubMed]
- Li, S.J.; Zhu, T.H. Purification of the toxin protein Pc from Arthrinium phaeospermum and its effect on the defence enzymes of Bambusa pervariabilis × Dendrocalamopsis grandis varieties. For. Pathol. 2014, 44, 96–106. [Google Scholar] [CrossRef]
- Li, S.; Fang, X.; Han, S.; Zhu, T.; Zhu, H. Differential proteome analysis of hybrid Bamboo (Bambusa pervariabilis × Dendrocalamopsis grandis) under fungal stress (Arthrinium phaeospermum). Sci. Rep. 2019, 9, 18681. [Google Scholar] [CrossRef]
- Peng, Q.; Fang, X.M.; Zong, X.Z.; He, Q.Q.; Zhu, T.H.; Han, S.; Li, S.J. Comparative transcriptome analysis of Bambusa pervariabilis × Dendrocalamopsis grandis against Arthrinium phaeospermum under protein AP-toxin induction. Gene 2020, 725, 144160. [Google Scholar] [CrossRef]
- Luo, F.; Fang, X.; Liu, H.; Zhu, T.; Han, S.; Peng, Q.; Li, S. Differential transcriptome analysis and identification of genes related to resistance to blight in three varieties of Bambusa pervariabilis × Dendrocalamopsis grandis. PeerJ 2021, 9, e12301. [Google Scholar] [CrossRef]
- Yan, P.; Yu, J.; Fang, X.; Li, S.; Han, S.; Lin, T.; Liu, Y.; Yang, C.; He, F.; Zhu, T.; et al. Identification of the interacting proteins of Bambusa pervariabilis × Dendrocalamopsis grandis in response to the transcription factor ApCtf1β in Arthrinium phaeospermum. Front. Plant Sci. 2022, 13, 991077. [Google Scholar]
- Luo, F.; Yan, P.; Xie, L.; Li, S.; Zhu, T.; Han, S.; Lin, T.; Li, S. Molecular Mechanisms of Phenylpropane-Synthesis-Related Genes Regulating the Shoot Blight Resistance of Bambusa pervariabilis × Dendrocalamopsis grandis. Int. J. Mol. Sci. 2022, 23, 6760. [Google Scholar] [CrossRef]
- Riechmann, J.L.; Heard, G.; Martin, G.; Reuber, L.; Jiang, C.Z.; Keddie, J.; Adam, L.; Pineda, O.; Ratcliffe, O.J.; Samaha, R.R.; et al. Arabidopsis transcription factors: Genome-wide comparative analysis among eukaryotes. Science 2000, 290, 2105–2110. [Google Scholar] [CrossRef]
- Robson, F.; Costa, M.M.; Hepworth, S.R.; Vizir, I.; Pineiro, M.; Reeves, P.H.; Putterill, J.; Coupland, G. Functional importance of conserved domains in the flowering-time gene CONSTANS demonstrated by analysis of mutant alleles and transgenic plants. Plant J. 2001, 28, 619–631. [Google Scholar] [CrossRef]
- Khanna, R.; Kronmiller, B.; Maszle, D.R. The Arabidopsis B-Box Zinc Finger Family. Plant Cell 2009, 21, 3416–3420. [Google Scholar] [CrossRef]
- Klug, A.; Schwabe, J.W. Protein motifs 5. Zinc fingers. FASEB J. 1995, 9, 597–604. [Google Scholar] [PubMed]
- Zobell, O.; Coupland, G.; Reiss, B. The family of CONSTANS-like genes in Physcomitrella patens. Plant Biol. 2005, 7, 266–275. [Google Scholar] [PubMed]
- Crocco, C.D.; Botto, J.F. BBX proteins in green plants: Insights into their evolution, structure feature and functional diversification. Gene 2013, 531, 44–52. [Google Scholar] [PubMed]
- Chu, Z.; Wang, X.; Li, Y.; Yu, H.; Li, J.; Lu, Y.; Li, H.; Ouyang, B. Genomic Organization, Phylogenetic and Expression Analysis of the B-BOX Gene Family in Tomato. Front. Plant Sci. 2016, 7, 1552. [Google Scholar]
- Liu, X.; Dai, Y.Q.; Li, R.; Yuan, L.; Chen, X.; Wang, X. Members of B-box Protein Family from Malus domestica Enhanced Abiotic Stresses Tolerance in Escherichia coli. Mol. Biotechnol. 2019, 61, 421–426. [Google Scholar]
- Putterill, J.; Robson, F.; Lee, K.; Simon, R.; Coupland, G. The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 1995, 80, 847–857. [Google Scholar]
- Datta, S.; Hettiarachchi, C.; Johansson, H.; Holm, M. SALT TOLERANCE HOMOLOG2, a B-box protein in Arabidopsis that activates transcription and positively regulates light-mediated development. Plant Cell 2007, 19, 3242–3255. [Google Scholar]
- Gangappa, S.N.; Botto, J.F. The BBX family of plant transcription factors. Trends Plant Sci. 2014, 19, 460–470. [Google Scholar]
- Gangappa, S.N.; Botto, J.F. The multifaceted roles of HY5 in plant growth and development. Mol. Plant 2016, 9, 1353–1365. [Google Scholar]
- Cao, S.; Kumimoto, R.W.; Gnesutta, N.; Calogero, A.M.; Mantovani, R.; Holt, B.F. A distal CCAAT/NUCLEAR FACTOR Y complex promotes chromatin looping at the FLOWERING LOCUS T promoter and regulates the timing of flowering in Arabidopsis. Plant Cell 2014, 26, 1009–1017. [Google Scholar]
- Sun, Z.B.; Qi, X.Y.; Wang, Z.L.; Li, P.; Wu, C.; Zhang, H.; Zhao, Y. Overexpression of TsGOLS2, a galactinol synthase, in Arabidopsis thaliana enhances tolerance to high salinity and osmotic stresses. Plant Physiol. Biochem. 2013, 69, 82–89. [Google Scholar] [PubMed]
- Gruber, M.; Wu, L.M.; Links, M.; Gjetvaj, B.; Durkin, J.; Lewis, C.; Sharpe, A.; Lydiate, D.; Hegedus, D. Analysis of expressed sequence tags in Brassica napus cotyledons damaged by crucifer flea beetle feeding. Genome 2012, 55, 118–133. [Google Scholar] [PubMed]
- Liu, H.; Dong, S.Y.; Sun, D.Y.; Liu, W.; Gu, F.; Liu, Y.; Guo, T.; Wang, H.; Wang, J.; Chen, Z. CONSTANS-Like 9 (OsCOL9) interacts with receptor for activated C-Kinase 1 (OsRACK1) to regulate blast resistance through salicylic acid and ethylene signaling pathways. PLoS ONE 2016, 11, e166249. [Google Scholar]
- Vaishak, K.; Yadukrishnan, P.; Bakshi, S.; Kushwaha, A.K.; Ramachandran, H.; Job, N.; Babu, D.; Datta, S. The B-box bridge between light and hormones in plants. J. Photochem. Photobiol. B Biol. 2019, 191, 164–174. [Google Scholar]
- Torok, M.; Etkin, L.D. Two B or not two B? Overview of the rapidly expanding B-box family of proteins. Differentiation 2001, 67, 63–71. [Google Scholar] [PubMed]
- Huang, J.; Zhao, X.; Weng, X.; Wang, L.; Xie, W. The Rice B-Box Zinc Finger Gene Family: Genomic Identification, Characterization, Expression Profiling and Diurnal Analysis. PLoS ONE 2012, 7, e48242. [Google Scholar]
- Wen, S.Y.; Zhang, Y.; Deng, Y.; Chen, G.; Yu, Y.; Wei, Q. Genomic identification and expression analysis of the BBX transcription factor gene family in Petunia hybrida. Mol. Biol. Rep. 2020, 47, 6027–6041. [Google Scholar] [PubMed]
- Singh, S.; Chhapekar, S.S.; Ma, Y.; Rameneni, J.J.; Oh, S.H.; Kim, J.; Lim, Y.P.; Choi, S.R. Genome-Wide Identification, Evolution, and Comparative Analysis of B-Box Genes in Brassica rapa, B. oleracea, and B. napus and Their Expression Profiling in B. rapa in Response to Multiple Hormones and Abiotic Stresses. Int. J. Mol. Sci. 2021, 22, 10367. [Google Scholar]
- Ma, R.; Chen, J.; Huang, B.; Huang, Z.; Zhang, Z. The BBX gene family in Moso bamboo (Phyllostachys edulis): Identification, characterization and expression profiles. BMC Genom. 2021, 22, 533. [Google Scholar]
- Zhao, J.; Li, H.; Huang, J.; Shi, T.; Meng, Z.; Chen, Q.; Deng, J. Genome-wide analysis of BBX gene family in Tartary buckwheat (Fagopyrum tataricum). PeerJ 2021, 9, e11939. [Google Scholar]
- Li, W.L.; Wang, J.C.; Sun, Q.; Li, W.; Yu, Y.; Zhao, M.; Meng, Z. Expression analysis of genes encoding double B-box zinc finger proteins in maize. Funct. Integr. Genom. 2017, 17, 653–666. [Google Scholar]
- Cui, X.; Wang, Y.X.; Liu, Z.W.; Wang, W.-L.; Li, H.; Zhuang, J. Transcriptome-wide identification and expression profile analysis of the bHLH family genes in Camellia sinensis. Funct. Integr. Genom. 2018, 18, 489–503. [Google Scholar]
- Cao, Y.; Han, Y.; Meng, D.; Li, D.; Jiao, C.; Jin, Q.; Lin, Y.; Cai, Y. B-BOX genes: Genome-wide identification, evolution and their contribution to pollen growth in pear (Pyrus bretschneideri Rehd.). BMC Plant Biol. 2017, 17, 156. [Google Scholar]
- Talar, U.; Kiełbowicz-Matuk, A.; Czarnecka, J.; Rorat, T. Genome-wide survey of B-box proteins in potato (Solanum tuberosum)—Identification, characterization and expression patterns during diurnal cycle, etiolation and de-etiolation. PLoS ONE 2017, 12, e0177471. [Google Scholar]
- Shalmani, A.; Jing, X.Q.; Shi, Y.; Muhammad, I.; Zhou, M.R.; Wei, X.Y.; Chen, Q.Q.; Li, W.Q.; Liu, W.T.; Chen, K.M. Characterization of B-BOX gene family and their expression profiles under hormonal, abiotic and metal stresses in Poaceae plants. BMC Genom. 2019, 20, 27. [Google Scholar]
- Liu, T.; Yu, E.; Hou, L.; Hua, P.; Zhao, M.; Wang, Y.; Hu, J.; Zhang, M.; Wang, K.; Wang, Y. Transcriptome-Based Identification, Characterization, Evolutionary Analysis, and Expression Pattern Analysis of the WRKY Gene Family and Salt Stress Response in Panax ginseng. Horticulturae 2022, 8, 756. [Google Scholar]
- Magadum, S.; Banerjee, U.; Murugan, P.; Gangapur, D.; Ravikesavan, R. Gene duplication as a major force in evolution. J. Genet. 2013, 92, 155–161. [Google Scholar]
- Cannon, S.B.; Mitra, A.; Baumgarten, A.; Young, N.D.; May, G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004, 4, 10. [Google Scholar] [CrossRef]
- Pan, F.; Wang, Y.; Liu, H.; Wu, M.; Chu, W.; Chen, D.; Xiang, Y. Genome-wide identification and expression analysis of SBP-like transcription factor genes in Moso Bamboo (Phyllostachys edulis). BMC Genom. 2017, 18, 486. [Google Scholar]
- Gangappa, S.N.; Crocco, C.D.; Johansson, H.; Datta, S.; Hettiarachchi, C.; Holm, M.; Botto, J.F. The Arabidopsis B-BOX protein BBX25 interacts with HY5, negatively regulating BBX22 expression to suppress seedling photomorphogenesis. Plant Cell 2013, 25, 1243–1257. [Google Scholar]
- Anukriti; Dhasmana, A.; Uniyal, S.; Somvanshi, P.; Bhardwaj, U.; Gupta, M.; Haque, S.; Lohani, M.; Kumar, D.; Ruokolainen, J.; et al. Investigation of Precise Molecular Mechanistic Action of Tobacco-Associated Carcinogen ‘NNK Induced Carcinogenesis: A System Biology Approach. Genes 2019, 10, 564–585. [Google Scholar]
- Xu, D.Q.; Li, J.G.; Gangappa, S.N.; Hettiarachchi, C.; Lin, F.; Andersson, M.X.; Jiang, Y.; Deng, X.W.; Holm, M. Convergence of light and ABA signaling on the ABI5 promoter. PLoS Genet. 2014, 10, e1004197. [Google Scholar] [CrossRef]
- Qi, Q.; Gibson, A.; Fu, X.; Zheng, M.; Kuehn, R.; Wang, Y.; Wang, Y.; Navarro, S.; Morrell, J.A.; Jiang, D.; et al. Involvement of the N-terminal B-box domain of Arabidopsis BBX32 protein in interaction with soybean BBX62 protein. J. Biol. Chem. 2012, 287, 31482–31493. [Google Scholar] [CrossRef]
- Cramer, G.R.; Urano, K.; Delrot, S.; Pezzotti, M.; Shinozaki, K. Effects of abiotic stress on plants: A systems biology perspective. BMC Plant Biol. 2011, 11, 163. [Google Scholar] [CrossRef]
- Lippuner, V.; Cyert, M.S.; Gasser, C.S. Two classes of plant cDNA clones differentially complement yeast calcineurin mutants and increase salt tolerance of wild-type yeast. J. Biol. Chem. 1996, 271, 12859–12866. [Google Scholar] [CrossRef]
- Nagaoka, S.; Takano, T. Salt tolerance-related protein STO binds to a Myb transcription factor homologue and confers salt tolerance in Arabidopsis. J. Exp. Bot. 2003, 54, 2231–2237. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Li, R.; Dai, Y.Q.; Chen, X.; Wang, X. Genome-wide identification and expression analysis of the B-box gene family in the Apple (Malus domestica Borkh.) genome. Mol. Genet. Genom. 2018, 293, 303–315. [Google Scholar] [CrossRef]
- Yang, Y.; Ma, C.; Xu, Y.; Wei, Q.; Imtiaz, M.; Lan, H.; Gao, S.; Cheng, L.; Wang, M.; Fei, Z. A zinc finger protein regulates flowering time and abiotic stress tolerance in chrysanthemum by modulating gibberellin biosynthesis. Plant Cell 2014, 26, 2038–2054. [Google Scholar] [CrossRef]
- Yang, S.; Zhu, H.; Huang, L.; Zhang, G.; Wang, L.; Jiang, X.; Zhong, Q. Transcriptome-wide and expression analysis of the NAC gene family in pepino (Solanum muricatum) during water deficit stress. PeerJ 2021, 9, e10966. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using realtime quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]

| Transcriptome ID | Gene | Protein Length (AA) | Molecular Weight (Da) | Theoretical pI | Instability Index | Aliphatic Index | Grand Average of Hydropathicity (GRAVY) | Signal Peptide | Subcellular Localization |
|---|---|---|---|---|---|---|---|---|---|
| PH01000042G1610 | BDBBX1 | 127 | 13,945.96 | 10.26 | 70.44 | 60.08 | −0.521 | NO | Nucleus |
| PH01000149G0140 | BDBBX2 | 227 | 24,305.66 | 4.82 | 59.77 | 80.84 | −0.013 | NO | Nucleus |
| PH01000192G1360 | BDBBX3 | 210 | 22,874.85 | 11.57 | 63.94 | 90.29 | −0.337 | NO | Nucleus |
| PH01000303G0990 | BDBBX4 | 309 | 32,565.29 | 6.41 | 55.53 | 58.25 | −0.518 | NO | Chloroplast.Nucleus |
| PH01000450G1030 | BDBBX5 | 102 | 10,539.61 | 8.66 | 74.10 | 51.08 | −0.52 | NO | Chloroplast. Cytoplasm |
| PH01000481G0620 | BDBBX6 | 383 | 41,494.94 | 7.59 | 41.16 | 67.44 | −0.295 | NO | Cell membrane.Nucleus |
| PH01000616G0330 | BDBBX7 | 83 | 8412.52 | 6.25 | 56.72 | 71.93 | −0.014 | NO | Nucleus |
| PH01000917G0300 | BDBBX8 | 160 | 16,681.41 | 10.50 | 73.87 | 62.62 | −0.358 | NO | Nucleus |
| PH01001451G0380 | BDBBX9 | 143 | 14,723.24 | 4.69 | 61.26 | 67.69 | −0.113 | NO | Nucleus |
| PH01001725G0310 | BDBBX10 | 270 | 28,776.28 | 7.48 | 50.37 | 73.89 | −0.087 | NO | Nucleus |
| PH01001870G0430 | BDBBX11 | 239 | 26,213.84 | 11.48 | 63.61 | 86.65 | −0.431 | NO | Nucleus |
| PH01002961G0180 | BDBBX12 | 90 | 10,098.74 | 9.23 | 56.06 | 58.78 | −0.371 | NO | Nucleus |
| PH01003421G0140 | BDBBX13 | 198 | 21,263.84 | 5.30 | 52.63 | 64.70 | −0.366 | NO | Nucleus |
| Phyllostachys_edulis_newGene_30773 | BDBBX14 | 241 | 26,221.19 | 7.86 | 41.89 | 81.08 | −0.126 | NO | Nucleus |
| Phyllostachys_edulis_newGene_38997 | BDBBX15 | 269 | 29,499.68 | 6.45 | 60.16 | 82.75 | −0.099 | NO | Nucleus |
| PH01000037G0060 | BDBBX16 | 194 | 20,980.48 | 9.49 | 79.35 | 55.46 | −0.699 | NO | Nucleus |
| PH01000780G0510 | BDBBX17 | 332 | 36,625.69 | 11.90 | 93.79 | 41.08 | −0.932 | NO | Chloroplas. |
| PH01002146G0170 | BDBBX18 | 409 | 44,318.82 | 6.43 | 63.00 | 58.58 | −0.593 | NO | Nucleus |
| PH01002727G0080 | BDBBX19 | 316 | 34,063.68 | 6.08 | 37.30 | 74.56 | −0.199 | NO | Nucleus |
| Phyllostachys_edulis_newGene_62371 | BDBBX20 | 298 | 32,384.84 | 6.81 | 59.12 | 74.03 | −0.345 | NO | Nucleus |
| PH01003160G0610 | BDBBX21 | 256 | 27,100.62 | 4.96 | 50.25 | 76.33 | −0.155 | NO | Nucleus |
| Motif | Motif Length (AA) | Motif Sequence | Function Annotation |
|---|---|---|---|
| 1 | 34 | VVCCADEAALCARCDADVHAANKLASRHQRLPLD | zf-B_box |
| 2 | 50 | MGEGREARLMRYREKRKNRRFEKTIRYASRKAYAEKRPRIKGRFAKRADH | CCT |
| 3 | 11 | MKIQCDACEGA | UnKnown |
| 4 | 47 | VQEKAAFIFCVEDRALLCRDCDEPIHVPGTLSGNHQRYLATGIRVGF | zf-B_box |
| 5 | 8 | PRCDVCQV | UnKnown |
| 6 | 21 | DLDVEDDDEKLDYRFPDFDPY | UnKnown |
| 7 | 50 | MMELHKYWGVGGRRCGTCEASPAAVHCRTCGGAYLCTACDARPAHARAGH | zf-B_box |
| 8 | 50 | NVEFARFPHADSVVPNGAGVGAVVELDFTCGLGAKPSYSSYTATSLAHSV | UnKnown |
| 9 | 18 | TGTLPGWAVEDLLFDSPA | UnKnown |
| 10 | 40 | DEGCWAIWEEPQVJSLEDJIVPTTSCHGFQPLLAPPSPKV | UnKnown |
| Gene ID | Phosphorylation Sites | Glycosylation Sites | |||||
|---|---|---|---|---|---|---|---|
| Serine | Threonine | Tyrosine | Position | Potential | Jury Agreement | N-Glyc Result | |
| BDBBX1 | 9 | 5 | / | None | |||
| BDBBX2 | 13 | 7 | 1 | 105 NASA | 0.5176 | (4/9) | + |
| 216 NGTS | 0.669 | (8/9) | + | ||||
| BDBBX3 | 2 | / | 1 | None | |||
| BDBBX4 | 27 | 6 | 2 | 299 NCSN | 0.5557 | (5/9) | + |
| BDBBX5 | 11 | 1 | 1 | None | |||
| BDBBX6 | 17 | 9 | 6 | None | |||
| BDBBX7 | 4 | / | 2 | None | |||
| BDBBX8 | 12 | 3 | 3 | None | |||
| BDBBX9 | 16 | 5 | 1 | 70 NSSS | 0.57 | (8/9) | + |
| BDBBX10 | 20 | 7 | 1 | 209 NRTR | 0.5787 | (6/9) | + |
| BDBBX11 | 5 | 2 | 3 | None | |||
| BDBBX12 | 5 | 1 | 1 | None | |||
| BDBBX13 | 6 | 8 | / | None | |||
| BDBBX14 | 14 | 9 | 2 | 234 NLTL | 0.6876 | (9/9) | ++ |
| BDBBX15 | 25 | 12 | 3 | 152 NSSS | 0.611 | (8/9) | + |
| BDBBX16 | 21 | 5 | 3 | None | |||
| BDBBX17 | 30 | 28 | / | 200 NSSA | 0.5469 | (6/9) | + |
| BDBBX18 | 29 | 8 | 1 | None | |||
| BDBBX19 | 10 | 5 | 6 | None | |||
| BDBBX20 | 18 | 15 | 1 | 179 NSTL | 0.6232 | (8/9) | + |
| 227 NSTE | 0.5468 | (6/9) | + | ||||
| BDBBX21 | 10 | 6 | 3 | None | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Y.; Wang, Y.; Liao, J.; Chen, Q.; Jin, W.; Li, S.; Zhu, T.; Li, S. Identification and Characterization of the BBX Gene Family in Bambusa pervariabilis × Dendrocalamopsis grandis and Their Potential Role under Adverse Environmental Stresses. Int. J. Mol. Sci. 2023, 24, 13465. https://doi.org/10.3390/ijms241713465
Liu Y, Wang Y, Liao J, Chen Q, Jin W, Li S, Zhu T, Li S. Identification and Characterization of the BBX Gene Family in Bambusa pervariabilis × Dendrocalamopsis grandis and Their Potential Role under Adverse Environmental Stresses. International Journal of Molecular Sciences. 2023; 24(17):13465. https://doi.org/10.3390/ijms241713465
Chicago/Turabian StyleLiu, Yi, Yaxuan Wang, Jiao Liao, Qian Chen, Wentao Jin, Shuying Li, Tianhui Zhu, and Shujiang Li. 2023. "Identification and Characterization of the BBX Gene Family in Bambusa pervariabilis × Dendrocalamopsis grandis and Their Potential Role under Adverse Environmental Stresses" International Journal of Molecular Sciences 24, no. 17: 13465. https://doi.org/10.3390/ijms241713465
APA StyleLiu, Y., Wang, Y., Liao, J., Chen, Q., Jin, W., Li, S., Zhu, T., & Li, S. (2023). Identification and Characterization of the BBX Gene Family in Bambusa pervariabilis × Dendrocalamopsis grandis and Their Potential Role under Adverse Environmental Stresses. International Journal of Molecular Sciences, 24(17), 13465. https://doi.org/10.3390/ijms241713465






