Identification of the HNSC88 Molecular Signature for Predicting Subtypes of Head and Neck Cancer
Abstract
:1. Introduction
2. Results
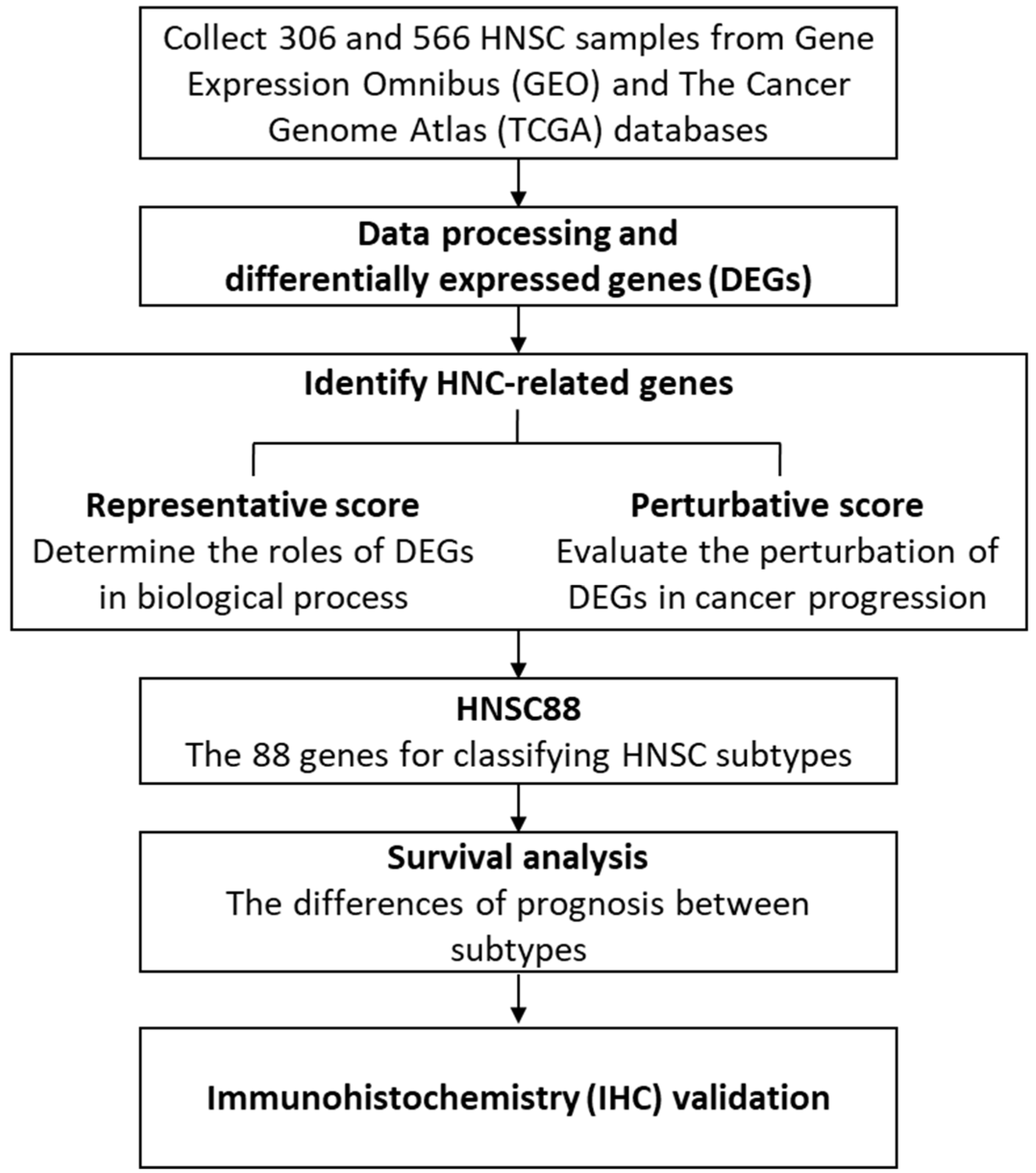
2.1. Identification of Molecular Signature for HNSC Using CS, RS, and PS Scoring
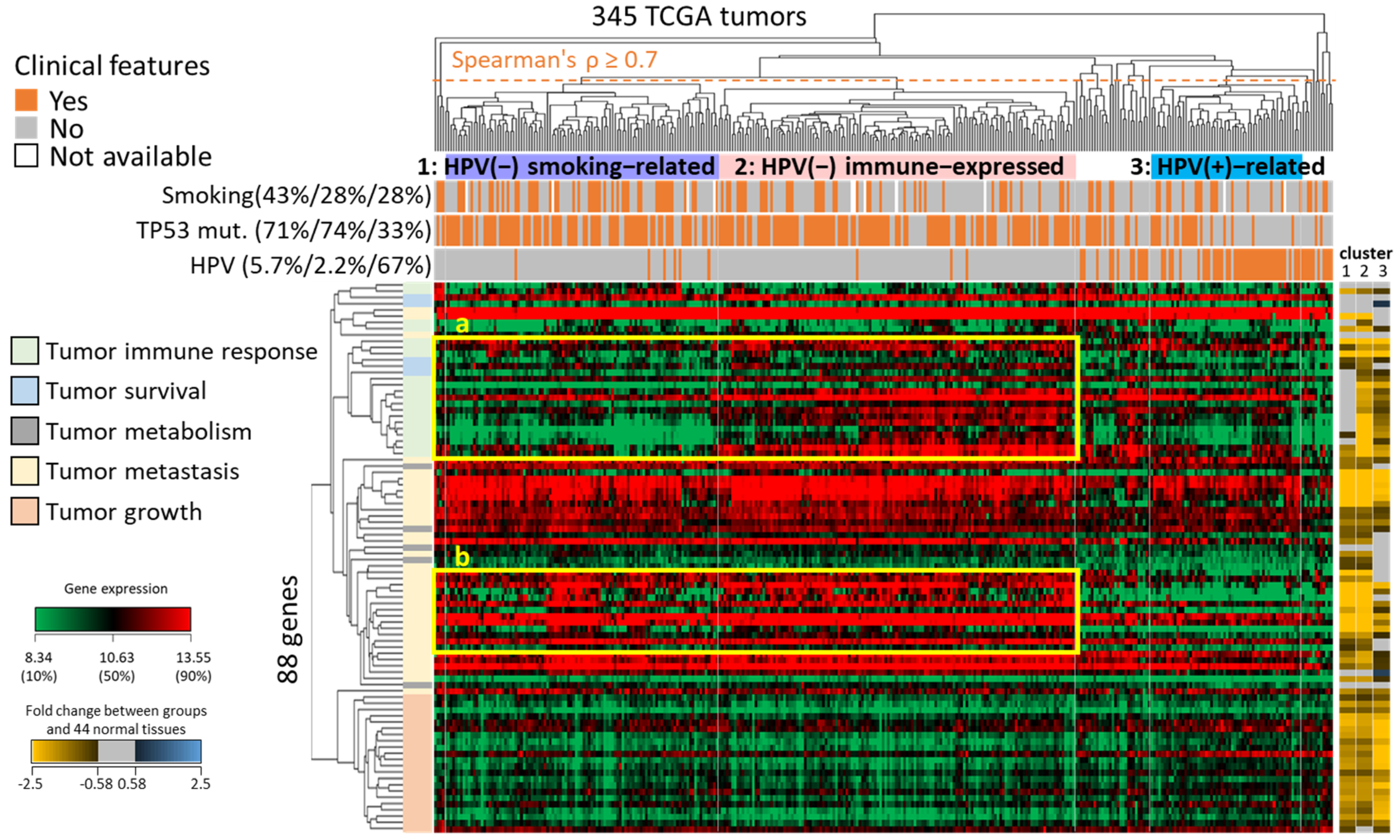
2.2. Unsupervised Clustering of HNSC Subtypes Based on the Expression of HNSC88
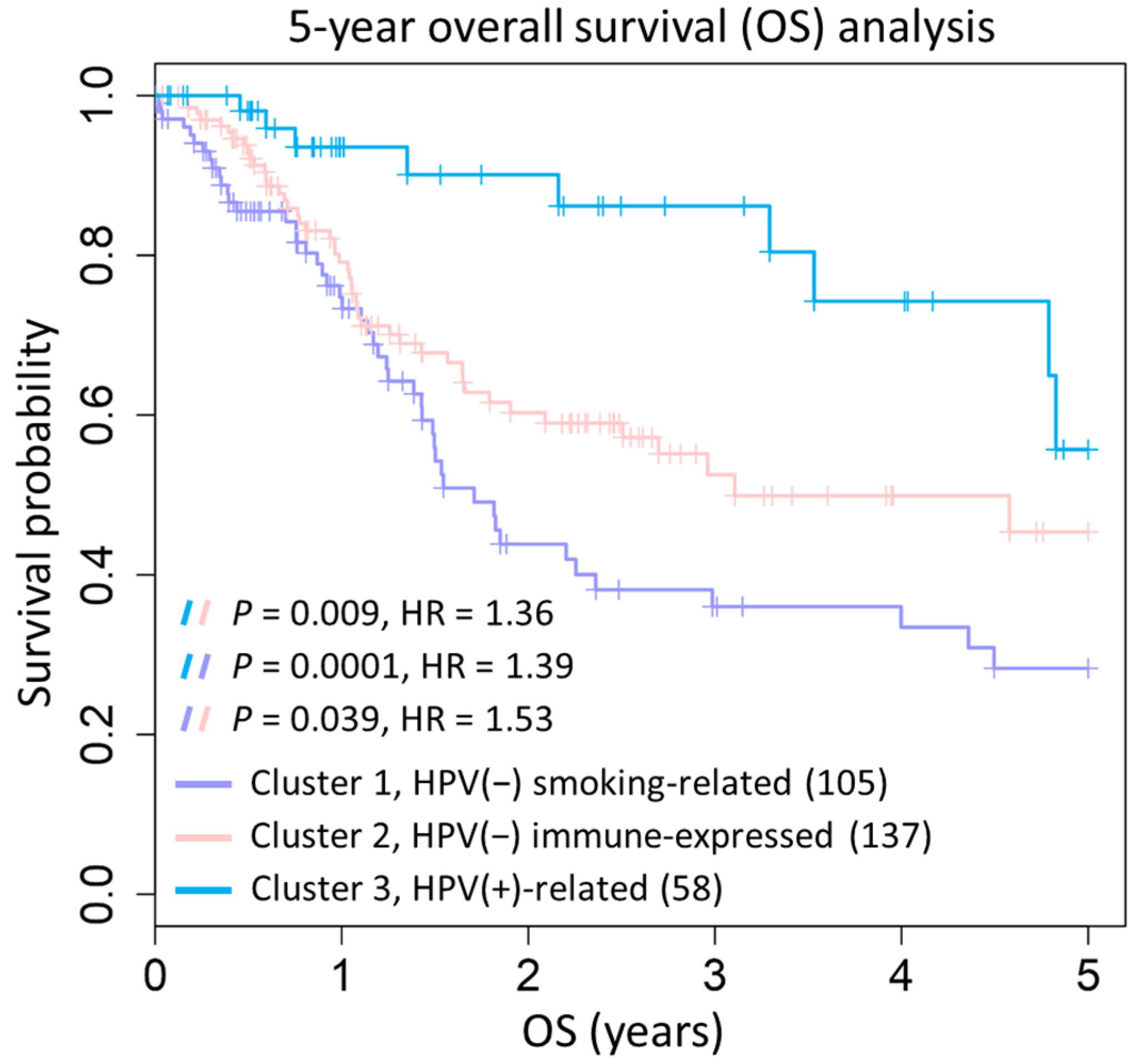
2.3. Prognostic Significance of Three HNSC Subtypes
2.4. Immunohistochemistry (IHC) Stain of Clinical HNSC Patients
3. Methods
3.1. Datasets
3.2. Identification of Differentially Expressed Genes (DEGs)
3.3. Functional Enrichment Analysis
3.4. The Scoring Methods for Identification of HNSC-Related Genes
3.5. Survival Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Leemans, C.R.; Braakhuis, B.J.M.; Brakenhoff, R.H. The molecular biology of head and neck cancer. Nat. Rev. Cancer 2011, 11, 9–22. [Google Scholar] [CrossRef] [PubMed]
- Leemans, C.R.; Snijders, P.J.F.; Brakenhoff, R.H. The molecular landscape of head and neck cancer. Nat. Rev. Cancer 2018, 18, 269–282. [Google Scholar] [CrossRef] [PubMed]
- Suh, Y.; Amelio, I.; Urbano, T.G.; Tavassoli, M. Clinical update on cancer: Molecular oncology of head and neck cancer. Cell Death Dis. 2014, 5, e1018. [Google Scholar] [CrossRef] [PubMed]
- Mroz, E.A.; Tward, A.M.; Hammon, R.J.; Ren, Y.; Rocco, J.W. Intra-tumor Genetic Heterogeneity and Mortality in Head and Neck Cancer: Analysis of Data from the Cancer Genome Atlas. PLoS Med. 2015, 12, e1001786. [Google Scholar] [CrossRef] [PubMed]
- Keren, S.; Shoude, Z.; Lu, Z.; Beibei, Y. Role of EGFR as a prognostic factor for survival in head and neck cancer: A meta-analysis. Tumor Biol. 2014, 35, 2285–2295. [Google Scholar] [CrossRef] [PubMed]
- Johnson, D.E.; Burtness, B.; Leemans, C.R.; Lui, V.W.Y.; Bauman, J.E.; Grandis, J.R. Head and Neck Squamous Cell Carcinoma. Nat. Rev. Dis. Primers 2020, 6, 92. [Google Scholar] [CrossRef] [PubMed]
- Madu, C.O.; Lu, Y. Novel diagnostic biomarkers for prostate cancer. J. Cancer 2010, 1, 150–177. [Google Scholar] [CrossRef]
- D’Amico, A.V.; Chen, M.-H.; Roehl, K.A.; Catalona, W.J. Preoperative PSA Velocity and the Risk of Death from Prostate Cancer after Radical Prostatectomy. N. Engl. J. Med. 2004, 351, 125–135. [Google Scholar] [CrossRef]
- Sørlie, T.; Perou, C.M.; Tibshirani, R.; Aas, T.; Geisler, S.; Johnsen, H.; Hastie, T.; Eisen, M.B.; van de Rijn, M.; Jeffrey, S.S.; et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl. Acad. Sci. USA 2001, 98, 10869–10874. [Google Scholar] [CrossRef]
- Hammond, M.E.H.; Hayes, D.F.; Dowsett, M.; Allred, D.C.; Hagerty, K.L.; Badve, S.; Fitzgibbons, P.L.; Francis, G.; Goldstein, N.S.; Hayes, M.; et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer (unabridged version). Arch. Pathol. Lab. Med. 2010, 134, e48–e72. [Google Scholar] [CrossRef] [PubMed]
- Parker, J.S.; Mullins, M.; Cheang, M.C.; Leung, S.; Voduc, D.; Vickery, T.; Davies, S.; Fauron, C.; He, X.; Hu, Z.; et al. Supervised Risk Predictor of Breast Cancer Based on Intrinsic Subtypes. J. Clin. Oncol. 2009, 27, 1160–1167. [Google Scholar] [CrossRef] [PubMed]
- Bastien, R.R.; Rodriguez-Lescure, A.; Ebbert, M.T.; Prat, A.; Munarriz, B.; Rowe, L.; Miller, P.; Ruiz-Borrego, M.; Anderson, D.; Lyons, B.; et al. Pam50 Breast Cancer Subtyping by Rt-Qpcr and Concordance with Standard Clinical Molecular Markers. BMC Med. Genom. 2012, 5, 44. [Google Scholar] [CrossRef] [PubMed]
- Van’t Veer, L.J.; Dai, H.; van de Vijver, M.J.; He, Y.D.; Hart, A.A.; Mao, M.; Peterse, H.L.; van der Kooy, K.; Marton, M.J.; Witteveen, A.T.; et al. Gene Expression Profiling Predicts Clinical Outcome of Breast Cancer. Nature 2002, 415, 530–536. [Google Scholar] [CrossRef] [PubMed]
- Walter, V.; Yin, X.; Wilkerson, M.D.; Cabanski, C.R.; Zhao, N.; Du, Y.; Ang, M.K.; Hayward, M.C.; Salazar, A.H.; Hoadley, K.A.; et al. Molecular Subtypes in Head and Neck Cancer Exhibit Distinct Patterns of Chromosomal Gain and Loss of Canonical Cancer Genes. PLoS ONE 2013, 8, e56823. [Google Scholar] [CrossRef]
- Tang, K.-W.; Alaei-Mahabadi, B.; Samuelsson, T.; Lindh, M.; Larsson, E. The landscape of viral expression and host gene fusion and adaptation in human cancer. Nat. Commun. 2013, 4, 2513. [Google Scholar] [CrossRef] [PubMed]
- Sopori, M. Effects of cigarette smoke on the immune system. Nat. Rev. Immunol. 2002, 2, 372–377. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Charlesworth, J.C.; Curran, J.E.; Johnson, M.P.; Göring, H.H.; Dyer, T.D.; Diego, V.P.; Kent, J.W.; Mahaney, M.C.; Almasy, L.; MacCluer, J.W.; et al. Transcriptomic epidemiology of smoking: The effect of smoking on gene expression in lymphocytes. BMC Med. Genom. 2010, 3, 29. [Google Scholar] [CrossRef]
- De la Iglesia, J.V.; Slebos, R.J.C.; Martin-Gomez, L.; Wang, X.; Teer, J.K.; Tan, A.C.; Gerke, T.A.; Aden-Buie, G.; van Veen, T.; Masannat, J.; et al. Effects of Tobacco Smoking on the Tumor Immune Microenvironment in Head and Neck Squamous Cell Carcinoma. Clin. Cancer Res. 2020, 26, 1474–1485. [Google Scholar] [CrossRef]
- Kumar, B.; Cordell, K.G.; Lee, J.S.; Worden, F.P.; Prince, M.E.; Tran, H.H.; Wolf, G.T.; Urba, S.G.; Chepeha, D.B.; Teknos, T.N.; et al. EGFR, p16, HPV Titer, Bcl-xL and p53, Sex, and Smoking as Indicators of Response to Therapy and Survival in Oropharyngeal Cancer. J. Clin. Oncol. 2008, 26, 3128–3137. [Google Scholar] [CrossRef] [PubMed]
- Karam-Hage, M.; Cinciripini, P.M.; Gritz, E.R. Tobacco use and cessation for cancer survivors: An overview for clinicians. CA Cancer J. Clin. 2014, 64, 272–290. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J. Diagnostic value of MCM2 immunocytochemical staining in cervical lesions and its relationship with HPV infection. Int. J. Clin. Exp. Pathol. 2015, 8, 875–880. [Google Scholar] [PubMed]
- Pietruszewska, W.; Bojanowska-Poźniak, K.; Kobos, J. Matrix metalloproteinases MMP1, MMP2, MMP9 and their tissue inhibitors TIMP1, TIMP2, TIMP3 in head and neck cancer: An immunohistochemical study. Otolaryngol. Polska 2016, 70, 29–40. [Google Scholar] [CrossRef] [PubMed]
- Van Tubergen, E.A.; Banerjee, R.; Liu, M.; Broek, R.V.; Light, E.; Kuo, S.; Feinberg, S.E.; Willis, A.L.; Wolf, G.; Carey, T.; et al. Inactivation or Loss of TTP Promotes Invasion in Head and Neck Cancer via Transcript Stabilization and Secretion of MMP9, MMP2, and IL-6. Clin. Cancer Res. 2013, 19, 1169–1179. [Google Scholar] [CrossRef] [PubMed]
- Muthuswamy, R.; Corman, J.M.; Dahl, K.; Chatta, G.S.; Kalinski, P. Functional reprogramming of human prostate cancer to promote local attraction of effector CD8+T cells. Prostate 2016, 76, 1095–1105. [Google Scholar] [CrossRef]
- Lomnytska, M.I.; Becker, S.; Bodin, I.; Olsson, A.; Hellman, K.; Hellström, A.-C.; Mints, M.; Hellman, U.; Auer, G.; Andersson, S. Differential expression of ANXA6, HSP27, PRDX2, NCF2, and TPM4 during uterine cervix carcinogenesis: Diagnostic and prognostic value. Br. J. Cancer 2011, 104, 110–119. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Chiappinelli, K.B.; Guzzetta, A.A.; Easwaran, H.; Yen, R.-W.C.; Vatapalli, R.; Topper, M.J.; Luo, J.; Connolly, R.M.; Azad, N.S.; et al. Immune regulation by low doses of the DNA methyltransferase inhibitor 5-azacitidine in common human epithelial cancers. Oncotarget 2014, 5, 587–598. [Google Scholar] [CrossRef]
- Wenzel, J.; Tomiuk, S.; Zahn, S.; Küsters, D.; Vahsen, A.; Wiechert, A.; Mikus, S.; Birth, M.; Scheler, M.; von Bubnoff, D.; et al. Transcriptional profiling identifies an interferon-associated host immune response in invasive squamous cell carcinoma of the skin. Int. J. Cancer 2008, 123, 2605–2615. [Google Scholar] [CrossRef]
- Zhu, C.; Chen, X.; Guan, G.; Zou, C.; Guo, Q.; Cheng, P.; Cheng, W.; Wu, A. IFI30 Is a Novel Immune-Related Target with Predicting Value of Prognosis and Treatment Response in Glioblastoma. OncoTargets Ther. 2020, 13, 1129–1143. [Google Scholar] [CrossRef]
- Italiano, D.; Lena, A.M.; Melino, G.; Candi, E. Identification of Ncf2/P67phox as a Novel P53 Target Gene. Cell Cycle 2012, 11, 4589–4596. [Google Scholar] [CrossRef]
- Chen, D.-T.; Nasir, A.; Culhane, A.; Venkataramu, C.; Fulp, W.; Rubio, R.; Wang, T.; Agrawal, D.; McCarthy, S.M.; Gruidl, M.; et al. Proliferative genes dominate malignancy-risk gene signature in histologically-normal breast tissue. Breast Cancer Res. Treat. 2010, 119, 335–346. [Google Scholar] [CrossRef] [PubMed]
- Pyeon, D.; Newton, M.A.; Lambert, P.F.; den Boon, J.A.; Sengupta, S.; Marsit, C.J.; Woodworth, C.D.; Connor, J.P.; Haugen, T.H.; Smith, E.M.; et al. Fundamental Differences in Cell Cycle Deregulation in Human Papillomavirus-Positive and Human Papillomavirus-Negative Head/Neck and Cervical Cancers. Cancer Res. 2007, 67, 4605–4619. [Google Scholar] [CrossRef]
- Ye, H.; Yu, T.; Temam, S.; Ziober, B.L.; Wang, J.; Schwartz, J.L.; Mao, L.; Wong, D.T.; Zhou, X. Transcriptomic dissection of tongue squamous cell carcinoma. BMC Genom. 2008, 9, 69. [Google Scholar] [CrossRef] [PubMed]
- Gautier, L.; Cope, L.; Bolstad, B.M.; Irizarry, R.A. affy—Analysis of Affymetrix Genechip Data at the Probe Level. Bioinformatics 2004, 20, 307–315. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Network. Comprehensive Genomic Characterization of Head and Neck Squamous Cell Carcinomas. Nature 2015, 517, 576–582. [Google Scholar] [CrossRef] [PubMed]
- Stark, C.; Breitkreutz, B.-J.; Chatr-Aryamontri, A.; Boucher, L.; Oughtred, R.; Livstone, M.S.; Nixon, J.; Van Auken, K.; Wang, X.; Shi, X.; et al. The BioGRID Interaction Database: 2011 update. Nucleic Acids Res. 2010, 39, D698–D704. [Google Scholar] [CrossRef]
- Xenarios, I.; Salwínski, L.; Duan, X.J.; Higney, P.; Kim, S.-M.; Eisenberg, D. DIP, the Database of Interacting Proteins: A research tool for studying cellular networks of protein interactions. Nucleic Acids Res. 2002, 30, 303–305. [Google Scholar] [CrossRef]
- Mewes, H.W.; Dietmann, S.; Frishman, D.; Gregory, R.; Mannhaupt, G.; Mayer, K.F.X.; Munsterkotter, M.; Ruepp, A.; Spannagl, M.; Stumpflen, V.; et al. MIPS: Analysis and annotation of genome information in 2007. Nucleic Acids Res. 2008, 36, D196–D201. [Google Scholar] [CrossRef]
- Ceol, A.; Aryamontri, A.C.; Licata, L.; Peluso, D.; Briganti, L.; Perfetto, L.; Castagnoli, L.; Cesareni, G. MINT, the molecular interaction database: 2009 update. Nucleic Acids Res. 2010, 38, D532–D539. [Google Scholar] [CrossRef]
- Aranda, B.; Achuthan, P.; Alam-Faruque, Y.; Armean, I.; Bridge, A.; Derow, C.; Feuermann, M.; Ghanbarian, A.T.; Kerrien, S.; Khadake, J.; et al. The IntAct molecular interaction database in 2010. Nucleic Acids Res. 2010, 38, D525–D531. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-C.; Lo, Y.-S.; Hsu, W.-C.; Yang, J.-M. 3D-partner: A web server to infer interacting partners and binding models. Nucleic Acids Res. 2007, 35, W561–W567. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.-C.; Lin, C.-Y.; Lo, Y.-S.; Yang, J.-M. PPISearch: A web server for searching homologous protein–protein interactions across multiple species. Nucleic Acids Res. 2009, 37, W369–W375. [Google Scholar] [CrossRef] [PubMed]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed]
- The Gene Ontology Consortium. The Gene Ontology Resource: 20 Years and Still Going Strong. Nucleic Acids Res. 2019, 47, D330–D338. [CrossRef] [PubMed]
- Kuperstein, I.; Bonnet, E.; Nguyen, H.-A.; Cohen, D.; Viara, E.; Grieco, L.; Fourquet, S.; Calzone, L.; Russo, C.; Kondratova, M.; et al. Atlas of Cancer Signalling Network: A systems biology resource for integrative analysis of cancer data with Google Maps. Oncogenesis 2015, 4, e160. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Zhu, L.; Guo, J.; Zhang, D.-Y.; Lin, K. Prediction of yeast protein-protein interaction network: Insights from the Gene Ontology and annotations. Nucleic Acids Res. 2006, 34, 2137–2150. [Google Scholar] [CrossRef] [PubMed]
- Mihály, Z.; Kormos, M.; Lánczky, A.; Dank, M.; Budczies, J.; Szász, M.A.; Győrffy, B. A meta-analysis of gene expression-based biomarkers predicting outcome after tamoxifen treatment in breast cancer. Breast Cancer Res. Treat. 2013, 140, 219–232. [Google Scholar] [CrossRef]
- Desrichard, A.; Kuo, F.; Chowell, D.; Lee, K.-W.; Riaz, N.; Wong, R.J.; Chan, T.A.; Morris, L.G.T. Tobacco Smoking-Associated Alterations in the Immune Microenvironment of Squamous Cell Carcinomas. J. Natl. Cancer Inst. 2018, 110, 1386–1392. [Google Scholar] [CrossRef]


| Cluster Group # | Members | GOBP | KEGG Pathways | Summary of Pathways | Cancer-Related Processes | The Hallmarks of Cancer (Summary of Processes) |
|---|---|---|---|---|---|---|
| G1 | APOBEC3A, CMPK2, DTX3L, HERC5, IFI30, STAT1 |
| - | - | - | - |
| G2 | CXCL1, IL1F9, IL8, MMP12, MMP9 |
|
|
|
|
|
| G3 | DDX60, GBP1, IFI35, IFI6, IFIT1, IFIT3, ISG15, RSAD2, UBE2L6 |
|
|
| - | - |
| G4 | CTSL1, NCF2, RGS2, SOD2, TLR2 |
|
|
| - | - |
| G5 | COL4A1, COL4A2, COL5A1, COL5A2, CTHRC1, PXDN |
|
|
|
|
|
| G6 | INHBA, LAMC2, MMP1, MMP10, MMP3, PLAU, PTHLH, TGFBI |
|
|
|
|
|
| G7 | ACVR1, CLIC4, DSG2, FAT1, FNDC3B, ITGAV, SNAI2 |
| - | - |
|
|
| G8 | CDC45, CDC6, CDK4, CHEK1, CKS1B, DTL, FEN1, FOXM1, GINS1, MCM2, RFC4, TPX2, UHRF1 |
|
|
|
|
|
| G9 | AURKA, BIRC5, CDC20, CEP55, ECT2, KIF2C, KIF4A, NUP155, TRIP13 |
| - | - | - | - |
| G10 | CDH3, CEBPB, DFNA5, GJA1, HOMER3, JUP, KLF10, KLF7, KRT17, MSN, MYO1B, PANX1, PRNP, RRAS2, SLC16A1 |
|
|
|
|
|
| G11 | ACOT9, FKBP9, GALNT2, UBE2Q2, VKORC1 |
| - | - | - | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chuang, Y.-H.; Lin, C.-Y.; Lee, J.-C.; Lee, C.-H.; Liu, C.-L.; Huang, S.-H.; Lee, J.-Y.; Lai, W.-S.; Yang, J.-M. Identification of the HNSC88 Molecular Signature for Predicting Subtypes of Head and Neck Cancer. Int. J. Mol. Sci. 2023, 24, 13068. https://doi.org/10.3390/ijms241713068
Chuang Y-H, Lin C-Y, Lee J-C, Lee C-H, Liu C-L, Huang S-H, Lee J-Y, Lai W-S, Yang J-M. Identification of the HNSC88 Molecular Signature for Predicting Subtypes of Head and Neck Cancer. International Journal of Molecular Sciences. 2023; 24(17):13068. https://doi.org/10.3390/ijms241713068
Chicago/Turabian StyleChuang, Yi-Hsuan, Chun-Yu Lin, Jih-Chin Lee, Chia-Hwa Lee, Chia-Lin Liu, Sing-Han Huang, Jung-Yu Lee, Wen-Sen Lai, and Jinn-Moon Yang. 2023. "Identification of the HNSC88 Molecular Signature for Predicting Subtypes of Head and Neck Cancer" International Journal of Molecular Sciences 24, no. 17: 13068. https://doi.org/10.3390/ijms241713068
APA StyleChuang, Y.-H., Lin, C.-Y., Lee, J.-C., Lee, C.-H., Liu, C.-L., Huang, S.-H., Lee, J.-Y., Lai, W.-S., & Yang, J.-M. (2023). Identification of the HNSC88 Molecular Signature for Predicting Subtypes of Head and Neck Cancer. International Journal of Molecular Sciences, 24(17), 13068. https://doi.org/10.3390/ijms241713068






