Proteomic Analysis of Mesenchymal Stromal Cells Secretome in Comparison to Leukocyte- and Platelet-Rich Fibrin
Abstract
:1. Introduction
2. Results
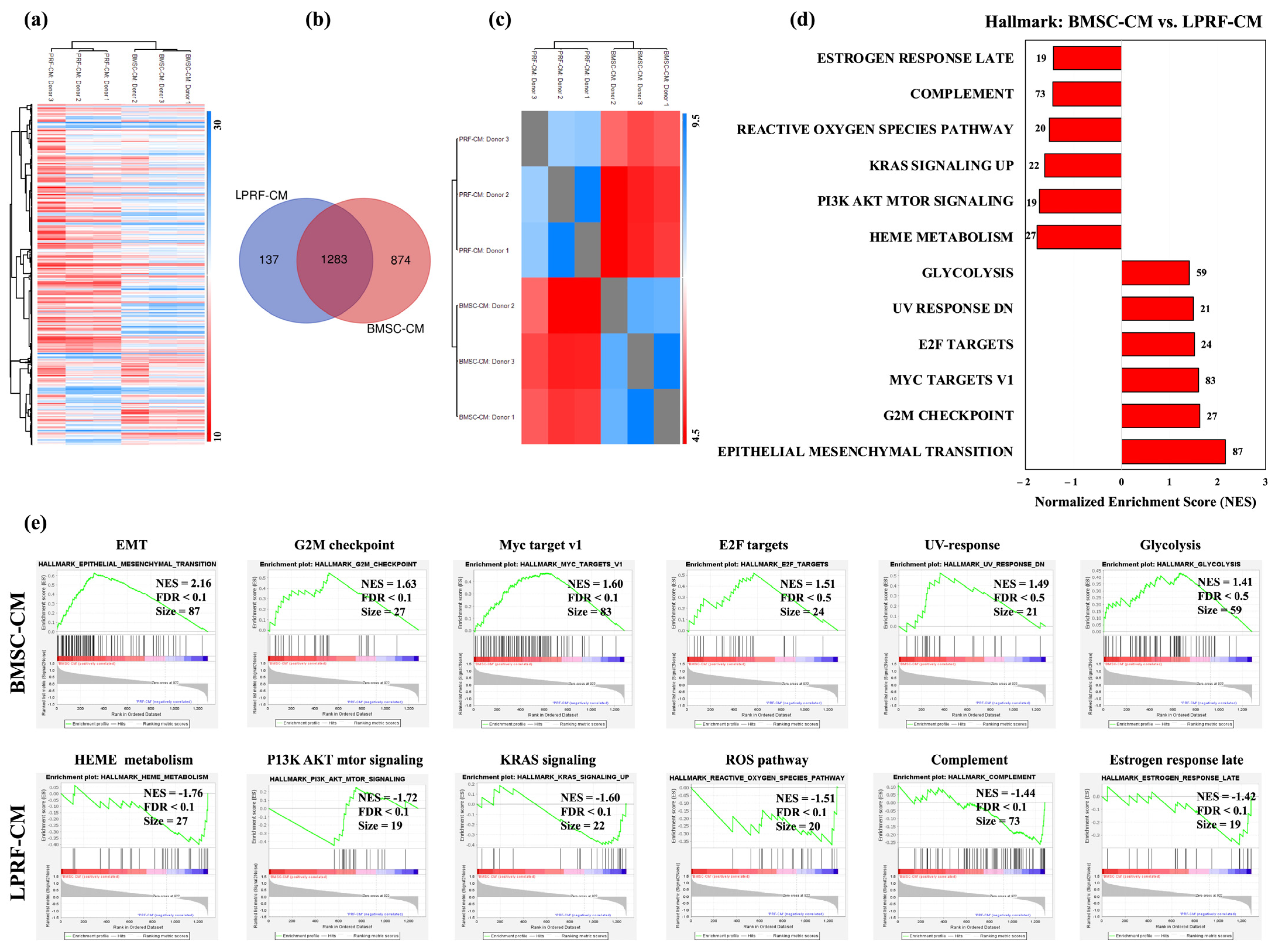
2.1. Proteomic Profiles of BMSC-CM and LPRF-CM
2.2. Similarities and Differences between BMSC and L-PRF Secretomes
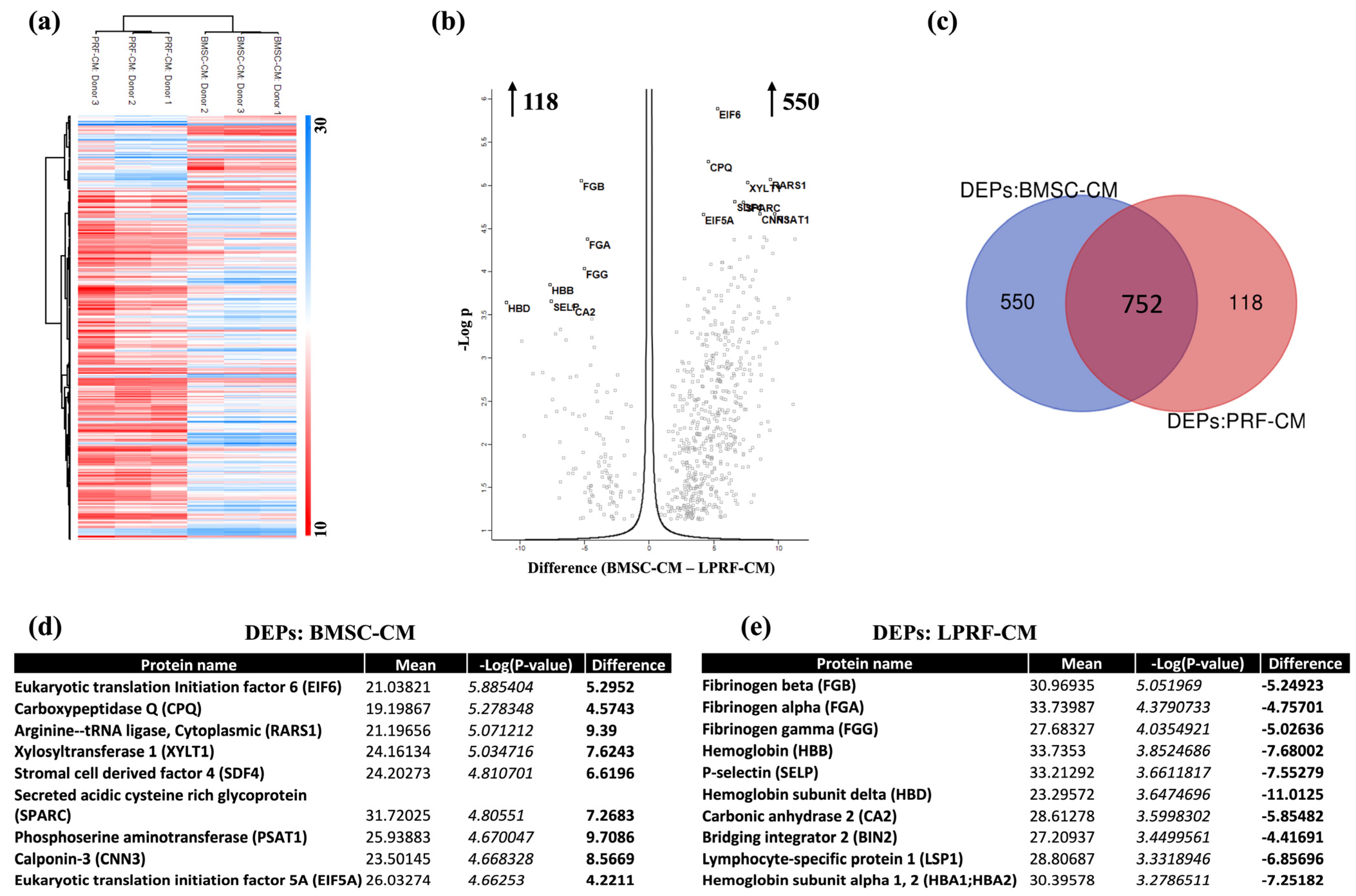
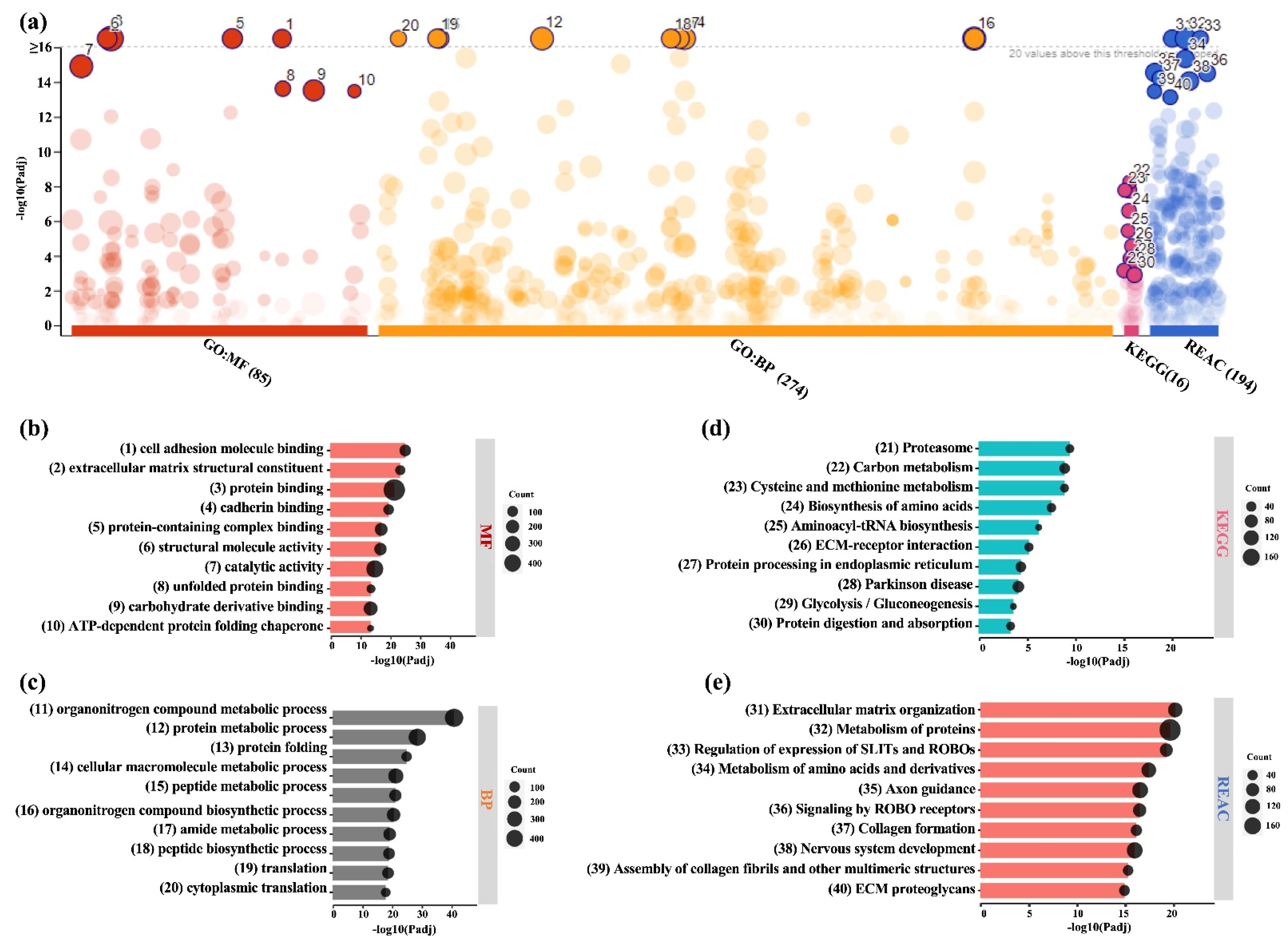
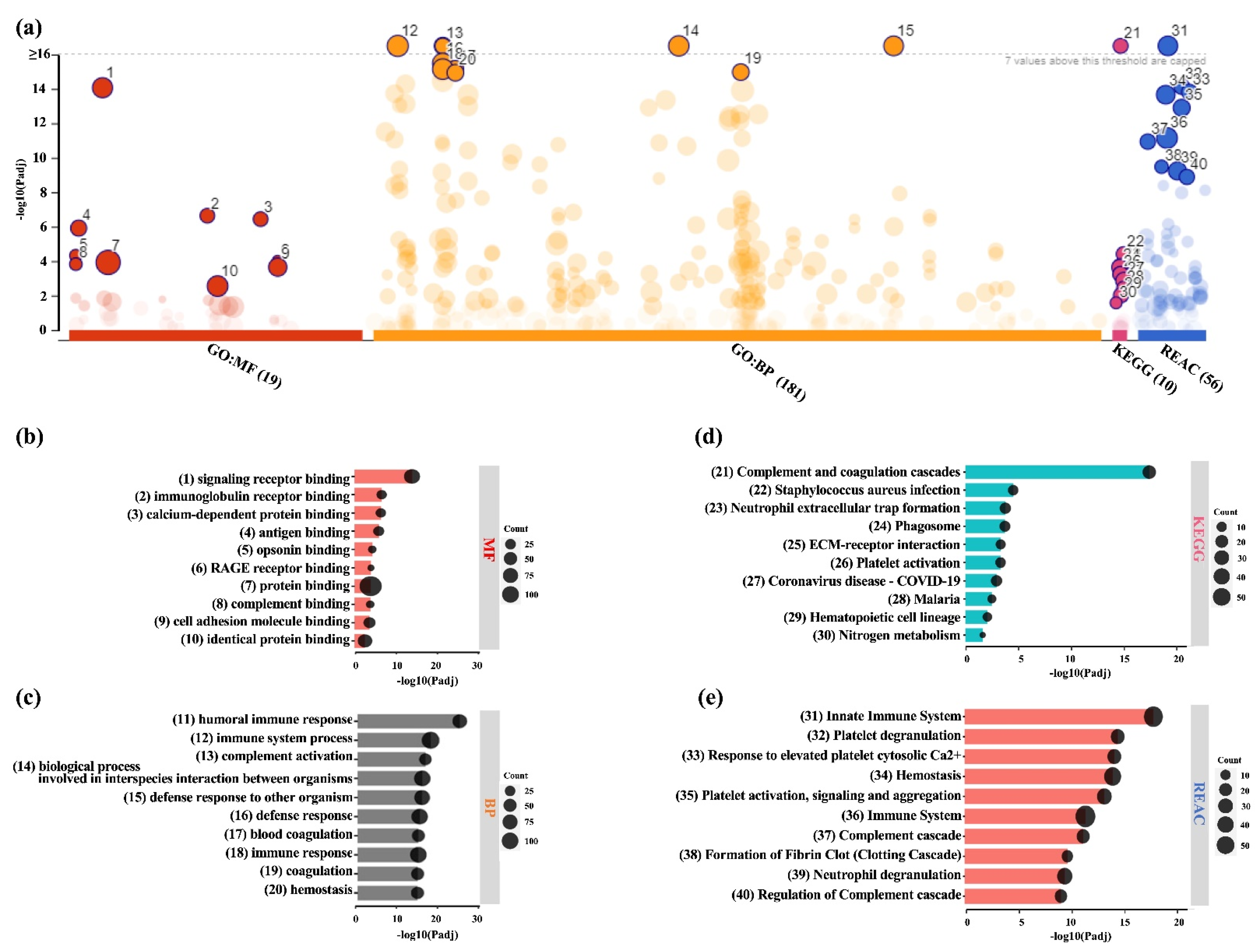
2.3. Analysis of DEPs in BMSC-CM and LPRF-CM
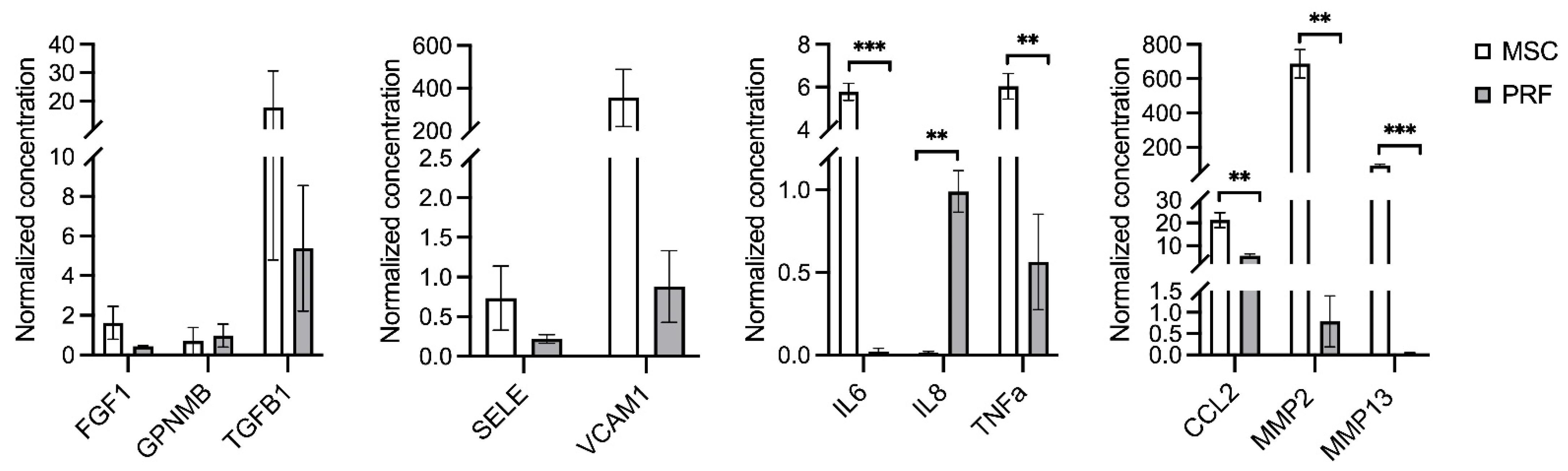
2.4. Multiplex Immunoassay
3. Discussion
4. Materials and Methods
4.1. Cell Culture and Preparation of BMSC-CM
4.2. Preparation of L-PRF and LPRF-CM
4.3. CM Ultrafiltration
4.4. Liquid Chromatography with Tandem Mass Spectrometry (LC-MS/MS)
4.5. Bioinformatic Analysis
4.6. Multiplex Immunoassay
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sanz, M.; Dahlin, C.; Apatzidou, D.; Artzi, Z.; Bozic, D.; Calciolari, E.; De Bruyn, H.; Dommisch, H.; Donos, N.; Eickholz, P.; et al. Biomaterials and regenerative technologies used in bone regeneration in the craniomaxillofacial region: Consensus report of group 2 of the 15th European Workshop on Periodontology on Bone Regeneration. J. Clin. Periodontol. 2019, 46 (Suppl. 21), 82–91. [Google Scholar] [CrossRef] [PubMed]
- Stavropoulos, A.; Bertl, K.; Spineli, L.M.; Sculean, A.; Cortellini, P.; Tonetti, M. Medium- and long-term clinical benefits of periodontal regenerative/reconstructive procedures in intrabony defects: Systematic review and network meta-analysis of randomized controlled clinical studies. J. Clin. Periodontol. 2021, 48, 410–430. [Google Scholar] [CrossRef] [PubMed]
- Atala, A.; Irvine, D.J.; Moses, M.; Shaunak, S. Wound Healing Versus Regeneration: Role of the Tissue Environment in Regenerative Medicine. MRS Bull. 2010, 35, 597–606. [Google Scholar] [CrossRef]
- Md Fadilah, N.I.; Mohd Abdul Kader Jailani, M.S.; Badrul Hisham, M.A.I.; Sunthar Raj, N.; Shamsuddin, S.A.; Ng, M.H.; Fauzi, M.B.; Maarof, M. Cell secretomes for wound healing and tissue regeneration: Next generation acellular based tissue engineered products. J. Tissue Eng. 2022, 13, 20417314221114273. [Google Scholar] [CrossRef]
- Gosain, A.; DiPietro, L.A. Aging and wound healing. World J. Surg. 2004, 28, 321–326. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.; Dipietro, L.A. Factors affecting wound healing. J. Dent. Res. 2010, 89, 219–229. [Google Scholar] [CrossRef] [PubMed]
- Farmani, A.R.; Nekoofar, M.H.; Ebrahimi Barough, S.; Azami, M.; Rezaei, N.; Najafipour, S.; Ai, J. Application of Platelet Rich Fibrin in Tissue Engineering: Focus on Bone Regeneration. Platelets 2021, 32, 183–188. [Google Scholar] [CrossRef]
- Guillamat-Prats, R. The Role of MSC in Wound Healing, Scarring and Regeneration. Cells 2021, 10, 1729. [Google Scholar] [CrossRef]
- Donos, N.; Dereka, X.; Calciolari, E. The use of bioactive factors to enhance bone regeneration: A narrative review. J. Clin. Periodontol. 2019, 46 (Suppl. 21), 124–161. [Google Scholar] [CrossRef]
- Avila-Ortiz, G.; Ambruster, J.; Barootchi, S.; Chambrone, L.; Chen, C.Y.; Dixon, D.R.; Geisinger, M.L.; Giannobile, W.V.; Goss, K.; Gunsolley, J.C.; et al. American Academy of Periodontology best evidence consensus statement on the use of biologics in clinical practice. J. Periodontol. 2022, 93, 1763–1770. [Google Scholar] [CrossRef]
- Castro, A.B.; Meschi, N.; Temmerman, A.; Pinto, N.; Lambrechts, P.; Teughels, W.; Quirynen, M. Regenerative potential of leucocyte- and platelet-rich fibrin. Part A: Intra-bony defects, furcation defects and periodontal plastic surgery. A systematic review and meta-analysis. J. Clin. Periodontol. 2017, 44, 67–82. [Google Scholar] [CrossRef] [PubMed]
- Castro, A.B.; Meschi, N.; Temmerman, A.; Pinto, N.; Lambrechts, P.; Teughels, W.; Quirynen, M. Regenerative potential of leucocyte- and platelet-rich fibrin. Part B: Sinus floor elevation, alveolar ridge preservation and implant therapy. A systematic review. J. Clin. Periodontol. 2017, 44, 225–234. [Google Scholar] [CrossRef] [PubMed]
- Dohan, D.M.; Choukroun, J.; Diss, A.; Dohan, S.L.; Dohan, A.J.; Mouhyi, J.; Gogly, B. Platelet-rich fibrin (PRF): A second-generation platelet concentrate. Part I: Technological concepts and evolution. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endod. 2006, 101, E37–E44. [Google Scholar] [CrossRef]
- Dohan Ehrenfest, D.M.; Del Corso, M.; Diss, A.; Mouhyi, J.; Charrier, J.B. Three-dimensional architecture and cell composition of a Choukroun’s platelet-rich fibrin clot and membrane. J. Periodontol. 2010, 81, 546–555. [Google Scholar] [CrossRef] [PubMed]
- Di Liddo, R.; Bertalot, T.; Borean, A.; Pirola, I.; Argentoni, A.; Schrenk, S.; Cenzi, C.; Capelli, S.; Conconi, M.T.; Parnigotto, P.P. Leucocyte and Platelet-rich Fibrin: A carrier of autologous multipotent cells for regenerative medicine. J. Cell. Mol. Med. 2018, 22, 1840–1854. [Google Scholar] [CrossRef]
- Di Summa, F.; Kargarpour, Z.; Nasirzade, J.; Stahli, A.; Mitulovic, G.; Panic-Jankovic, T.; Koller, V.; Kaltenbach, C.; Muller, H.; Panahipour, L.; et al. TGFbeta activity released from platelet-rich fibrin adsorbs to titanium surface and collagen membranes. Sci. Rep. 2020, 10, 10203. [Google Scholar] [CrossRef]
- Castro, A.B.; Andrade, C.; Li, X.; Pinto, N.; Teughels, W.; Quirynen, M. Impact of g force and timing on the characteristics of platelet-rich fibrin matrices. Sci. Rep. 2021, 11, 6038. [Google Scholar] [CrossRef]
- Hermida-Nogueira, L.; Barrachina, M.N.; Moran, L.A.; Bravo, S.; Diz, P.; Garcia, A.; Blanco, J. Deciphering the secretome of leukocyte-platelet rich fibrin: Towards a better understanding of its wound healing properties. Sci. Rep. 2020, 10, 14571. [Google Scholar] [CrossRef]
- Strauss, F.J.; Nasirzade, J.; Kargarpoor, Z.; Stahli, A.; Gruber, R. Effect of platelet-rich fibrin on cell proliferation, migration, differentiation, inflammation, and osteoclastogenesis: A systematic review of in vitro studies. Clin. Oral. Investig. 2020, 24, 569–584. [Google Scholar] [CrossRef]
- Dohan Ehrenfest, D.M.; Doglioli, P.; de Peppo, G.M.; Del Corso, M.; Charrier, J.B. Choukroun’s platelet-rich fibrin (PRF) stimulates in vitro proliferation and differentiation of human oral bone mesenchymal stem cell in a dose-dependent way. Arch. Oral Biol. 2010, 55, 185–194. [Google Scholar] [CrossRef]
- Strauss, F.J.; Stahli, A.; Gruber, R. The use of platelet-rich fibrin to enhance the outcomes of implant therapy: A systematic review. Clin. Oral Implant. Res. 2018, 29 (Suppl. 18), 6–19. [Google Scholar] [CrossRef] [PubMed]
- Del Fabbro, M.; Karanxha, L.; Panda, S.; Bucchi, C.; Nadathur Doraiswamy, J.; Sankari, M.; Ramamoorthi, S.; Varghese, S.; Taschieri, S. Autologous platelet concentrates for treating periodontal infrabony defects. Cochrane Database Syst. Rev. 2018, 11, CD011423. [Google Scholar] [CrossRef] [PubMed]
- Miron, R.J.; Moraschini, V.; Fujioka-Kobayashi, M.; Zhang, Y.; Kawase, T.; Cosgarea, R.; Jepsen, S.; Bishara, M.; Canullo, L.; Shirakata, Y.; et al. Use of platelet-rich fibrin for the treatment of periodontal intrabony defects: A systematic review and meta-analysis. Clin. Oral Investig. 2021, 25, 2461–2478. [Google Scholar] [CrossRef] [PubMed]
- Shanbhag, S.; Suliman, S.; Pandis, N.; Stavropoulos, A.; Sanz, M.; Mustafa, K. Cell therapy for orofacial bone regeneration: A systematic review and meta-analysis. J. Clin. Periodontol. 2019, 46 (Suppl. 21), 162–182. [Google Scholar] [CrossRef] [PubMed]
- Sandor, G.K.; Numminen, J.; Wolff, J.; Thesleff, T.; Miettinen, A.; Tuovinen, V.J.; Ohman, J. Adipose stem cells used to reconstruct 13 cases with cranio-maxillofacial hard-tissue defects. Stem Cells Transl. Med. 2014, 3, 530–540. [Google Scholar] [CrossRef]
- Gjerde, C.; Mustafa, K.; Hellem, S.; Rojewski, M.; Gjengedal, H.; Yassin, M.A.; Feng, X.; Skaale, S.; Berge, T.; Rosen, A.; et al. Cell therapy induced regeneration of severely atrophied mandibular bone in a clinical trial. Stem Cell Res. Ther. 2018, 9, 213. [Google Scholar] [CrossRef]
- Haumer, A.; Bourgine, P.E.; Occhetta, P.; Born, G.; Tasso, R.; Martin, I. Delivery of cellular factors to regulate bone healing. Adv. Drug Deliv. Rev. 2018, 129, 285–294. [Google Scholar] [CrossRef]
- Caplan, A.I.; Dennis, J.E. Mesenchymal stem cells as trophic mediators. J. Cell. Biochem. 2006, 98, 1076–1084. [Google Scholar] [CrossRef]
- Gnecchi, M.; Danieli, P.; Malpasso, G.; Ciuffreda, M.C. Paracrine Mechanisms of Mesenchymal Stem Cells in Tissue Repair. Methods Mol. Biol. 2016, 1416, 123–146. [Google Scholar] [CrossRef]
- Weiss, A.R.R.; Dahlke, M.H. Immunomodulation by Mesenchymal Stem Cells (MSCs): Mechanisms of Action of Living, Apoptotic, and Dead MSCs. Front. Immunol. 2019, 10, 1191. [Google Scholar] [CrossRef]
- Harrell, C.R.; Jovicic, N.; Djonov, V.; Arsenijevic, N.; Volarevic, V. Mesenchymal Stem Cell-Derived Exosomes and Other Extracellular Vesicles as New Remedies in the Therapy of Inflammatory Diseases. Cells 2019, 8, 1605. [Google Scholar] [CrossRef] [PubMed]
- Saleem, R.; Mohamed-Ahmed, S.; Elnour, R.; Berggreen, E.; Mustafa, K.; Al-Sharabi, N. Conditioned Medium from Bone Marrow Mesenchymal Stem Cells Restored Oxidative Stress-Related Impaired Osteogenic Differentiation. Int. J. Mol. Sci. 2021, 22, 13458. [Google Scholar] [CrossRef]
- Bari, E.; Perteghella, S.; Di Silvestre, D.; Sorlini, M.; Catenacci, L.; Sorrenti, M.; Marrubini, G.; Rossi, R.; Tripodo, G.; Mauri, P.; et al. Pilot Production of Mesenchymal Stem/Stromal Freeze-Dried Secretome for Cell-Free Regenerative Nanomedicine: A Validated GMP-Compliant Process. Cells 2018, 7, 190. [Google Scholar] [CrossRef] [PubMed]
- Veronesi, F.; Borsari, V.; Sartori, M.; Orciani, M.; Mattioli-Belmonte, M.; Fini, M. The use of cell conditioned medium for musculoskeletal tissue regeneration. J. Cell. Physiol. 2018, 233, 4423–4442. [Google Scholar] [CrossRef] [PubMed]
- Benavides-Castellanos, M.P.; Garzon-Orjuela, N.; Linero, I. Effectiveness of mesenchymal stem cell-conditioned medium in bone regeneration in animal and human models: A systematic review and meta-analysis. Cell Regen. 2020, 9, 5. [Google Scholar] [CrossRef]
- Qiu, J.; Wang, X.; Zhou, H.; Zhang, C.; Wang, Y.; Huang, J.; Liu, M.; Yang, P.; Song, A. Enhancement of periodontal tissue regeneration by conditioned media from gingiva-derived or periodontal ligament-derived mesenchymal stem cells: A comparative study in rats. Stem Cell Res. Ther. 2020, 11, 42. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Haensel, D.; Dai, X. Epithelial-to-mesenchymal transition in cutaneous wound healing: Where we are and where we are heading. Dev. Dyn. 2018, 247, 473–480. [Google Scholar] [CrossRef] [PubMed]
- Wilson, A.; Murphy, M.J.; Oskarsson, T.; Kaloulis, K.; Bettess, M.D.; Oser, G.M.; Pasche, A.C.; Knabenhans, C.; Macdonald, H.R.; Trumpp, A. c-Myc controls the balance between hematopoietic stem cell self-renewal and differentiation. Genes Dev. 2004, 18, 2747–2763. [Google Scholar] [CrossRef]
- Castilho, R.M.; Squarize, C.H.; Gutkind, J.S. Exploiting PI3K/mTOR signaling to accelerate epithelial wound healing. Oral. Dis. 2013, 19, 551–558. [Google Scholar] [CrossRef]
- Dunnill, C.; Patton, T.; Brennan, J.; Barrett, J.; Dryden, M.; Cooke, J.; Leaper, D.; Georgopoulos, N.T. Reactive oxygen species (ROS) and wound healing: The functional role of ROS and emerging ROS-modulating technologies for augmentation of the healing process. Int. Wound J. 2017, 14, 89–96. [Google Scholar] [CrossRef] [PubMed]
- Kehl, D.; Generali, M.; Mallone, A.; Heller, M.; Uldry, A.C.; Cheng, P.; Gantenbein, B.; Hoerstrup, S.P.; Weber, B. Proteomic analysis of human mesenchymal stromal cell secretomes: A systematic comparison of the angiogenic potential. npj Regen. Med. 2019, 4, 8. [Google Scholar] [CrossRef]
- Shin, S.; Lee, J.; Kwon, Y.; Park, K.S.; Jeong, J.H.; Choi, S.J.; Bang, S.I.; Chang, J.W.; Lee, C. Comparative Proteomic Analysis of the Mesenchymal Stem Cells Secretome from Adipose, Bone Marrow, Placenta and Wharton’s Jelly. Int. J. Mol. Sci. 2021, 22, 845. [Google Scholar] [CrossRef] [PubMed]
- Pranskunas, M.; Simoliunas, E.; Alksne, M.; Kaupinis, A.; Juodzbalys, G. Periosteum-Derived Mesenchymal Stem Cells Secretome-Cell-Free Strategy for Endogenous Bone Regeneration: Proteomic Analysis in Vitro. J. Oral Maxillofac. Res. 2021, 12, e2. [Google Scholar] [CrossRef] [PubMed]
- Anitua, E.; Prado, R.; Azkargorta, M.; Rodriguez-Suarez, E.; Iloro, I.; Casado-Vela, J.; Elortza, F.; Orive, G. High-throughput proteomic characterization of plasma rich in growth factors (PRGF-Endoret)-derived fibrin clot interactome. J. Tissue Eng. Regen. Med. 2015, 9, E1–E12. [Google Scholar] [CrossRef] [PubMed]
- Ghanaati, S.; Herrera-Vizcaino, C.; Al-Maawi, S.; Lorenz, J.; Miron, R.J.; Nelson, K.; Schwarz, F.; Choukroun, J.; Sader, R. Fifteen Years of Platelet Rich Fibrin in Dentistry and Oromaxillofacial Surgery: How High is the Level of Scientific Evidence? J. Oral Implantol. 2018, 44, 471–492. [Google Scholar] [CrossRef]
- Masuki, H.; Okudera, T.; Watanebe, T.; Suzuki, M.; Nishiyama, K.; Okudera, H.; Nakata, K.; Uematsu, K.; Su, C.Y.; Kawase, T. Growth factor and pro-inflammatory cytokine contents in platelet-rich plasma (PRP), plasma rich in growth factors (PRGF), advanced platelet-rich fibrin (A-PRF), and concentrated growth factors (CGF). Int. J. Implant Dent. 2016, 2, 19. [Google Scholar] [CrossRef]
- Yaprak, E.; Kasap, M.; Akpinar, G.; Islek, E.E.; Sinanoglu, A. Abundant proteins in platelet-rich fibrin and their potential contribution to wound healing: An explorative proteomics study and review of the literature. J. Dent. Sci. 2018, 13, 386–395. [Google Scholar] [CrossRef]
- Dos Santos Canellas, J.V.; Soares, B.N.; Ritto, F.G.; Vettore, M.V.; Júnior, G.M.V.; Fischer, R.G.; Medeiros, P.J.D. What grafting materials produce greater alveolar ridge preservation after tooth extraction? A systematic review and network meta-analysis. J. Cranio-Maxillofac. Surg. 2021, 49, 1064–1071. [Google Scholar] [CrossRef]
- Ortega-Mejia, H.; Estrugo-Devesa, A.; Saka-Herran, C.; Ayuso-Montero, R.; Lopez-Lopez, J.; Velasco-Ortega, E. Platelet-Rich Plasma in Maxillary Sinus Augmentation: Systematic Review. Materials 2020, 13, 622. [Google Scholar] [CrossRef]
- Werner, S.; Grose, R. Regulation of Wound Healing by Growth Factors and Cytokines. Physiol. Rev. 2003, 83, 835–870. [Google Scholar] [CrossRef] [PubMed]
- Barrientos, S.; Stojadinovic, O.; Golinko, M.S.; Brem, H.; Tomic-Canic, M. Growth factors and cytokines in wound healing. Wound Repair Regen. 2008, 16, 585–601. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Yang, J.; Fang, J.; Zhou, Y.; Candi, E.; Wang, J.; Hua, D.; Shao, C.; Shi, Y. The secretion profile of mesenchymal stem cells and potential applications in treating human diseases. Signal Transduct. Target. Ther. 2022, 7, 92. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, J.R.; Teixeira, G.Q.; Santos, S.G.; Barbosa, M.A.; Almeida-Porada, G.; Goncalves, R.M. Mesenchymal Stromal Cell Secretome: Influencing Therapeutic Potential by Cellular Pre-conditioning. Front. Immunol. 2018, 9, 2837. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Park, Y.S.; Choi, N.Y.; Kim, Y.I.; Koh, Y.G. Proteomic Analysis Reveals Commonly Secreted Proteins of Mesenchymal Stem Cells Derived from Bone Marrow, Adipose Tissue, and Synovial Membrane to Show Potential for Cartilage Regeneration in Knee Osteoarthritis. Stem Cells Int. 2021, 2021, 6694299. [Google Scholar] [CrossRef]
- Peshkova, M.; Korneev, A.; Suleimanov, S.; Vlasova, I.I.; Svistunov, A.; Kosheleva, N.; Timashev, P. MSCs’ conditioned media cytokine and growth factor profiles and their impact on macrophage polarization. Stem Cell Res. Ther. 2023, 14, 142. [Google Scholar] [CrossRef]
- Ratajczak, J.; Vangansewinkel, T.; Gervois, P.; Merckx, G.; Hilkens, P.; Quirynen, M.; Lambrichts, I.; Bronckaers, A. Angiogenic Properties of ‘Leukocyte- and Platelet-Rich Fibrin’. Sci. Rep. 2018, 8, 14632. [Google Scholar] [CrossRef]
- Shekatkar, M.R.; Kheur, S.M.; Kharat, A.H.; Deshpande, S.S.; Sanap, A.P.; Kheur, M.G.; Bhonde, R.R. Assessment of angiogenic potential of mesenchymal stem cells derived conditioned medium from various oral sources. J. Clin. Transl. Res. 2022, 8, 323–338. [Google Scholar]
- Kalra, H.; Simpson, R.J.; Ji, H.; Aikawa, E.; Altevogt, P.; Askenase, P.; Bond, V.C.; Borras, F.E.; Breakefield, X.; Budnik, V.; et al. Vesiclepedia: A compendium for extracellular vesicles with continuous community annotation. PLoS Biol. 2012, 10, e1001450. [Google Scholar] [CrossRef]
- Wang, D.; Cao, H.; Hua, W.; Gao, L.; Yuan, Y.; Zhou, X.; Zeng, Z. Mesenchymal Stem Cell-Derived Extracellular Vesicles for Bone Defect Repair. Membranes 2022, 12, 716. [Google Scholar] [CrossRef]
- Wu, J.; Piao, Y.; Liu, Q.; Yang, X. Platelet-rich plasma-derived extracellular vesicles: A superior alternative in regenerative medicine? Cell Prolif. 2021, 54, e13123. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Patil, S.; Gao, Y.G.; Qian, A. The Bone Extracellular Matrix in Bone Formation and Regeneration. Front. Pharmacol. 2020, 11, 757. [Google Scholar] [CrossRef] [PubMed]
- Butler, W.T.; Birkedal-Hansen, H.; Beegle, W.F.; Taylor, R.E.; Chung, E. Proteins of the periodontium. Identification of collagens with the [alpha1(I)]2alpha2 and [alpha1(III)]3 structures in bovine periodontal ligament. J. Biol. Chem. 1975, 250, 8907–8912. [Google Scholar] [CrossRef] [PubMed]
- Paiva, K.B.S.; Granjeiro, J.M. Matrix Metalloproteinases in Bone Resorption, Remodeling, and Repair. Prog. Mol. Biol. Transl. Sci. 2017, 148, 203–303. [Google Scholar] [CrossRef]
- Mulholland, B.S.; Forwood, M.R.; Morrison, N.A. Monocyte Chemoattractant Protein-1 (MCP-1/CCL2) Drives Activation of Bone Remodelling and Skeletal Metastasis. Curr. Osteoporos. Rep. 2019, 17, 538–547. [Google Scholar] [CrossRef]
- Boyce, B.F.; Xing, L. Functions of RANKL/RANK/OPG in bone modeling and remodeling. Arch. Biochem. Biophys. 2008, 473, 139–146. [Google Scholar] [CrossRef]
- Wu, M.; Chen, G.; Li, Y.P. TGF-beta and BMP signaling in osteoblast, skeletal development, and bone formation, homeostasis and disease. Bone Res. 2016, 4, 16009. [Google Scholar] [CrossRef]
- Asharani, P.V.; Keupp, K.; Semler, O.; Wang, W.; Li, Y.; Thiele, H.; Yigit, G.; Pohl, E.; Becker, J.; Frommolt, P.; et al. Attenuated BMP1 function compromises osteogenesis, leading to bone fragility in humans and zebrafish. Am. J. Hum. Genet. 2012, 90, 661–674. [Google Scholar] [CrossRef]
- Salmon, C.R.; Tomazela, D.M.; Ruiz, K.G.; Foster, B.L.; Paes Leme, A.F.; Sallum, E.A.; Somerman, M.J.; Nociti, F.H., Jr. Proteomic analysis of human dental cementum and alveolar bone. J. Proteom. 2013, 91, 544–555. [Google Scholar] [CrossRef]
- Shanbhag, S.; Kampleitner, C.; Al-Sharabi, N.; Mohamed-Ahmed, S.; Apaza Alccayhuaman, K.A.; Heimel, P.; Tangl, S.; Beinlich, A.; Rana, N.; Sanz, M.; et al. Functionalizing Collagen Membranes with MSC-Conditioned Media Promotes Guided Bone Regeneration in Rat Calvarial Defects. Cells 2023, 12, 767. [Google Scholar] [CrossRef]
- Ankrum, J.A.; Ong, J.F.; Karp, J.M. Mesenchymal stem cells: Immune evasive, not immune privileged. Nat. Biotechnol. 2014, 32, 252–260. [Google Scholar] [CrossRef] [PubMed]
- Scopelliti, F.; Cattani, C.; Dimartino, V.; Mirisola, C.; Cavani, A. Platelet Derivatives and the Immunomodulation of Wound Healing. Int. J. Mol. Sci. 2022, 23, 8370. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.Y.; Nair, M.G. Macrophages in wound healing: Activation and plasticity. Immunol. Cell Biol. 2019, 97, 258–267. [Google Scholar] [CrossRef] [PubMed]
- Nasirzade, J.; Kargarpour, Z.; Hasannia, S.; Strauss, F.J.; Gruber, R. Platelet-rich fibrin elicits an anti-inflammatory response in macrophages in vitro. J. Periodontol. 2020, 91, 244–252. [Google Scholar] [CrossRef]
- Zhang, J.; Yin, C.; Zhao, Q.; Zhao, Z.; Wang, J.; Miron, R.J.; Zhang, Y. Anti-inflammation effects of injectable platelet-rich fibrin via macrophages and dendritic cells. J. Biomed. Mater. Res. Part A 2020, 108, 61–68. [Google Scholar] [CrossRef]
- Gao, F.; Chiu, S.M.; Motan, D.A.; Zhang, Z.; Chen, L.; Ji, H.L.; Tse, H.F.; Fu, Q.L.; Lian, Q. Mesenchymal stem cells and immunomodulation: Current status and future prospects. Cell Death Dis. 2016, 7, e2062. [Google Scholar] [CrossRef]
- Chen, J.; Li, Y.; Hao, H.; Li, C.; Du, Y.; Hu, Y.; Li, J.; Liang, Z.; Li, C.; Liu, J.; et al. Mesenchymal Stem Cell Conditioned Medium Promotes Proliferation and Migration of Alveolar Epithelial Cells under Septic Conditions In Vitro via the JNK-P38 Signaling Pathway. Cell. Physiol. Biochem. 2015, 37, 1830–1846. [Google Scholar] [CrossRef]
- Winkel, A.; Jaimes, Y.; Melzer, C.; Dillschneider, P.; Hartwig, H.; Stiesch, M.; von der Ohe, J.; Strauss, S.; Vogt, P.M.; Hamm, A.; et al. Cell culture media notably influence properties of human mesenchymal stroma/stem-like cells from different tissues. Cytotherapy 2020, 22, 653–668. [Google Scholar] [CrossRef]
- Mead, B.; Chamling, X.; Zack, D.J.; Ahmed, Z.; Tomarev, S. TNFalpha-Mediated Priming of Mesenchymal Stem Cells Enhances Their Neuroprotective Effect on Retinal Ganglion Cells. Investig. Ophthalmol. Vis. Sci. 2020, 61, 6. [Google Scholar] [CrossRef]
- Shanbhag, S.; Mohamed-Ahmed, S.; Lunde, T.H.F.; Suliman, S.; Bolstad, A.I.; Hervig, T.; Mustafa, K. Influence of platelet storage time on human platelet lysates and platelet lysate-expanded mesenchymal stromal cells for bone tissue engineering. Stem Cell Res. Ther. 2020, 11, 351. [Google Scholar] [CrossRef]
- Aasebo, E.; Brenner, A.K.; Hernandez-Valladares, M.; Birkeland, E.; Berven, F.S.; Selheim, F.; Bruserud, O. Proteomic Comparison of Bone Marrow Derived Osteoblasts and Mesenchymal Stem Cells. Int. J. Mol. Sci. 2021, 22, 5665. [Google Scholar] [CrossRef] [PubMed]
- Tyanova, S.; Temu, T.; Sinitcyn, P.; Carlson, A.; Hein, M.Y.; Geiger, T.; Mann, M.; Cox, J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 2016, 13, 731–740. [Google Scholar] [CrossRef] [PubMed]
- Mi, H.; Muruganujan, A.; Ebert, D.; Huang, X.; Thomas, P.D. PANTHER version 14: More genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 2019, 47, D419–D426. [Google Scholar] [CrossRef] [PubMed]
- Raudvere, U.; Kolberg, L.; Kuzmin, I.; Arak, T.; Adler, P.; Peterson, H.; Vilo, J. g:Profiler: A web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 2019, 47, W191–W198. [Google Scholar] [CrossRef] [PubMed]

| Category | Common Proteins | Upregulated Proteins | Exclusive Proteins | ||
|---|---|---|---|---|---|
| BMSC-CM | LPRF-CM | BMSC-CM | LPRF-CM | ||
| GF-related | FGF1 *, TGFB1, LTBP1, MST1/HGFL, MET/HGFR, HGFAC, HGS, CCN5/CTGFL, PDGFRL, PDGFB, PDGFC, PDGFD, IGF2, IGFALS, CLEC11A/SCGF, KITLG/SCF, HDGFL2, EPS15, EPS15L1, MEGF8, ENAM, GPNMB * | TGFBI, TGFB2, LTBP2, LTBP4, BMP1, EGFR, IGFBP2, IGFBP3, IGFBP4, IGFBP7, CCN2/CTGF, IGF2R, HDGF, MYDGF, CSF1/MCSF, SDF1/CXCL12 | TGFB1I1, GRB2 | TGFBR2, LTBP3, FGF2 *, FGF7, FGFRL1, PDGFRA, IGFBP5, CCN1/IGFBP10, CCN3/IGFBP9, EPS8, PTN/HBNF | EGF, PDGFA |
| Cell function-related | PA2G4, NECTIN1, NECTIN2, VCL, CCAR1, BCAM, CKAP5, PCNA | CTNNB1, CDH11, TUBB4B, TUBB, TUBA1A, TUBB3, TUBA1C, PA2G4, TUBB1, PLEC, SDF2, SDF4, SERPINF1 | PTK2 | WNT5A, WNT5B, CCN4/WISP1, CKAP4, GAP43, CADM1, CDH1, CDH4, CDH13, TUBB2B, TUBB6 | F11R, CHL1 |
| Angiogen-esis-related | ANG, ANGPT1, ANGPTL4, ANGPTL6, AGT, VASP, ENG, SELE * | VEGFC, VCAM1, EPCR/PROCR, SVEP1 | VWF, PECAM1, ECE1 | VEGFA, FLT4/VEGFR3, ANGPTL2, EDF1, AAMP | ICAM2, ICAM3, VWA5A |
| ECM-related | SPP1/OPN, CLEC3B, ECM1, FN1, FBLN1, FBLN5, FAM20C/DMP4, THBS1, GC, HABP2, CRTAP, EFEMP1, EFEMP2, MXRA8, CHSY3 | SPARC/ON, SMOC1, POSTN/OSF2, OGN, BGN, DCN, THBS2, THBS5, MMP1, MMP2, MMP3, TIMP1, TIMP2, TIMP3, CCBE1, COLGALT1, ACTL6A, HAPLN1, ARPC1B, COL1A1, COL1A2, COL3A1, COL4A1, COL4A2, COL5A1, COL5A2, COL6A1, COL6A2, COL6A3, COL8A2, COL11A1, COL12A1, COL14A1, LAMA4, LAMB1, LAMB2, LAMC1, LAMC2, VIM, PCOLCE, MXRA5, PLOD1, PLOD2, CEMIP, MYH10 | MMP8, MMP9, CRTAC1 | ALPL, OMD, MMP13, MMP14, ARPC1A, HAPLN3, MXRA7, MYH2, COL5A3, COL7A1, COL8A1, COL16A1, CTHRC1, SAFB, SAFB2 | ECM2 |
| Inflamma-tion/immune-related | MIF, CCL5/RANTES, CCL18/MIP4, MARCO, CTSD, LGALSL, LGALS3 | CCL2/MCP1, TNFA *, TNFAIP6, IL1RAP, ILF2, TNFRSF11B/OPG, OSTF1, CTSB, CTSZ, CTSL, PTGR1, LGALS1, LGALS3BP, MRC2 | CXCL8, IL6ST, PRG2, ARG1, CD36 | IL6, LIF, TNFRSF1A, EDA2R, IKBKG, CTSF, CTSO, PTGES3 | CD14, CD40/TNFRSF5, CLEC1B, CTSG, CTSS, AIF1, HLA-DRA, TNFRSF14, CXCL5, CLEC2B |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Al-Sharabi, N.; Gruber, R.; Sanz, M.; Mohamed-Ahmed, S.; Kristoffersen, E.K.; Mustafa, K.; Shanbhag, S. Proteomic Analysis of Mesenchymal Stromal Cells Secretome in Comparison to Leukocyte- and Platelet-Rich Fibrin. Int. J. Mol. Sci. 2023, 24, 13057. https://doi.org/10.3390/ijms241713057
Al-Sharabi N, Gruber R, Sanz M, Mohamed-Ahmed S, Kristoffersen EK, Mustafa K, Shanbhag S. Proteomic Analysis of Mesenchymal Stromal Cells Secretome in Comparison to Leukocyte- and Platelet-Rich Fibrin. International Journal of Molecular Sciences. 2023; 24(17):13057. https://doi.org/10.3390/ijms241713057
Chicago/Turabian StyleAl-Sharabi, Niyaz, Reinhard Gruber, Mariano Sanz, Samih Mohamed-Ahmed, Einar K. Kristoffersen, Kamal Mustafa, and Siddharth Shanbhag. 2023. "Proteomic Analysis of Mesenchymal Stromal Cells Secretome in Comparison to Leukocyte- and Platelet-Rich Fibrin" International Journal of Molecular Sciences 24, no. 17: 13057. https://doi.org/10.3390/ijms241713057
APA StyleAl-Sharabi, N., Gruber, R., Sanz, M., Mohamed-Ahmed, S., Kristoffersen, E. K., Mustafa, K., & Shanbhag, S. (2023). Proteomic Analysis of Mesenchymal Stromal Cells Secretome in Comparison to Leukocyte- and Platelet-Rich Fibrin. International Journal of Molecular Sciences, 24(17), 13057. https://doi.org/10.3390/ijms241713057







