CRISPR/Cas-Based Techniques for Live-Cell Imaging and Bioanalysis
Abstract
:1. Introduction
1.1. The Mechanism of the CRISPR/Cas9 System
1.2. The Mechanism of the CRISPR/Cas12 System
1.3. The Mechanism of CRISPR/Cas13 System
2. Application of CRISPR in Live-Cell Imaging
| Application | Imaging System | Cas Protein | Effector Molecule(s) | Advantage(s) | Limitation(s) | Ref. |
|---|---|---|---|---|---|---|
| Chromatin/genomic loci imaging | FP-based | dCas 9 | dCas9-EGPF | Simple and versatile tool for imaging | Low SNR, poor labeling efficiency | [31] |
| dCas 9 | dSaCas9-EGFP dSpCas9-mCherry | Achieved two-color CRISPR imaging | Relatively weak fluorescence signal and low signal-to-noise ratio | [34] | ||
| dCas 9 | sgRNA-aptamer-FP | Simultaneous labeling of six genetic loci | Complex sgRNA engineering, efficient delivery system needed | [35] | ||
| dCas 9 | dCas9-Suntag-scFv-GCN 4-sfGFP | Amplification of fluorescence signal intensity to improve signal-to-noise ratio enables labeling of low-repeat motifs | Large array size affects Cas9 functionality | [36] | ||
| dCas 9 | dCas9-SuTag10×-sfGFP-LEXY | Simultaneous labeling of nine duplicate genomic loci with a significantly improved signal-to-noise ratio | Large array size affects Cas9 functionality | [37] | ||
| Organic dye-based | dCas 9 | dCas9-HaloTag | Economical, efficient | Altered cell physiology | [38] | |
| dCas 9 | sgRNA-MTS-MB | Different combinations of fluorophore/bursting agent pairs and MB/MTS sequences can be flexibly selected | MBs are expensive | [39] | ||
| dCas 9 | 3sgMUC4-dual-MTS-MB | Dynamic imaging of non-repetitive genomic motifs with only three unique sgRNAs Higher sensitivity | MBs are expensive No simultaneous visualization of multiple genomic motifs | [40] | ||
| Quantum dot (QD)-based imaging | dCas 9 | dCas9-QD | Enables tracking of individual viruses | Large size, difficult cell delivery | [41] | |
| dCas 9 | SA-QDs | Efficient labeling of internal viruses without altering the virus envelope and capsid | Large size, difficult cell delivery | [42] | ||
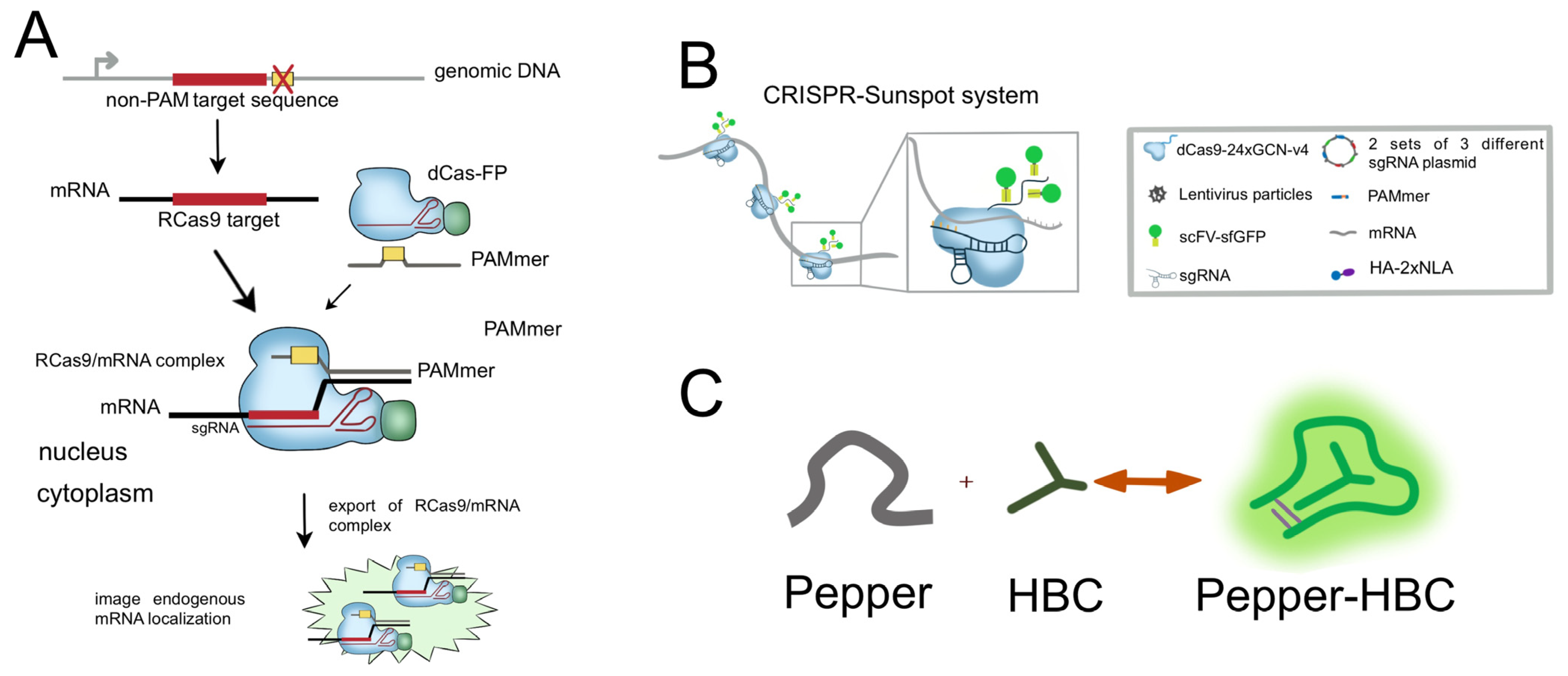
| RNA imaging | FP-based | Rcas9 | dCas9-FP | Imaging of RNA | Limit to imaging of low-abundance mRNA | [43] |
| FP-based | RCas9 | dCas9-EGFP | Imaging of RNA | Limit to imaging of low-abundance mRNA | [44] | |
| FP-based | dCas9 | dCas9-EGFP | Allow efficient imaging of low-abundance mRNA | Large array size | [45] | |
| Organic dye-based | dCas9 | sgRNA-Pepper530 | High fluorescence intensity and turn-on rate | Detailed structure-activity of Pepper to be analyzed | [46] | |
| FP-based | dCas13a | dCas13-EGFP | Imaging of RNA | No current guidelines to design efficient gRNAs targeting an RNA of interest | [47] | |
| FP-based | dCas13a | dCas13a-msfGFP-ZF-KRAB | Optimized S/N ratio | Imaging of intranuclear RNA is not possible | [48] | |
| Organic dye-based | dCas13b | CRISPR-TRA-tag | Modular design flexibility Smaller and more accurate fluorescent signals | Large array size | [49] |

2.1. CRISPR/Cas for Imaging of Chromatin/Genomic Loci
2.1.1. FP-Based CRISPR/dCas9 Systems
2.1.2. Organic Dye-Based CRISPR/dCas9 Systems
2.1.3. Quantum-Dots-Based CRISPR/dCas9 Systems
2.2. CRISPR/Cas for RNA Imaging
2.2.1. CRISPR/Cas9-Based RNA Imaging System
2.2.2. CRISPR/Cas13-Based RNA Imaging System
2.2.3. RNA Imaging Based on other Cas Platforms
2.3. CRISPR/Cas for Protein Imaging
2.3.1. Fluorescent Protein Labeling
2.3.2. Near-Infrared Imaging
2.4. Deficiencies and Shortcomings of CRISPR/Cas for Live-Cell Imaging
3. Research Progress of CRISPR/Cas for Bioanalysis
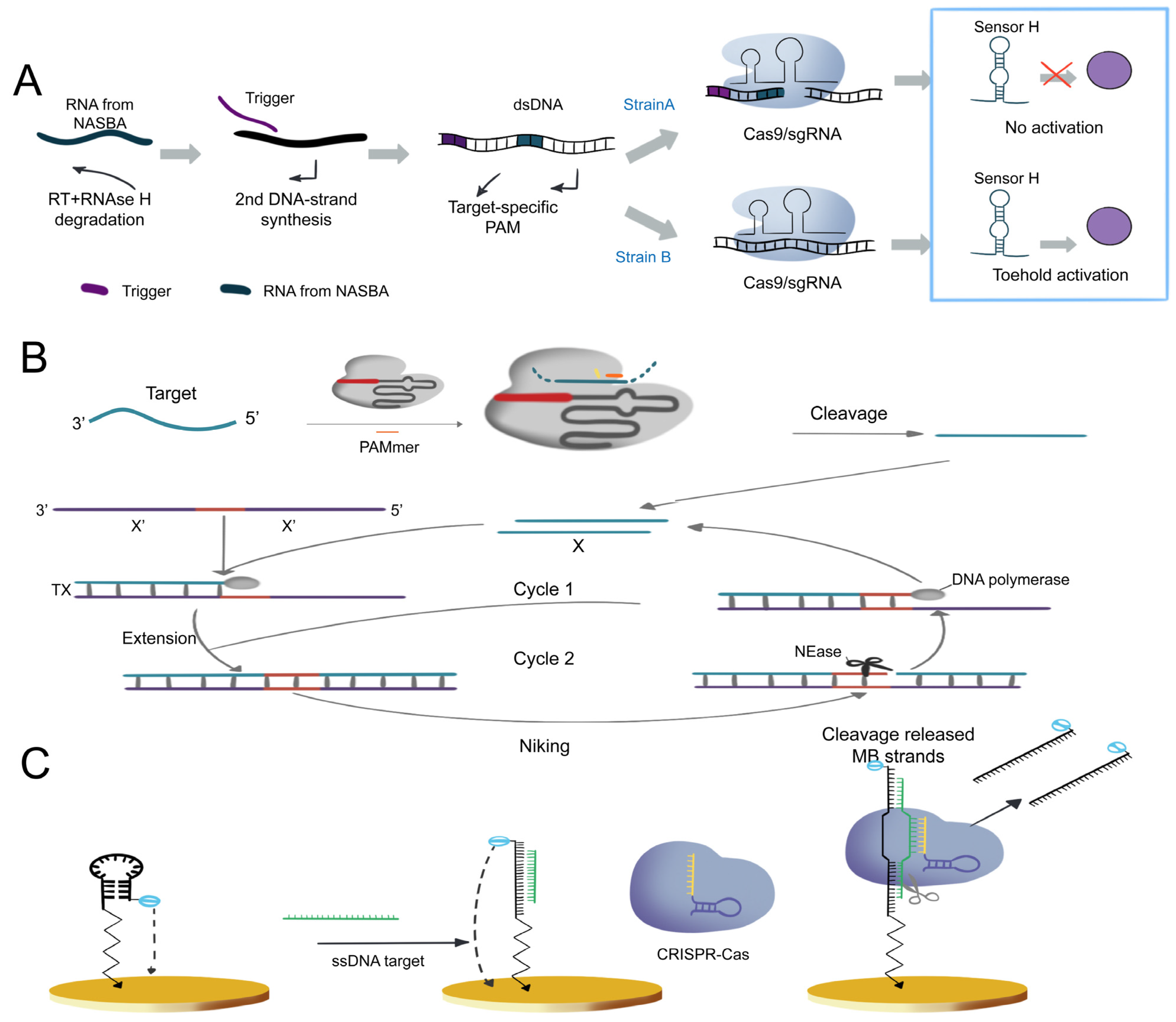
3.1. Nucleic Acid Analysis
3.1.1. Cas9-Based Nucleic Acid Detection System
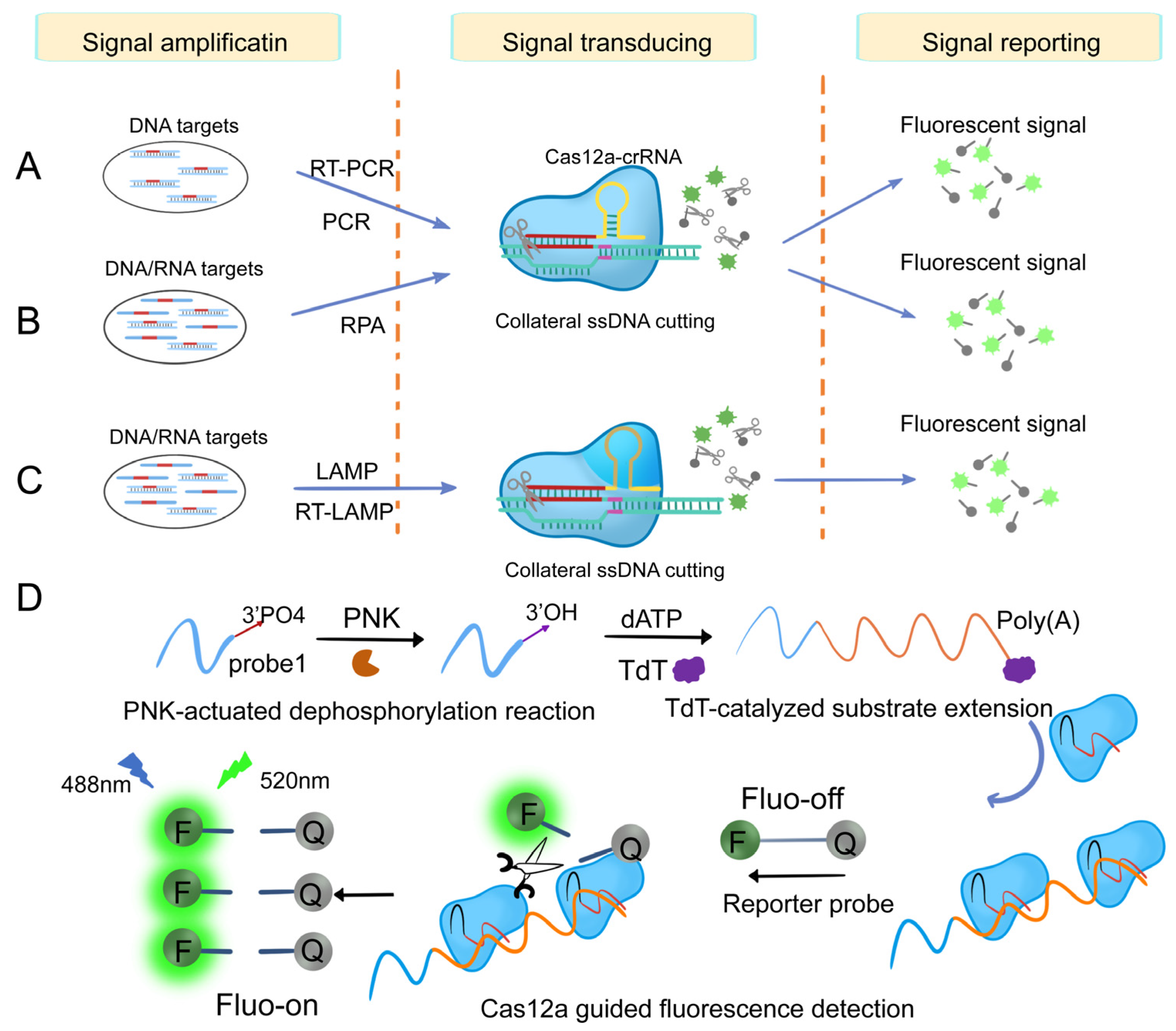
3.1.2. Cas12-Based Nucleic Acid Detection System
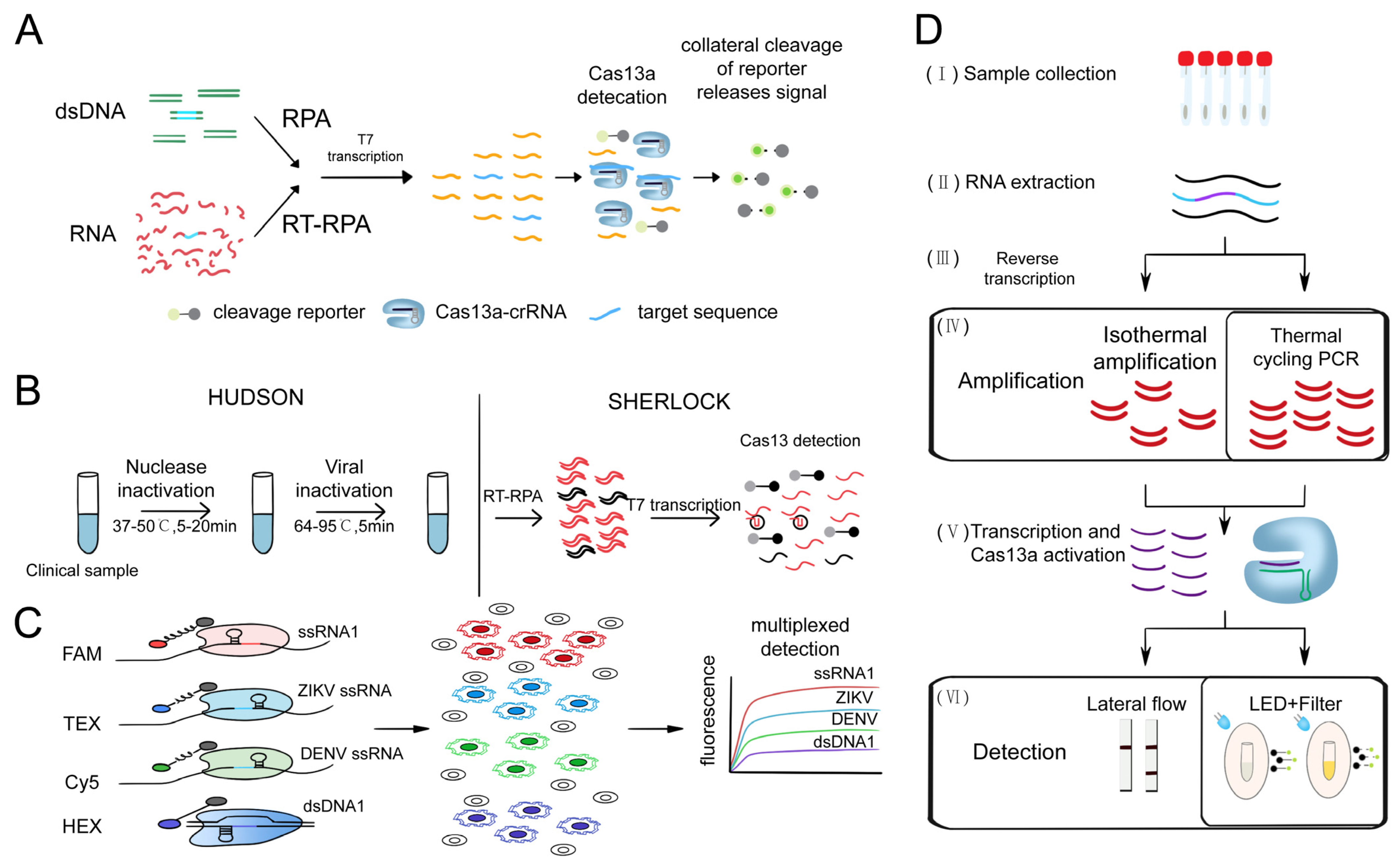
3.1.3. Cas13-Based Nucleic Acid Detection System
3.1.4. Other Cas-Based Nucleic Acid Detection Systems
3.2. Protein Analysis
3.3. Small Molecule Analysis
3.4. Flaws and Shortcomings of CRISPR/Cas Systems for Bioanalysis
4. Outlook
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| ACTB | Actin beta |
| aM | Artificial microorganisms |
| AMP | Adenosine monophosphate |
| APC | Antigen-presenting cell |
| ASD | Activating sequence discriminator |
| ATP | Adenosine-5′-triphosphate |
| BA-CASLFA | Bivalent aptamer-assisted CRISPR/Cas12a-mediated transversal flow assay |
| BPE-ECL | bipolar electrode-electrochemiluminescence |
| CAS-EXPER | CRISPR/Cas9-triggered isothermal exponential amplification reaction |
| C2c2 | Class 2 type VI-A CRISPR/Cas effector |
| Cas | CRISPR-associated proteins |
| CFU | Colony-forming units |
| CLISA | CRISPR/Cas signal amplification linked immunosorbent assay |
| CRISPR | Clustered regularly interspaced short palindromic repeats |
| CREST | Cas13-based robust, equitable, scalable testing |
| crRNA | CRISPR RNA |
| dCas | Deactivated CRISPR-associated proteins |
| DENV | Dengue virus |
| DETECTR | DNA endonuclease-targeted CRISPR transporter |
| DNA | Deoxyribonucleic acid |
| DNase | Deoxyribonuclease I |
| dSaCas9 | Deactivated Staphylococcus aureus Cas9 |
| dsDNA | Double-stranded DNA |
| dSpCas9 | Deactivated Streptococcus pyogenes Cas9 |
| ECL | enhanced chemiluminescence |
| E-CRISPR | Electrochemical CRISPR |
| E-DNA | Electrochemical DNA |
| EGFP | Enhanced green fluorescent protein |
| ELISA | Enzyme-linked immunosorbent assay |
| EXPAR | Exponential amplification reaction |
| Fc-ssDNA | Fragment crystallizable region ssDNA |
| FPs | Farnesyl diphosphate synthase |
| F-Q | Fluorophore quencher |
| GCN | General control noninducible |
| GFP | Green fluorescence protein |
| HA | Humic acid |
| HARRY | Highly sensitive aptamer-regulated Cas12f R-loop for bioanalysis |
| HDR | Homology-directed repair |
| HIV | Human immunodeficiency virus |
| HNH | His-Asn-His |
| HOLMES | One-hour low-cost multipurpose highly efficient system |
| HUDSON | Heating unextracted diagnostic samples to obliterate nucleases |
| KRAB | KRAB-containing zinc finger proteins |
| LAMP | Loop-mediated isothermal amplification |
| LbCas12a | Lachnospiraceae bacterium Cas12a |
| lncRNAs | Long non-coding RNAs |
| LRET | Luminescence resonance energy transfer |
| LwaCas13a | Leptotrichia wadei Cas13a |
| MB | Molecular beacon |
| MCP | MS2 coat protein |
| miRNAs | MicroRNAs |
| MNPs | Magnetic nanoparticles |
| MS | Methylation-sensitive |
| MTS | MB target sequence |
| MUC | Mucin |
| mRNA | Messenger RNA |
| NASBA | Nucleic acid sequence-based amplification |
| NASBACC | Nucleic acid sequence-based amplification—CRISPR cleavage |
| NIR | Near-infrared |
| NLS | Nuclear localization signal |
| PAM | Protospacer-adjacent motif |
| PAMmer | PAM-presenting DNA oligonucleotide |
| PCR | Polymerase chain reaction |
| POCT | Point-of-care testing |
| PM | Peritrophic matrix |
| PNK | Polynucleotide kinase |
| PRV | Pseudorabies virus |
| QD | Quantum dot |
| RACE | Rapid amplification of cDNA end |
| RCA | Rolling circle amplification |
| RNA | Ribonucleic acid |
| RN | Ribonucleic |
| RPA | Recombinase polymerase amplification |
| RT | Reverse transcription |
| SaCas9 | Staphylococcus aureus Cas9 |
| SA | Streptavidin |
| scFv | Single-chain antibody fragment |
| SDA | Strand displacement amplification |
| sfGFP | Superfolder GFP |
| SgRNA | Single guide RNA |
| sgMUC | Small guide mucin |
| SHERLOCK | Specific high-sensitivity enzymatic reporter unlocking |
| SNPs | Single-nucleotide polymorphisms |
| SNR | Signal-to-noise ratio |
| SpCas9 | Streptococcus pyogenes Cas9 |
| ssDNA | Single-stranded DNA |
| ssRNA | Single-stranded RNA |
| SunTag | Supernova tagging system |
| SVT | Single-virus tracking |
| T4 PNK | T4 polynucleotide kinase |
| TdT | Terminal deoxynucleotidyl transferase |
| TFRC | Transferrin receptor protein 1 |
| TGF-β1 | Transforming growth factor beta 1 |
| tracrRNA | Transactivating CRISPR RNA |
| UCNP | Upconversion nanoparticle |
| UDG | Uracil-DNA glycosylase |
| UV | Ultraviolet radiation |
| VEGF | Vascular endothelial growth factor |
| ZIF | Zeolite imidazolate framework |
| ZIKV | Zika virus |
| ZF | Zinc finger |
References
- Hsu, P.D.; Lander, E.S.; Zhang, F. Development and applications of CRISPR-Cas9 for genome engineering. Cell 2014, 157, 1262–1278. [Google Scholar] [CrossRef]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef]
- Makarova, K.S.; Wolf, Y.I.; Koonin, E.V. The basic building blocks and evolution of CRISPR-CAS systems. Biochem. Soc. Trans. 2013, 41, 1392–1400. [Google Scholar] [CrossRef]
- Xu, Y.; Li, Z. CRISPR-Cas systems: Overview, innovations and applications in human disease research and gene therapy. Comput. Struct. Biotechnol. J. 2020, 18, 2401–2415. [Google Scholar] [CrossRef]
- Glotzer, J.B.; Saffrich, R.; Glotzer, M.; Ephrussi, A. Cytoplasmic flows localize injected oskar RNA in Drosophila oocytes. Curr. Biol. 1997, 7, 326–337. [Google Scholar] [CrossRef]
- Guan, Y.; Ma, Y.; Li, Q.; Sun, Z.; Ma, L.; Wu, L.; Wang, L.; Zeng, L.; Shao, Y.; Chen, Y.; et al. CRISPR/Cas9-mediated somatic correction of a novel coagulator factor IX gene mutation ameliorates hemophilia in mouse. EMBO Mol. Med. 2016, 8, 477–488. [Google Scholar] [CrossRef]
- Kumar, V.; Jain, M. The CRISPR–Cas system for plant genome editing: Advances and opportunities. J. Exp. Bot. 2015, 66, 47–57. [Google Scholar] [CrossRef]
- Goell, J.H.; Hilton, I.B. CRISPR/Cas-Based Epigenome Editing: Advances, Applications, and Clinical Utility. Trends Biotechnol. 2021, 39, 678–691. [Google Scholar] [CrossRef]
- Doudna, J.A.; Charpentier, E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 2014, 346, 1258096. [Google Scholar] [CrossRef]
- Kazi, T.A.; Biswas, S.R. Chapter Four—CRISPR/dCas system as the modulator of gene expression. In Progress in Molecular Biology and Translational Science; Advances in CRISPR/Cas and Related Technologies; Ghosh, D., Ed.; Academic Press: Cambridge, MA, USA, 2021; Volume 178, pp. 99–122. [Google Scholar]
- Wu, X.; Mao, S.; Ying, Y.; Krueger, C.J.; Chen, A.K. Progress and Challenges for Live-cell Imaging of Genomic Loci Using CRISPR-based Platforms. Genom. Proteom. Bioinform. 2019, 17, 119–128. [Google Scholar] [CrossRef]
- O’Connell, M.R. Molecular Mechanisms of RNA Targeting by Cas13-containing Type VI CRISPR-Cas Systems. J. Mol. Biol. 2019, 431, 66–87. [Google Scholar] [CrossRef]
- Burmistrz, M.; Pyrc, K. CRISPR-Cas Systems in Prokaryotes. Pol. J. Microbiol. 2015, 64, 193–202. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Dhingra, Y.; Sashital, D.G. The Cas4-Cas1-Cas2 complex mediates precise prespacer processing during CRISPR adaptation. eLife 2019, 8, e44248. [Google Scholar] [CrossRef] [PubMed]
- Modell, J.W.; Jiang, W.; Marraffini, L.A. CRISPR–Cas systems exploit viral DNA injection to establish and maintain adaptive immunity. Nature 2017, 544, 101–104. [Google Scholar] [CrossRef] [PubMed]
- Sander, J.D.; Joung, J.K. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat. Biotechnol. 2014, 32, 347–355. [Google Scholar] [CrossRef] [PubMed]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.; Jiang, W.; Marraffini, L.A.; et al. Multiplex Genome Engineering Using CRISPR/Cas Systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef]
- Chen, B.; Guan, J.; Huang, B. Imaging Specific Genomic DNA in Living Cells. Annu. Rev. Biophys. 2016, 45, 1–23. [Google Scholar] [CrossRef]
- Gilbert, L.A.; Larson, M.H.; Morsut, L.; Liu, Z.; Brar, G.A.; Torres, S.E.; Stern-Ginossar, N.; Brandman, O.; Whitehead, E.H.; Doudna, J.A.; et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 2013, 154, 442–451. [Google Scholar] [CrossRef]
- Konermann, S.; Brigham, M.D.; Trevino, A.E.; Joung, J.; Abudayyeh, O.O.; Barcena, C.; Hsu, P.D.; Habib, N.; Gootenberg, J.S.; Nishimasu, H.; et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature 2015, 517, 583–588. [Google Scholar] [CrossRef]
- Wang, X.; Huang, X.; Fang, X.; Zhang, Y.; Wang, W. CRISPR-Cas9 System as a Versatile Tool for Genome Engineering in Human Cells. Mol. Ther. Nucleic Acids 2016, 5, e388. [Google Scholar] [CrossRef]
- Xue, H.-Y.; Ji, L.-J.; Gao, A.-M.; Liu, P.; He, J.-D.; Lu, X.-J. CRISPR-Cas9 for medical genetic screens: Applications and future perspectives. J. Med. Genet. 2016, 53, 91–97. [Google Scholar] [CrossRef] [PubMed]
- Singh, D.; Mallon, J.; Poddar, A.; Wang, Y.; Tippana, R.; Yang, O.; Bailey, S.; Ha, T. Real-time observation of DNA target interrogation and product release by the RNA-guided endonuclease CRISPR Cpf1 (Cas12a). Proc. Natl. Acad. Sci. USA 2018, 115, 5444–5449. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.S.; Ma, E.; Harrington, L.B.; Da Costa, M.; Tian, X.; Palefsky, J.M.; Doudna, J.A. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science 2018, 360, 436–439. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Sun, S.; Moonen, D.; Lee, C.; Lee, A.Y.-F.; Schaffer, D.V.; He, L. CRISPR-READI: Efficient Generation of Knockin Mice by CRISPR RNP Electroporation and AAV Donor Infection. Cell Rep. 2019, 27, 3780–3789. [Google Scholar] [CrossRef] [PubMed]
- Kordyś, M.; Sen, R.; Warkocki, Z. Applications of the versatile CRISPR-Cas13 RNA targeting system. WIREs RNA 2022, 13, e1694. [Google Scholar] [CrossRef]
- Savage, D.F. Cas14: Big Advances from Small CRISPR Proteins. Biochemistry 2019, 58, 1024–1025. [Google Scholar] [CrossRef]
- Liu, L.; Pei, D.-S. Insights Gained from RNA Editing Targeted by the CRISPR-Cas13 Family. Int. J. Mol. Sci. 2022, 23, 11400. [Google Scholar] [CrossRef]
- Aman, R.; Mahas, A.; Mahfouz, M. Nucleic Acid Detection Using CRISPR/Cas Biosensing Technologies. ACS Synth. Biol. 2020, 9, 1226–1233. [Google Scholar] [CrossRef]
- Gootenberg, J.S.; Abudayyeh, O.O.; Lee, J.W.; Essletzbichler, P.; Dy, A.J.; Joung, J.; Verdine, V.; Donghia, N.; Daringer, N.M.; Freije, C.A.; et al. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science 2017, 356, 438–442. [Google Scholar] [CrossRef]
- Chen, B.; Gilbert, L.A.; Cimini, B.A.; Schnitzbauer, J.; Zhang, W.; Li, G.-W.; Park, J.; Blackburn, E.H.; Weissman, J.S.; Qi, L.S.; et al. Dynamic Imaging of Genomic Loci in Living Human Cells by an Optimized CRISPR/Cas System. Cell 2013, 155, 1479–1491. [Google Scholar] [CrossRef]
- Singh, V.; Jain, M. Recent advancements in CRISPR-Cas toolbox for imaging applications. Crit. Rev. Biotechnol. 2022, 42, 508–531. [Google Scholar] [CrossRef] [PubMed]
- He, S.; Bhatt, R.; Brown, C.; Brown, E.A.; Buhr, D.L.; Chantranuvatana, K.; Danaher, P.; Dunaway, D.; Garrison, R.G.; Geiss, G.; et al. High-plex imaging of RNA and proteins at subcellular resolution in fixed tissue by spatial molecular imaging. Nat. Biotechnol. 2022, 40, 1794–1806. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Hu, J.; Almeida, R.; Liu, H.; Balakrishnan, S.; Covill-Cooke, C.; Lim, W.A.; Huang, B. Expanding the CRISPR imaging toolset with Staphylococcus aureus Cas9 for simultaneous imaging of multiple genomic loci. Nucleic Acids Res. 2016, 44, e75. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Tu, L.-C.; Naseri, A.; Huisman, M.; Zhang, S.; Grunwald, D.; Pederson, T. Multiplexed labeling of genomic loci with dCas9 and engineered sgRNAs using CRISPRainbow. Nat. Biotechnol. 2016, 34, 528–530. [Google Scholar] [CrossRef]
- Ye, H.; Rong, Z.; Lin, Y. Live cell imaging of genomic loci using dCas9-SunTag system and a bright fluorescent protein. Protein Cell 2017, 8, 853–855. [Google Scholar] [CrossRef]
- Hou, Y.; Wang, D.; Lu, S.; Guo, D.; Li, M.; Cui, M.; Zhang, X.-E. Optogenetic Control of Background Fluorescence Reduction for CRISPR-Based Genome Imaging. Anal. Chem. 2022, 94, 8724–8731. [Google Scholar] [CrossRef] [PubMed]
- Knight, S.C.; Xie, L.; Deng, W.; Guglielmi, B.; Witkowsky, L.B.; Bosanac, L.; Zhang, E.T.; Beheiry, M.E.; Masson, J.-B.; Dahan, M.; et al. Dynamics of CRISPR-Cas9 genome interrogation in living cells. Science 2015, 350, 823–826. [Google Scholar] [CrossRef]
- Wu, X.; Mao, S.; Yang, Y.; Rushdi, M.N.; Krueger, C.J.; Chen, A.K. A CRISPR/molecular beacon hybrid system for live-cell genomic imaging. Nucleic Acids Res. 2018, 46, e80. [Google Scholar] [CrossRef]
- Mao, S.; Ying, Y.; Wu, X.; Krueger, C.J.; Chen, A.K. CRISPR/dual-FRET molecular beacon for sensitive live-cell imaging of non-repetitive genomic loci. Nucleic Acids Res. 2019, 47, e131. [Google Scholar] [CrossRef]
- Ma, Y.; Wang, M.; Li, W.; Zhang, Z.; Zhang, X.; Wu, G.; Tan, T.; Cui, Z.; Zhang, X.-E. Live Visualization of HIV-1 Proviral DNA Using a Dual-Color-Labeled CRISPR System. Anal. Chem. 2017, 89, 12896–12901. [Google Scholar] [CrossRef]
- Yang, Y.-B.; Tang, Y.-D.; Hu, Y.; Yu, F.; Xiong, J.-Y.; Sun, M.-X.; Lyu, C.; Peng, J.-M.; Tian, Z.-J.; Cai, X.-H.; et al. Single Virus Tracking with Quantum Dots Packaged into Enveloped Viruses Using CRISPR. Nano Lett. 2020, 20, 1417–1427. [Google Scholar] [CrossRef] [PubMed]
- Nelles, D.A.; Fang, M.Y.; O’Connell, M.R.; Xu, J.L.; Markmiller, S.J.; Doudna, J.A.; Yeo, G.W. Programmable RNA Tracking in Live Cells with CRISPR/Cas9. Cell 2016, 165, 2. [Google Scholar] [CrossRef] [PubMed]
- Batra, R.; Nelles, D.A.; Pirie, E.; Blue, S.M.; Marina, R.J.; Wang, H.; Chaim, I.A.; Thomas, J.D.; Zhang, N.; Nguyen, V.; et al. Elimination of Toxic Microsatellite Repeat Expansion RNA by RNA-Targeting Cas9. Cell 2017, 170, 899–912.e10. [Google Scholar] [CrossRef] [PubMed]
- Sun, N.-H.; Chen, D.-Y.; Ye, L.-P.; Sheng, G.; Gong, J.-J.; Chen, B.-H.; Lu, Y.-M.; Han, F. CRISPR-Sunspot: Imaging of endogenous low-abundance RNA at the single-molecule level in live cells. Theranostics 2020, 10, 10993–11012. [Google Scholar] [CrossRef]
- Chen, X.; Zhang, D.; Su, N.; Bao, B.; Xie, X.; Zuo, F.; Yang, L.; Wang, H.; Jiang, L.; Lin, Q.; et al. Visualizing RNA dynamics in live cells with bright and stable fluorescent RNAs. Nat. Biotechnol. 2019, 37, 1287–1293. [Google Scholar] [CrossRef]
- Yang, L.-Z.; Wang, Y.; Li, S.-Q.; Yao, R.-W.; Luan, P.-F.; Wu, H.; Carmichael, G.G.; Chen, L.-L. Dynamic Imaging of RNA in Living Cells by CRISPR-Cas13 Systems. Mol. Cell 2019, 76, 981–997.e7. [Google Scholar] [CrossRef]
- Abudayyeh, O.O.; Gootenberg, J.S.; Essletzbichler, P.; Han, S.; Joung, J.; Belanto, J.J.; Verdine, V.; Cox, D.B.T.; Kellner, M.J.; Regev, A.; et al. RNA targeting with CRISPR-Cas13. Nature 2017, 550, 280–284. [Google Scholar] [CrossRef]
- Tang, H.; Peng, J.; Jiang, X.; Peng, S.; Wang, F.; Weng, X.; Zhou, X. A CRISPR-Cas and Tat Peptide with Fluorescent RNA Aptamer System for Signal Amplification in RNA Imaging. Biosensors 2023, 13, 293. [Google Scholar] [CrossRef]
- Hong, Y.; Lu, G.; Duan, J.; Liu, W.; Zhang, Y. Comparison and optimization of CRISPR/dCas9/gRNA genome-labeling systems for live cell imaging. Genome Biol. 2018, 19, 39. [Google Scholar] [CrossRef]
- Baum, M. Retrieving the intracellular topology from multi-scale protein mobility mapping in living cells. Nat. Commun. 2014, 5, 4494. [Google Scholar] [CrossRef]
- Tanenbaum, M.E.; Gilbert, L.A.; Qi, L.S.; Weissman, J.S.; Vale, R.D. A protein-tagging system for signal amplification in gene expression and fluorescence imaging. Cell 2014, 159, 635–646. [Google Scholar] [CrossRef] [PubMed]
- Resch-Genger, U.; Grabolle, M.; Cavaliere-Jaricot, S.; Nitschke, R.; Nann, T. Quantum dots versus organic dyes as fluorescent labels. Nat. Methods 2008, 5, 763–775. [Google Scholar] [CrossRef] [PubMed]
- Grimm, J.B.; English, B.P.; Chen, J.; Slaughter, J.P.; Zhang, Z.; Revyakin, A.; Patel, R.; Macklin, J.J.; Normanno, D.; Singer, R.H.; et al. A general method to improve fluorophores for live-cell and single-molecule microscopy. Nat. Methods 2015, 12, 244–250. [Google Scholar] [CrossRef]
- Tyagi, S.; Kramer, F.R. Molecular beacons: Probes that fluoresce upon hybridization. Nat. Biotechnol. 1996, 14, 3. [Google Scholar] [CrossRef]
- Wang, Z.-G.; Liu, S.-L.; Pang, D.-W. Quantum Dots: A Promising Fluorescent Label for Probing Virus Trafficking. Acc. Chem. Res. 2021, 54, 2991–3002. [Google Scholar] [CrossRef]
- Viushkov, V.S.; Lomov, N.A.; Rubtsov, M.A.; Vassetzky, Y.S. Visualizing the Genome: Experimental Approaches for Live-Cell Chromatin Imaging. Cells 2022, 11, 4086. [Google Scholar] [CrossRef] [PubMed]
- Cech, T.R. The RNA Worlds in Context. Cold Spring Harb. Perspect. Biol. 2012, 4, a006742. [Google Scholar] [CrossRef]
- Le, P.; Ahmed, N.; Yeo, G.W. Illuminating RNA biology through imaging. Nat. Cell Biol. 2022, 24, 815–824. [Google Scholar] [CrossRef]
- Li, X.; Yin, F.; Xu, X.; Liu, L.; Xue, Q.; Tong, L.; Jiang, W.; Li, C. A facile DNA/RNA nanoflower for sensitive imaging of telomerase RNA in living cells based on “zipper lock-and-key” strategy. Biosens. Bioelectron. 2020, 147, 111788. [Google Scholar] [CrossRef]
- Du, J.; Dartawan, R.; Rice, W.; Gao, F.; Zhou, J.H.; Sheng, J. Fluorescent Platforms for RNA Chemical Biology Research. Genes 2022, 13, 1348. [Google Scholar] [CrossRef]
- Han, S.; Zhao, B.S.; Myers, S.A.; Carr, S.A.; He, C.; Ting, A.Y. RNA–protein interaction mapping via MS2- or Cas13-based APEX targeting. Proc. Natl. Acad. Sci. USA 2020, 117, 22068–22079. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Zhang, L.; Huang, X. Genome modification by CRISPR/Cas9. FEBS J. 2014, 281, 5186–5193. [Google Scholar] [CrossRef] [PubMed]
- O’Connell, M.R.; Oakes, B.L.; Sternberg, S.H.; East-Seletsky, A.; Kaplan, M.; Doudna, J.A. Programmable RNA recognition and cleavage by CRISPR/Cas9. Nature 2014, 516, 263–266. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.-Z.; Gao, B.-Q.; Huang, Y.; Wang, Y.; Yang, L.; Chen, L.-L. Multi-color RNA imaging with CRISPR-Cas13b systems in living cells. Cell Insight 2022, 1, 100044. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.; Peng, J.; Peng, S.; Wang, Q.; Jiang, X.; Xue, X.; Tao, Y.; Xiang, L.; Ji, Q.; Liu, S.-M.; et al. Live-cell RNA imaging using the CRISPR-dCas13 system with modified sgRNAs appended with fluorescent RNA aptamers. Chem. Sci. 2022, 13, 14032–14040. [Google Scholar] [CrossRef]
- de la Fuente, J.M.; Berry, C.C. Tat peptide as an efficient molecule to translocate gold nanoparticles into the cell nucleus. Bioconjug. Chem. 2005, 16, 1176–1180. [Google Scholar] [CrossRef]
- Lo, S.L.; Wang, S. An endosomolytic Tat peptide produced by incorporation of histidine and cysteine residues as a nonviral vector for DNA transfection. Biomaterials 2008, 29, 2408–2414. [Google Scholar] [CrossRef]
- Fusco, D.; Accornero, N.; Lavoie, B.; Shenoy, S.M.; Blanchard, J.-M.; Singer, R.H.; Bertrand, E. Single mRNA Molecules Demonstrate Probabilistic Movement in Living Mammalian Cells. Curr. Biol. 2003, 13, 161–167. [Google Scholar] [CrossRef]
- Gao, F.; Zheng, K.; Li, Y.-B.; Jiang, F.; Han, C.-Y. A Cas6-based RNA tracking platform functioning in a fluorescence-activation mode. Nucleic Acids Res. 2022, 50, e46. [Google Scholar] [CrossRef]
- Wang, Q.; Liu, X.; Zhou, J.; Yang, C.; Wang, G.; Tan, Y.; Wu, Y.; Zhang, S.; Yi, K.; Kang, C. The CRISPR-Cas13a Gene-Editing System Induces Collateral Cleavage of RNA in Glioma Cells. Adv. Sci. 2019, 6, 1901299. [Google Scholar] [CrossRef]
- Özcan, A.; Krajeski, R.; Ioannidi, E.; Lee, B.; Gardner, A.; Makarova, K.S.; Koonin, E.V.; Abudayyeh, O.O.; Gootenberg, J.S. Programmable RNA targeting with the single-protein CRISPR effector Cas7-11. Nature 2021, 597, 720–725. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Wang, R.; Lei, C.; Nie, Z. CRISPR-Cas System for RNA Detection and Imaging. Chem. Res. Chin. Univ. 2020, 36, 157–163. [Google Scholar] [CrossRef]
- Hekstra, D.R. Emerging Time-Resolved X-Ray Diffraction Approaches for Protein Dynamics. Annu. Rev. Biophys. 2023, 52, 255–274. [Google Scholar] [CrossRef]
- Samaras, P.; Schmidt, T.; Frejno, M.; Gessulat, S.; Reinecke, M.; Jarzab, A.; Zecha, J.; Mergner, J.; Giansanti, P.; Ehrlich, H.-C.; et al. ProteomicsDB: A multi-omics and multi-organism resource for life science research. Nucleic Acids Res. 2020, 48, D1153–D1163. [Google Scholar] [CrossRef] [PubMed]
- Mikuni, T.; Nishiyama, J.; Sun, Y.; Kamasawa, N.; Yasuda, R. High-Throughput, High-Resolution Mapping of Protein Localization in Mammalian Brain by In Vivo Genome Editing. Cell 2016, 165, 1803–1817. [Google Scholar] [CrossRef] [PubMed]
- Kamiyama, D.; Sekine, S.; Barsi-Rhyne, B.; Hu, J.; Chen, B.; Gilbert, L.A.; Ishikawa, H.; Leonetti, M.D.; Marshall, W.F.; Weissman, J.S.; et al. Versatile protein tagging in cells with split fluorescent protein. Nat. Commun. 2016, 7, 11046. [Google Scholar] [CrossRef]
- Schwinn, M.K.; Machleidt, T.; Zimmerman, K.; Eggers, C.T.; Dixon, A.S.; Hurst, R.; Hall, M.P.; Encell, L.P.; Binkowski, B.F.; Wood, K.V. CRISPR-Mediated Tagging of Endogenous Proteins with a Luminescent Peptide. ACS Chem. Biol. 2018, 13, 467–474. [Google Scholar] [CrossRef]
- Willems, J.; de Jong, A.P.H.; Scheefhals, N.; MacGillavry, H.D. ORANGE: A CRISPR/Cas9-based genome editing toolbox for epitope tagging of endogenous proteins in neurons. bioRxiv 2019, 18, 700187. [Google Scholar] [CrossRef]
- Baldering, T.N.; Karathanasis, C.; Harwardt, M.-L.I.E.; Freund, P.; Meurer, M.; Rahm, J.V.; Knop, M.; Dietz, M.S.; Heilemann, M. CRISPR/Cas12a-mediated labeling of MET receptor enables quantitative single-molecule imaging of endogenous protein organization and dynamics. iScience 2021, 24, 101895. [Google Scholar] [CrossRef]
- Chen, B.; Shi, H.; Zhang, J.; Zhou, C.; Han, M.; Jiang, W.; Lai, Y.; Tu, X.; Li, H. CRISPR-based RNA-binding protein mapping in live cells. Biochem. Biophys. Res. Commun. 2021, 583, 79–85. [Google Scholar] [CrossRef]
- He, S.; Song, J.; Qu, J.; Cheng, Z. Crucial breakthrough of second near-infrared biological window fluorophores: Design and synthesis toward multimodal imaging and theranostics. Chem. Soc. Rev. 2018, 47, 4258–4278. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Chen, G.; Zhang, Y.; Wu, F.; Wang, Q. Advanced Fluorescence Imaging Technology in the Near-Infrared-II Window for Biomedical Applications. J. Am. Chem. Soc. 2020, 142, 14789–14804. [Google Scholar] [CrossRef] [PubMed]
- Butkevich, A.N.; Ta, H.; Ratz, M.; Stoldt, S.; Jakobs, S.; Belov, V.N.; Hell, S.W. Two-Color 810 nm STED Nanoscopy of Living Cells with Endogenous SNAP-Tagged Fusion Proteins. ACS Chem. Biol. 2018, 13, 475–480. [Google Scholar] [CrossRef] [PubMed]
- Smargon, A.A.; Cox, D.B.T.; Pyzocha, N.K.; Zheng, K.; Slaymaker, I.M.; Gootenberg, J.S.; Abudayyeh, O.A.; Essletzbichler, P.; Shmakov, S.; Makarova, K.S.; et al. Cas13b Is a Type VI-B CRISPR-Associated RNA-Guided RNase Differentially Regulated by Accessory Proteins Csx27 and Csx28. Mol. Cell 2017, 65, 618–630.e7. [Google Scholar] [CrossRef] [PubMed]
- Higgs, P.G.; Lehman, N. The RNA World: Molecular cooperation at the origins of life. Nat. Rev. Genet. 2015, 16, 7–17. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Tian, P.; Tan, T. CRISPR-Cas13 technology portfolio and alliance with other genetic tools. Biotechnol. Adv. 2022, 61, 108047. [Google Scholar] [CrossRef]
- Okeke, I.N.; Ihekweazu, C. The importance of molecular diagnostics for infectious diseases in low-resource settings. Nat. Rev. Microbiol. 2021, 19, 547–548. [Google Scholar] [CrossRef]
- Gootenberg, J.S.; Abudayyeh, O.O.; Kellner, M.J.; Joung, J.; Collins, J.J.; Zhang, F. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science 2018, 360, 439–444. [Google Scholar] [CrossRef]
- Li, S.-Y.; Cheng, Q.-X.; Wang, J.-M.; Li, X.-Y.; Zhang, Z.-L.; Gao, S.; Cao, R.-B.; Zhao, G.-P.; Wang, J. CRISPR-Cas12a-assisted nucleic acid detection. Cell Discov. 2018, 4, 1. [Google Scholar] [CrossRef]
- Myhrvold, C.; Freije, C.A.; Gootenberg, J.S.; Abudayyeh, O.O.; Metsky, H.C.; Durbin, A.F.; Kellner, M.J.; Tan, A.L.; Paul, L.M.; Parham, L.A.; et al. Field-deployable viral diagnostics using CRISPR-Cas13. Science 2018, 360, 444–448. [Google Scholar] [CrossRef]
- Pardee, K.; Green, A.A.; Takahashi, M.K.; Braff, D.; Lambert, G.; Lee, J.W.; Ferrante, T.; Ma, D.; Donghia, N.; Fan, M.; et al. Rapid, Low-Cost Detection of Zika Virus Using Programmable Biomolecular Components. Cell 2016, 165, 1255–1266. [Google Scholar] [CrossRef] [PubMed]
- Rauch, J.N.; Valois, E.; Solley, S.C.; Braig, F.; Lach, R.S.; Audouard, M.; Ponce-Rojas, J.C.; Costello, M.S.; Baxter, N.J.; Kosik, K.S.; et al. A Scalable, Easy-to-Deploy Protocol for Cas13-Based Detection of SARS-CoV-2 Genetic Material. J. Clin. Microbiol. 2021, 59, e02402–e02420. [Google Scholar] [CrossRef] [PubMed]
- Weng, Z.; You, Z.; Li, H.; Wu, G.; Song, Y.; Sun, H.; Fradlin, A.; Neal-Harris, C.; Lin, M.; Gao, X.; et al. CRISPR-Cas12a Biosensor Array for Ultrasensitive Detection of Unamplified DNA with Single-Nucleotide Polymorphic Discrimination. ACS Sens. 2023, 8, 1489–1499. [Google Scholar] [CrossRef] [PubMed]
- Li, S.-Y.; Cheng, Q.-X.; Liu, J.-K.; Nie, X.-Q.; Zhao, G.-P.; Wang, J. CRISPR-Cas12a has both cis- and trans-cleavage activities on single-stranded DNA. Cell Res. 2018, 28, 491–493. [Google Scholar] [CrossRef]
- Sun, X.; Wang, Y.; Zhang, L.; Liu, S.; Zhang, M.; Wang, J.; Ning, B.; Peng, Y.; He, J.; Hu, Y.; et al. CRISPR-Cas9 Triggered Two-Step Isothermal Amplification Method for E. coli O157:H7 Detection Based on a Metal-Organic Framework Platform. Anal. Chem. 2020, 92, 3032–3041. [Google Scholar] [CrossRef]
- Wang, R.; Chen, R.; Qian, C.; Pang, Y.; Wu, J.; Li, F. Ultrafast visual nucleic acid detection with CRISPR/Cas12a and rapid PCR in single capillary. Sens. Actuators B Chem. 2021, 326, 128618. [Google Scholar] [CrossRef]
- Zhang, X.; Zheng, C.; Ding, L.; Wu, Y.; Xu, H.; Sun, Y.; Zeng, Y.; Liu, X.; Liu, J. CRISPR-Cas12a coupled with terminal deoxynucleotidyl transferase mediated isothermal amplification for sensitive detection of polynucleotide kinase activity. Sens. Actuators B Chem. 2021, 330, 129317. [Google Scholar] [CrossRef]
- Niu, C.; Wang, C.; Li, F.; Zheng, X.; Xing, X.; Zhang, C. Aptamer assisted CRISPR-Cas12a strategy for small molecule diagnostics. Biosens. Bioelectron. 2021, 183, 113196. [Google Scholar] [CrossRef]
- Peng, L.; Zhou, J.; Liu, G.; Yin, L.; Ren, S.; Man, S.; Ma, L. CRISPR-Cas12a based aptasensor for sensitive and selective ATP detection. Sens. Actuators B Chem. 2020, 320, 128164. [Google Scholar] [CrossRef]
- Wang, X.; Chen, X.; Chu, C.; Deng, Y.; Yang, M.; Ji, Z.; Xu, F.; Huo, D.; Luo, Y.; Hou, C. Four-stage signal amplification for trace ATP detection using allosteric probe-conjugated strand displacement and CRISPR/Cpf1 trans-cleavage (ASD-Cpf1). Sens. Actuators B Chem. 2020, 323, 128653. [Google Scholar] [CrossRef]
- Huang, M.; Zhou, X.; Wang, H.; Xing, D. Clustered Regularly Interspaced Short Palindromic Repeats/Cas9 Triggered Isothermal Amplification for Site-Specific Nucleic Acid Detection. Anal. Chem. 2018, 90, 2193–2200. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Zhao, X.; Chen, X.; Qiu, X.; Qing, G.; Zhang, H.; Zhang, L.; Hu, X.; He, Z.; Zhong, D.; et al. Rolling Circular Amplification (RCA)-Assisted CRISPR/Cas9 Cleavage (RACE) for Highly Specific Detection of Multiple Extracellular Vesicle MicroRNAs. Anal. Chem. 2020, 92, 2176–2185. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Jin, T.; Dai, Y.; Liu, C.C. Surpassing the detection limit and accuracy of the electrochemical DNA sensor through the application of CRISPR Cas systems. Biosens. Bioelectron. 2020, 155, 112100. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Li, S.; Wu, N.; Wu, J.; Wang, G.; Zhao, G.; Wang, J. HOLMESv2: A CRISPR-Cas12b-Assisted Platform for Nucleic Acid Detection and DNA Methylation Quantitation. ACS Synth. Biol. 2019, 8, 2228–2237. [Google Scholar] [CrossRef]
- Li, C.-Y.; Liu, J.-X.; Liu, Y.-H.; Gao, J.-L.; Zheng, B.; Liu, D. An exceptional and universal DNA walker amplified “one-to-many” CRISPR/Cas12a-mediated fluorescent biosensor for ultrasensitive detection of non-DNA biomarkers. Sens. Actuators B Chem. 2022, 361, 131743. [Google Scholar] [CrossRef]
- Harrington, L.B.; Burstein, D.; Chen, J.S.; Paez-Espino, D.; Ma, E.; Witte, I.P.; Cofsky, J.C.; Kyrpides, N.C.; Banfield, J.F.; Doudna, J.A. Programmed DNA destruction by miniature CRISPR-Cas14 enzymes. Science 2018, 362, 839–842. [Google Scholar] [CrossRef]
- Du, Y.-C.; Wang, S.-Y.; Wang, Y.-X.; Ma, J.-Y.; Wang, D.-X.; Tang, A.-N.; Kong, D.-M. Terminal deoxynucleotidyl transferase combined CRISPR-Cas12a amplification strategy for ultrasensitive detection of uracil-DNA glycosylase with zero background. Biosens. Bioelectron. 2021, 171, 112734. [Google Scholar] [CrossRef]
- Makarova, K.S.; Koonin, E.V. Annotation and Classification of CRISPR-Cas Systems. Methods Mol. Biol. Clifton NJ 2015, 1311, 47–75. [Google Scholar] [CrossRef]
- Dai, Y.; Somoza, R.A.; Wang, L.; Welter, J.F.; Li, Y.; Caplan, A.I.; Liu, C.C. Exploring the Trans-Cleavage Activity of CRISPR-Cas12a (cpf1) for the Development of a Universal Electrochemical Biosensor. Angew. Chem.-Int. Ed. 2019, 58, 17399–17405. [Google Scholar] [CrossRef]
- Zhao, Q.; Ren, X.; Wang, S.; Fan, D.; Zhang, N.; Liu, L.; Li, Y.; Wei, Q. CRISPR-Cas12a accessory cleavage activity triggering electrochemiluminescence biosensor for adenosine triphosphate detection. Sens. Actuators B Chem. 2022, 371, 132553. [Google Scholar] [CrossRef]
- Li, X.; Chen, X.; Mao, M.; Peng, C.; Wang, Z. Accelerated CRISPR/Cas12a-based small molecule detection using bivalent aptamer. Biosens. Bioelectron. 2022, 217, 114725. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, D.; Zeng, Y.; Sun, Y.; Qi, P. Simultaneous ultrasensitive ADP and ATP quantification based on CRISPR/Cas12a integrated ZIF-90@Ag3AuS2@Fe3O4 nanocomposites. Biosens. Bioelectron. 2022, 218, 114784. [Google Scholar] [CrossRef] [PubMed]
- Politza, A.J.; Nouri, R.; Guan, W. Digital CRISPR systems for the next generation of nucleic acid quantification. TrAC Trends Anal. Chem. 2023, 159, 116917. [Google Scholar] [CrossRef]
- Zhou, Y.; Wang, P.; Tian, F.; Gao, G.; Huang, L.; Wei, W.; Xie, X.S. Painting a specific chromosome with CRISPR/Cas9 for live-cell imaging. Cell Res. 2017, 27, 298–301. [Google Scholar] [CrossRef]
- Anderson, K.R.; Haeussler, M.; Watanabe, C.; Janakiraman, V.; Lund, J.; Modrusan, Z.; Stinson, J.; Bei, Q.; Buechler, A.; Yu, C.; et al. CRISPR off-target analysis in genetically engineered rats and mice. Nat. Methods 2018, 15, 512–514. [Google Scholar] [CrossRef]
- Zhang, Y.; Qian, L.; Wei, W.; Wang, Y.; Wang, B.; Lin, P.; Liu, W.; Xu, L.; Li, X.; Liu, D.; et al. Paired Design of dCas9 as a Systematic Platform for the Detection of Featured Nucleic Acid Sequences in Pathogenic Strains. ACS Synth. Biol. 2016, 6, 211–216. [Google Scholar] [CrossRef] [PubMed]
- Compton, J. Nucleic acid sequence-based amplification. Nature 1991, 350, 91–92. [Google Scholar] [CrossRef]
- Van Ness, J.; Van Ness, L.K.; Galas, D.J. Isothermal reactions for the amplification of oligonucleotides. Proc. Natl. Acad. Sci. USA 2003, 100, 4504–4509. [Google Scholar] [CrossRef]
- Fire, A.; Xu, S.Q. Rolling replication of short DNA circles. Proc. Natl. Acad. Sci. USA 1995, 92, 4641–4645. [Google Scholar] [CrossRef]
- Fan, C.; Plaxco, K.W.; Heeger, A.J. Electrochemical interrogation of conformational changes as a reagentless method for the sequence-specific detection of DNA. Proc. Natl. Acad. Sci. USA 2003, 100, 9134–9137. [Google Scholar] [CrossRef]
- Guk, K.; Yi, S.; Kim, H.; Bae, Y.; Yong, D.; Kim, S.; Lee, K.-S.; Lim, E.-K.; Kang, T.; Jung, J. Hybrid CRISPR/Cas protein for one-pot detection of DNA and RNA. Biosens. Bioelectron. 2022, 219, 114819. [Google Scholar] [CrossRef]
- Abudayyeh, O.O.; Gootenberg, J.S.; Konermann, S.; Joung, J.; Slaymaker, I.M.; Cox, D.B.T.; Shmakov, S.; Makarova, K.S.; Semenova, E.; Minakhin, L.; et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science 2016, 353, aaf5573. [Google Scholar] [CrossRef] [PubMed]
- Makarova, K.S.; Wolf, Y.I.; Iranzo, J.; Shmakov, S.A.; Alkhnbashi, O.S.; Brouns, S.J.J.; Charpentier, E.; Cheng, D.; Haft, D.H.; Horvath, P.; et al. Evolutionary classification of CRISPR-Cas systems: A burst of class 2 and derived variants. Nat. Rev. Microbiol. 2020, 18, 67–83. [Google Scholar] [CrossRef]
- Aquino-Jarquin, G. CRISPR-Cas14 is now part of the artillery for gene editing and molecular diagnostic. Nanomed. Nanotechnol. Biol. Med. 2019, 18, 428–431. [Google Scholar] [CrossRef]
- Zhou, B.; Yang, R.; Sohail, M.; Kong, X.; Zhang, X.; Fu, N.; Li, B. CRISPR/Cas14 provides a promising platform in facile and versatile aptasensing with improved sensitivity. Talanta 2023, 254, 124120. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.-C.; Cui, Y.-X.; Li, X.-Y.; Sun, G.-Y.; Zhang, Y.-P.; Tang, A.-N.; Kim, K.; Kong, D.-M. Terminal Deoxynucleotidyl Transferase and T7 Exonuclease-Aided Amplification Strategy for Ultrasensitive Detection of Uracil-DNA Glycosylase. Anal. Chem. 2018, 90, 8629–8634. [Google Scholar] [CrossRef]
- Du, Y.-C.; Jiang, H.-X.; Huo, Y.-F.; Han, G.-M.; Kong, D.-M. Optimization of strand displacement amplification-sensitized G-quadruplex DNAzyme-based sensing system and its application in activity detection of uracil-DNA glycosylase. Biosens. Bioelectron. 2016, 77, 971–977. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.-C.; Zhu, L.-N.; Kong, D.-M. Label-free thioflavin T/G-quadruplex-based real-time strand displacement amplification for biosensing applications. Biosens. Bioelectron. 2016, 86, 811–817. [Google Scholar] [CrossRef]
- Lu, Y.; Zhao, H.; Fan, G.-C.; Luo, X. Coupling photoelectrochemical and electrochemical strategies in one probe electrode: Toward sensitive and reliable dual-signal bioassay for uracil-DNA glycosylase activity. Biosens. Bioelectron. 2019, 142, 111569. [Google Scholar] [CrossRef] [PubMed]
- Dong, L.; Zhang, X.; Li, Y.; E, F.; Zhang, J.; Cheng, Y. Highly Sensitive Detection of Uracil-DNA Glycosylase Activity Based on Self-Initiating Multiple Rolling Circle Amplification. ACS Omega 2019, 4, 3881–3886. [Google Scholar] [CrossRef]
- Fowler, J.D.; Suo, Z. Biochemical, Structural, and Physiological Characterization of Terminal Deoxynucleotidyl Transferase. Chem. Rev. 2006, 106, 2092–2110. [Google Scholar] [CrossRef]
- Chen, X.; Cao, G.; Wang, X.; Ji, Z.; Xu, F.; Huo, D.; Luo, X.; Hou, C. Terminal deoxynucleotidyl transferase induced activators to unlock the trans-cleavage of CRISPR/Cpf 1 (TdT-IU- CRISPR/Cpf 1): An ultrasensitive biosensor for Dam MTase activity detection. Biosens. Bioelectron. 2020, 163, 112271. [Google Scholar] [CrossRef]
- Chen, Q.; Tian, T.; Xiong, E.; Wang, P.; Zhou, X. CRISPR/Cas13a Signal Amplification Linked Immunosorbent Assay for Femtomolar Protein Detection. Anal. Chem. 2020, 92, 573–577. [Google Scholar] [CrossRef] [PubMed]
- Li, C.-Y.; Cao, D.; Qi, C.-B.; Kang, Y.-F.; Song, C.-Y.; Xu, D.-D.; Zheng, B.; Pang, D.-W.; Tang, H.-W. Combining Holographic Optical Tweezers with Upconversion Luminescence Encoding: Imaging-Based Stable Suspension Array for Sensitive Responding of Dual Cancer Biomarkers. Anal. Chem. 2018, 90, 2639–2647. [Google Scholar] [CrossRef] [PubMed]
- Li, C.-Y.; Zheng, B.; Liu, Y.-H.; Gao, J.-L.; Zheng, M.-Q.; Pang, D.-W.; Tang, H.-W. A boosting upconversion luminescent resonance energy transfer and biomimetic periodic chip integrated CRISPR/Cas12a biosensor for functional DNA regulated transduction of non-nucleic acid targets. Biosens. Bioelectron. 2020, 169, 112650. [Google Scholar] [CrossRef]
- Zhang, Z.; Shikha, S.; Liu, J.; Zhang, J.; Mei, Q.; Zhang, Y. Upconversion Nanoprobes: Recent Advances in Sensing Applications. Anal. Chem. 2019, 91, 548–568. [Google Scholar] [CrossRef] [PubMed]
- Nimse, S.B.; Sonawane, M.D.; Song, K.-S.; Kim, T. Biomarker detection technologies and future directions. Anal. 2016, 141, 740–755. [Google Scholar] [CrossRef]
- Li, C.-Y.; Zheng, B.; Li, J.-T.; Gao, J.; Liu, Y.-H.; Pang, D.-W.; Tang, H.-W. Holographic Optical Tweezers and Boosting Upconversion Luminescent Resonance Energy Transfer Combined Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)/Cas12a Biosensors. ACS Nano 2021, 15, 8142–8154. [Google Scholar] [CrossRef]
- Stockwell, B.R. Exploring biology with small organic molecules. Nature 2004, 432, 846–854. [Google Scholar] [CrossRef]
- Olatunde, A.; Bahattab, O.; Rauf, A.; Muhammad, N.; Al-Awthan, Y.S.; Tufail, T.; Imran, M.; Mubarak, M.S. Chapter 3—The importance of biological macromolecules in biomedicine. In Biological Macromolecules; Nayak, A.K., Dhara, A.K., Pal, D., Eds.; Academic Press: Cambridge, MA, USA, 2022; pp. 53–68. ISBN 978-0-323-85759-8. [Google Scholar]
- Xu, Z.-H.; Zhao, Z.-Y.; Wang, H.; Wang, S.-M.; Chen, H.-Y.; Xu, J.-J. CRISPR-Cas12a-based efficient electrochemiluminescence biosensor for ATP detection. Anal. Chim. Acta 2021, 1188, 339180. [Google Scholar] [CrossRef]
- Ahmed, M.Z.; Badani, P.; Reddy, R.; Mishra, G. Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)/Cas Advancement in Molecular Diagnostics and Signal Readout Approaches. J. Mol. Diagn. JMD 2021, 23, 1433–1442. [Google Scholar] [CrossRef]
- Broughton, J.P.; Deng, X.; Yu, G.; Fasching, C.L.; Servellita, V.; Singh, J.; Miao, X.; Streithorst, J.A.; Granados, A.; Sotomayor-Gonzalez, A.; et al. CRISPR-Cas12-based detection of SARS-CoV-2. Nat. Biotechnol. 2020, 38, 870–874. [Google Scholar] [CrossRef]
- Azhar, M.; Phutela, R.; Kumar, M.; Ansari, A.H.; Rauthan, R.; Gulati, S.; Sharma, N.; Sinha, D.; Sharma, S.; Singh, S.; et al. Rapid and accurate nucleobase detection using FnCas9 and its application in COVID-19 diagnosis. Biosens. Bioelectron. 2021, 183, 113207. [Google Scholar] [CrossRef]
- Patchsung, M.; Jantarug, K.; Pattama, A.; Aphicho, K.; Suraritdechachai, S.; Meesawat, P.; Sappakhaw, K.; Leelahakorn, N.; Ruenkam, T.; Wongsatit, T.; et al. Clinical validation of a Cas13-based assay for the detection of SARS-CoV-2 RNA. Nat. Biomed. Eng. 2020, 4, 1140–1149. [Google Scholar] [CrossRef]
- Chaibun, T.; Puenpa, J.; Ngamdee, T.; Boonapatcharoen, N.; Athamanolap, P.; O’Mullane, A.P.; Vongpunsawad, S.; Poovorawan, Y.; Lee, S.Y.; Lertanantawong, B. Rapid electrochemical detection of coronavirus SARS-CoV-2. Nat. Commun. 2021, 12, 802. [Google Scholar] [CrossRef] [PubMed]
- Fozouni, P.; Son, S.; Díaz de León Derby, M.; Knott, G.J.; Gray, C.N.; D’Ambrosio, M.V.; Zhao, C.; Switz, N.A.; Kumar, G.R.; Stephens, S.I.; et al. Amplification-free detection of SARS-CoV-2 with CRISPR-Cas13a and mobile phone microscopy. Cell 2021, 184, 323–333.e9. [Google Scholar] [CrossRef] [PubMed]
- Katzmeier, F.; Aufinger, L.; Dupin, A.; Quintero, J.; Lenz, M.; Bauer, L.; Klumpe, S.; Sherpa, D.; Dürr, B.; Honemann, M.; et al. A low-cost fluorescence reader for in vitro transcription and nucleic acid detection with Cas13a. PLoS ONE 2019, 14, e0220091. [Google Scholar] [CrossRef]
- Kleinstiver, B.P.; Prew, M.S.; Tsai, S.Q.; Topkar, V.V.; Nguyen, N.T.; Zheng, Z.; Gonzales, A.P.W.; Li, Z.; Peterson, R.T.; Yeh, J.-R.J.; et al. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature 2015, 523, 481–485. [Google Scholar] [CrossRef]
- Barrangou, R.; Doudna, J.A. Applications of CRISPR technologies in research and beyond. Nat. Biotechnol. 2016, 34, 933–941. [Google Scholar] [CrossRef]



| Classification | Method | Cas Protein Based | Amplification | Analysis | Readout | Sensitivity | Quantification | Time | Portability | Ref. |
|---|---|---|---|---|---|---|---|---|---|---|
| nucleic acid analysis | NASBACC | Cas9 | NASBA | DNA | Fluorescence | aM | N | ~3 h | N | [92] |
| CAS-EXPAR | Cas9 | EXPAR | IncRNA | Fluorescence | aM | N | ~1 h | N | [102] | |
| RACE | Cas9 | RCA | RNA | colorimetric | fM | Y | ~4 h | N | [103] | |
| E-CRISPR | Cas9/Cas12a | N | DNA | electrochemistry | pM | Y | ~1 h | N | [104] | |
| HOLMES | Cas12a | PCR; RT-PCR | DNA/RNA | Fluorescence | aM | N | ~1 h | N | [90] | |
| HOLMESv2 | Cas12b | LAMP | DNA/RNA | Fluorescence | aM | Y | ~1 h | Y | [105] | |
| DETECTR | Cas12a | RPA | DNA | Fluorescence | aM | N | ~2 h | N | [24] | |
| Ultrafast visual detection platform | Cas12a | PCR | DNA | Fluorescence | 1.28 copies | N | ~10 min | Y | [97] | |
| CRISPR/hybrid Cas | Cas12a, Cas13a | N | DNA/RNA | Fluorescence | 10 viral copies/μL | Y | <2 h | N | [106] | |
| SHERLOCK | LwCas13a | RPA | DNA/RNA | Fluorescence | aM | N | 2–5 h | Y | [30] | |
| HUDSON + SHERLOCK | LwCas13a | RPA | RNA/DNA | Fluorescence | aM | N | ~2 h | Y | [91] | |
| DETECTR-Cas12f | Cas12f | RPA | DNA | Fluorescence | aM | N | ~2 h | N | [107] | |
| protein analysis | TdT-combined CRISPR/Cas12a amplification | Cas12 | N | UDG | Fluorescence | uM | Y | ~4 h | N | [108] |
| CLISA | Cas13 | N | IL-6, VEGF | Fluorescence | ng/Ml | Y | - | N | [109] | |
| E-CRISPR | Cas12a | LAMP | Protein | electrochemistry | nM | Y | ~1 h | Y | [110] | |
| ATP analysis | CRISPR-LbCas12a biosensor | LbCas12a | N | ATP | Fluorescence | μM | Y | 40 min | Y | [100] |
| ASD-Cas12a | LbCas12a | SDA | ATP | Fluorescence | μM | Y | ~20 min | N | [101] | |
| Molecular Radar | Cas12a | N | ATP | Fluorescence | nM | Y | ~25 min | Y | [99] | |
| BPE-ECL | Cas12a | N | ATP | electrochemistry | nM | Y | ~2 h | N | [111] | |
| BA-CASLFA | Cas12a | N | ATP | Fluorescence | μM | Y | ~26 min | N | [112] | |
| dsDNA-ZIF-90@Ag3AuS2@Fe3O4 nanoplatform | Cas12a | N | ATP, ADP | Fluorescence | nM | Y | ~30 min | N | [113] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, S.; Dai, R.; Zhang, Z.; Zhang, H.; Zhang, M.; Li, Z.; Zhao, K.; Xiong, W.; Cheng, S.; Wang, B.; et al. CRISPR/Cas-Based Techniques for Live-Cell Imaging and Bioanalysis. Int. J. Mol. Sci. 2023, 24, 13447. https://doi.org/10.3390/ijms241713447
Huang S, Dai R, Zhang Z, Zhang H, Zhang M, Li Z, Zhao K, Xiong W, Cheng S, Wang B, et al. CRISPR/Cas-Based Techniques for Live-Cell Imaging and Bioanalysis. International Journal of Molecular Sciences. 2023; 24(17):13447. https://doi.org/10.3390/ijms241713447
Chicago/Turabian StyleHuang, Shuo, Rui Dai, Zhiqi Zhang, Han Zhang, Meng Zhang, Zhangjun Li, Kangrui Zhao, Wenjun Xiong, Siyu Cheng, Buhua Wang, and et al. 2023. "CRISPR/Cas-Based Techniques for Live-Cell Imaging and Bioanalysis" International Journal of Molecular Sciences 24, no. 17: 13447. https://doi.org/10.3390/ijms241713447
APA StyleHuang, S., Dai, R., Zhang, Z., Zhang, H., Zhang, M., Li, Z., Zhao, K., Xiong, W., Cheng, S., Wang, B., & Wan, Y. (2023). CRISPR/Cas-Based Techniques for Live-Cell Imaging and Bioanalysis. International Journal of Molecular Sciences, 24(17), 13447. https://doi.org/10.3390/ijms241713447





