Effect of T-DNA Integration on Growth of Transgenic Populus × euramericana cv. Neva Underlying Field Stands
Abstract
1. Introduction
2. Results
2.1. Growth Traits of Transgenic Poplar in Field
2.2. Genotype by Environment Interaction Effect
2.3. NGS Sequencing Quality Control and Data Comparison
2.4. PCR Verification of T-DNA Insertion Sites
2.5. T-DNA Insertion Location and Adjacent Genes
2.6. RNA-seq and Differentially Expressed Genes (DEGs) Analysis
2.7. Gene Ontology (GO) Enrichment Analysis
2.8. Kyoto Encyclopedia of Genes and Genomes (KEGG) Metabolic Pathway Analysis
2.9. K-Means Clustering of DEGs Expression Patterns
2.10. Analysis of Genes Expression Related to Growth and Development
2.11. Analysis of Differentially Expressed Transcription Factors (TFs)
2.12. Verification of RNA-seq Data by Quantitative Reverse-Transcription PCR (qRT-PCR)
3. Discussion
3.1. T-DNA Integration Analysis
3.2. T-DNA Integration and Gene Expression
4. Conclusions
5. Materials and Methods
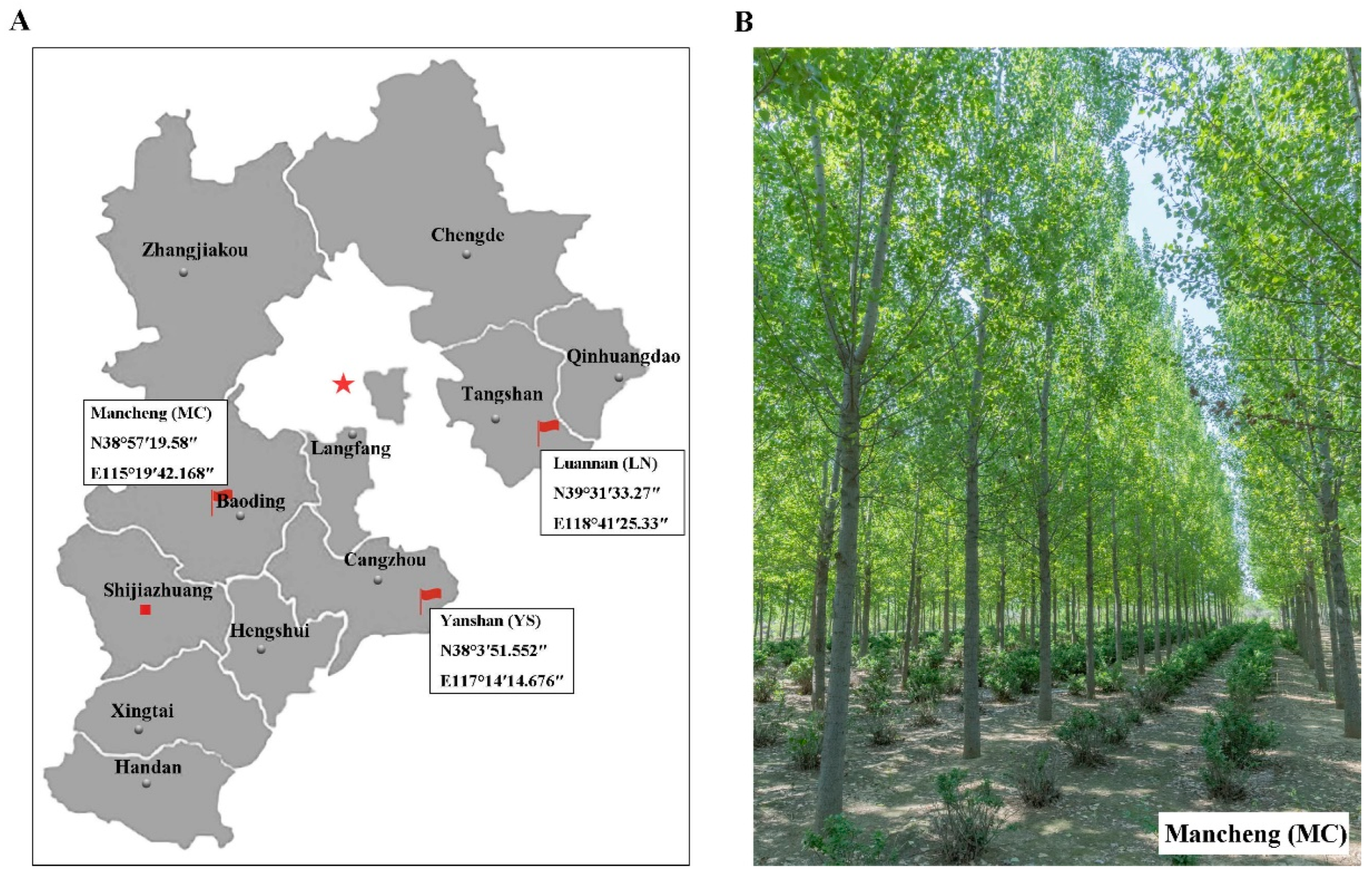
5.1. Plant Materials and Study Sites
5.2. Growth Characteristics
5.3. NGS Resequencing
5.4. T-DNA Integration Verification and Flanking Sequence Analysis
5.5. RNA-seq and Data Quality Control
5.6. DEGs Identification and Functional Analysis
5.7. qRT-PCR
5.8. Statistical Analysis Methods
5.9. Availability of Data and Material
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Wang, Y.; Liu, Q.; Song, X.; Yang, X.; Han, L.; Romeis, J.; Li, Y. Unintended changes in transgenic maize cause no nontarget effects. Plants People Planet 2022, 4, 392–402. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, Q.; Liu, Q.; Meissle, M.; Yang, Y.; Wang, Y.; Hua, H.; Chen, X.; Peng, Y.; Romeis, J. Bt rice in China—Focusing the nontarget risk assessment. Plant Biotechnol. J. 2017, 15, 1340–1345. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Yang, X.; Tzin, V.; Peng, Y.; Romeis, J.; Li, Y. Plant breeding involving genetic engineering does not result in unacceptable unintended effects in rice relative to conventional cross-breeding. Plant J. 2020, 103, 2236–2249. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Q.; Niu, F.; Sun, X.; Hu, Z.; Li, X.; Ma, Y.; Zhang, H. RNA-seq analysis of unintended effects in transgenic wheat overexpressing the transcription factor GmDREB1. Crop J. 2017, 5, 207–218. [Google Scholar] [CrossRef]
- Jiang, X.; Xiao, G. Detection of Unintended Effects in Genetically Modified Herbicide-tolerant (GMHT) Rice in Comparison with Non-target Phenotypic Characteristics. Afr. J. Agric. Res. 2010, 5, 1082–1088. [Google Scholar]
- Lucker, J.; Bouwmeester, H.J.; Schwab, W.; Blaas, J.; van der Plas, L.H.; Verhoeven, H.A. Expression of Clarkia S-linalool synthase in transgenic petunia plants results in the accumulation of S-linalyl-beta-D-glucopyranoside. Plant J. 2001, 27, 315–324. [Google Scholar] [CrossRef]
- Sheng, L.; Zang, S.; Wang, J.; Wei, T.; Xu, Y.; Feng, L. Overexpression of a Rosa rugosa Thunb. NUDX gene enhances biosynthesis of scent volatiles in petunia. PeerJ 2021, 9, e11098. [Google Scholar] [CrossRef]
- Filipecki, M.; Malepszy, S. Unintended consequences of plant transformation: A molecular insight. J. Appl. Genet. 2006, 47, 277–286. [Google Scholar] [CrossRef]
- Ouakfaoui, S.E.; Miki, B. The stability of the Arabidopsis transcriptome in transgenic plants expressing the marker genes nptII and uidA. Plant J. 2005, 41, 791–800. [Google Scholar] [CrossRef]
- Matzke, A.J.; Matzke, M.A. Position effects and epigenetic silencing of plant transgenes. Curr. Opin. Plant Biol. 1998, 1, 142–148. [Google Scholar] [CrossRef]
- Stam, M.; Mol, J.N.M.; Kooter, J.M. Review article: The silence of genes in transgenic plants. Ann. Bot. 1997, 79, 3–12. [Google Scholar] [CrossRef]
- Herman, R.A.; Price, W.D. Unintended compositional changes in genetically modified (GM) crops: 20 years of research. J. Agric. Food Chem. 2013, 61, 11695–11701. [Google Scholar] [CrossRef] [PubMed]
- Metzdorff, S.B.; Kok, E.J.; Knuthsen, P.; Pedersen, J. Evaluation of a non-targeted “Omic” approach in the safety assessment of genetically modified plants. Plant Biol. 2006, 8, 662–672. [Google Scholar] [CrossRef]
- Davies, H. A role for “omics” technologies in food safety assessment. Food Control 2010, 21, 1601–1610. [Google Scholar] [CrossRef]
- Kumar, R.; Bohra, A.; Pandey, A.K.; Pandey, M.K.; Kumar, A. Metabolomics for plant improvement: Status and prospects. Front. Plant Sci. 2017, 8, 1302. [Google Scholar] [CrossRef]
- Biselli, C.; Vietto, L.; Rosso, L.; Cattivelli, L.; Nervo, G.; Fricano, A. Advanced breeding for biotic stress resistance in poplar. Plants 2022, 11, 2032. [Google Scholar] [CrossRef]
- Ren, Y.; Zhou, X.; Dong, Y.; Zhang, J.; Wang, J.; Yang, M. Exogenous gene expression and insect resistance in dual Bt toxin Populus × euramericana ‘Neva’ transgenic plants. Front. Plant Sci. 2021, 12, 660226. [Google Scholar] [CrossRef]
- Yang, R.L.; Wang, A.X.; Zhang, J.; Dong, Y.; Yang, M.S.; Wang, J.M. Genetic transformation and expression of transgenic lines of Populus x euramericana with insect-resistance and salt-tolerance genes. Genet. Mol. Res. 2016, 15, r8635. [Google Scholar] [CrossRef]
- Abdeen, A.; Schnell, J.; Miki, B. Transcriptome analysis reveals absence of unintended effects in drought-tolerant transgenic plants overexpressing the transcription factor ABF3. BMC Genom. 2010, 11, 69. [Google Scholar] [CrossRef]
- Yang, H.; Peng, Y.; Tian, J.; Wang, J.; Hu, J.; Song, Q.; Wang, Z. Review: Biosafety assessment of Bt rice and other Bt crops using spiders as example for non-target arthropods in China. Plant Cell Rep. 2017, 36, 505–517. [Google Scholar] [CrossRef]
- Zuo, L.; Yang, R.; Zhen, Z.; Liu, J.; Huang, L.; Yang, M. A 5-year field study showed no apparent effect of the Bt transgenic 741 poplar on the arthropod community and soil bacterial diversity. Sci. Rep. 2018, 8, 1956. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Zhen, Z.; Cui, Z.; Liu, J.; Wang, S.; Yang, M.; Wu, J. Growth and arthropod community characteristics of transgenic poplar 741 in an experimental forest. Ind. Crops Prod. 2021, 162, 113284. [Google Scholar] [CrossRef]
- Choi, H.; Lemaux, P.; Cho, M.J. Use of fluorescence in situ hybridization for gross mapping of transgenes and screening for homozygous plants in transgenic barley (Hordeum vulgare L.). Theor. Appl. Genet. 2002, 106, 92–100. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Yang, Y.; Wang, F.; Wang, G.; Wang, C.; Wang, W.; Chen, K.; Gu, C.; Yu, Q.; Jiang, J. Growth adaptability and foreign gene stability of TaLEA transgenic Populus simonii × nigra. Ann. For. Sci. 2021, 78, 42. [Google Scholar] [CrossRef]
- Francis, K.E.; Spiker, S. Identification of Arabidopsis thaliana transformants without selection reveals a high occurrence of silenced T-DNA integrations. Plant J. 2005, 41, 464–477. [Google Scholar] [CrossRef]
- Kim, S.; Gelvin, S.B. Genome-wide analysis of Agrobacterium T-DNA integration sites in the Arabidopsis genome generated under non-selective conditions. Plant J. 2007, 51, 779–791. [Google Scholar] [CrossRef]
- Li, Y.; Rosso, M.G.; Ülker, B.; Weisshaar, B. Analysis of T-DNA insertion site distribution patterns in Arabidopsis thaliana reveals special features of genes without insertions. Genomics 2006, 87, 645–652. [Google Scholar] [CrossRef][Green Version]
- Kharb, P.; Chaudhary, R.; Tuteja, N.; Kaushik, P. A genotype-independent, simple, effective and efficient in planta Agrobacterium-mediated genetic transformation protocol. Methods Protoc. 2022, 5, 69. [Google Scholar] [CrossRef]
- Travella, S.; Ross, S.M.; Harden, J.; Everett, C.; Snape, J.W.; Harwood, W.A. A comparison of transgenic barley lines produced by particle bombardment and Agrobacterium-mediated techniques. Plant Cell Rep. 2005, 23, 780–789. [Google Scholar] [CrossRef]
- Zhou, X.; Ren, Y.; Wang, S.; Chen, X.; Zhang, C.; Yang, M.; Dong, Y. T-DNA integration and its effect on gene expression in dual Bt gene transgenic Populus × euramericana cv. Neva. Ind. Crops Prod. 2022, 178, 114636. [Google Scholar] [CrossRef]
- Yang, L.; Xu, S.; Pan, A.; Yin, C.; Zhang, K.; Wang, Z.; Zhou, Z.; Zhang, D. Event specific qualitative and quantitative polymerase chain reaction detection of genetically modified MON863 maize based on the 5′-transgene integration sequence. J. Agric. Food Chem. 2005, 53, 9312–9318. [Google Scholar] [CrossRef] [PubMed]
- Sims, D.; Sudbery, I.; Ilott, N.E.; Heger, A.; Ponting, C.P. Sequencing depth and coverage: Key considerations in genomic analyses. Nat. Rev. Genet. 2014, 15, 121–132. [Google Scholar] [CrossRef] [PubMed]
- Pucker, B.; Kleinbölting, N.; Weisshaar, B. Large scale genomic rearrangements in selected Arabidopsis thaliana T-DNA lines are caused by T-DNA insertion mutagenesis. BMC Genom. 2021, 22, 599. [Google Scholar] [CrossRef] [PubMed]
- Lusk, R.W. Diverse and widespread contamination evident in the unmapped depths of high throughput sequencing data. PLoS ONE 2014, 9, e110808. [Google Scholar] [CrossRef]
- Pfeiffer, F.; Gröber, C.; Blank, M.; Händler, K.; Beyer, M.; Schultze, J.L.; Mayer, G. Systematic evaluation of error rates and causes in short samples in next-generation sequencing. Sci. Rep. 2018, 8, 10950. [Google Scholar] [CrossRef]
- Xu, L.N.; Dong, Y.; Zhang, J.; Wang, R.X.; Liu, H.M.; Yang, Q.; Yang, M.S. Effect of dual Bt-expression transformation vectors on transgene expression in tobacco. Genet. Mol. Res. 2016, 15, gmr.15038293. [Google Scholar] [CrossRef]
- Liu, W.; Zhao, H.; Miao, C.; Jin, W. Integrated proteomics and metabolomics analysis of transgenic and gene-stacked maize line seeds. Gm Crops Food 2021, 12, 361–375. [Google Scholar] [CrossRef]
- Wang, X.J.; Zhang, X.; Yang, J.T.; Wang, Z.X. Effect on transcriptome and metabolome of stacked transgenic maize containing insecticidal cry and glyphosate tolerance epsps genes. Plant J. 2018, 93, 1007–1016. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, X.; Yang, J.; Liu, X.; Song, Y.; Wang, Z. Genetic variation assessment of stacked-trait transgenic maize via conventional breeding. BMC Plant Biol. 2019, 19, 346. [Google Scholar] [CrossRef]
- Klocko, A.L.; Meilan, R.; James, R.R.; Viswanath, V.; Ma, C.; Payne, P.; Miller, L.; Skinner, J.S.; Oppert, B.; Cardineau, G.A.; et al. Bt-Cry3Aa transgene expression reduces insect damage and improves growth in field-grown hybrid poplar. Can. J. For. Res. 2014, 44, 28–35. [Google Scholar] [CrossRef]
- Podevin, N.; du Jardin, P. Possible consequences of the overlap between the CaMV 35S promoter regions in plant transformation vectors used and the viral gene VI in transgenic plants. GM Crops Food 2012, 3, 296–300. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; An, Y.; Zhou, Y.; Liu, C.; Yin, W.; Xia, X. Comparative transcriptome analyses define genes and gene modules differing between two Populus genotypes with contrasting stem growth rates. Biotechnol. Biofuels 2020, 13, 139. [Google Scholar] [CrossRef] [PubMed]
- Quan, M.; Du, Q.; Xiao, L.; Lu, W.; Wang, L.; Xie, J.; Song, Y.; Xu, B.; Zhang, D. Genetic architecture underlying the lignin biosynthesis pathway involves noncoding RNAs and transcription factors for growth and wood properties in Populus. Plant Biotechnol. J. 2019, 17, 302–315. [Google Scholar] [CrossRef] [PubMed]
- Cassan-Wang, H.; Goué, N.; Saidi, M.N.; Legay, S.; Sivadon, P.; Goffner, D.; Grima-Pettenati, J. Identification of novel transcription factors regulating secondary cell wall formation in Arabidopsis. Front. Plant Sci. 2013, 4, 189. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Ge, H.; Zang, C.; Li, X.; Grierson, D.; Chen, K.; Yin, X. EjODO1, a MYB Transcription Factor, Regulating Lignin Biosynthesis in Developing Loquat (Eriobotrya japonica) Fruit. Front. Plant Sci. 2016, 7, 1360. [Google Scholar] [CrossRef]
- Zhang, C.; Xu, Y.; Lu, Y.; Yu, H.; Gu, M.; Liu, Q. The WRKY transcription factor OsWRKY78 regulates stem elongation and seed development in rice. Planta 2011, 234, 541–554. [Google Scholar] [CrossRef]
- Yu, F.; Huaxia, Y.; Lu, W.; Wu, C.; Cao, X.; Guo, X. GhWRKY15, a member of the WRKY transcription factor family identified from cotton (Gossypium hirsutum L.), is involved in disease resistance and plant development. BMC Plant Biol. 2012, 12, 144. [Google Scholar] [CrossRef]
- Abel, S.; Theologis, A. Early genes and auxin action. Plant Physiol. 1996, 111, 9–17. [Google Scholar] [CrossRef]
- Chen, S.; Zhong, K.; Li, Y.; Bai, C.; Xue, Z.; Wu, Y. Evolutionary analysis of the melon (Cucumis melo L.) GH3 gene family and identification of GH3 genes related to fruit growth and development. Plants 2023, 12, 1382. [Google Scholar] [CrossRef]
- Nakazawa, M.; Yabe, N.; Ichikawa, T.; Yamamoto, Y.Y.; Yoshizumi, T.; Hasunuma, K.; Matsui, M. DFL1, an auxin-responsive GH3 gene homologue, negatively regulates shoot cell elongation and lateral root formation, and positively regulates the light response of hypocotyl length. Plant J. 2001, 25, 213–221. [Google Scholar] [CrossRef]
- Dewitte, W.; Riou-Khamlichi, C.; Scofield, S.; Healy, J.M.S.; Jacqmard, A.; Kilby, N.J.; Murray, J.A.H. Altered cell cycle distribution, hyperplasia, and inhibited differentiation in Arabidopsis caused by the D-Type cyclin CYCD3. Plant Cell 2003, 15, 79–92. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Zhao, J.; Song, J.; Jameson, P.E. Cytokinin glucosyl transferases, key regulators of cytokinin homeostasis, have potential value for wheat improvement. Plant Biotechnol. J. 2021, 19, 878–896. [Google Scholar] [CrossRef] [PubMed]
- Kajita, S.; Hishiyama, S.; Tomimura, Y.; Katayama, Y.; Omori, S. Structural characterization of modified lignin in transgenic tobacco plants in which the activity of 4-Coumarate: Coenzyme a ligase 1s depressed. Plant Physiol. 1997, 114, 871–879. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Zhang, J.; Dong, Y.; Zhang, X.; Yang, M.; Gao, B. Genetic transformation and expression of Cry1Ac-Cry3A-NTHK1 genes in Populus × euramericana “Neva”. Acta Physiol. Plant. 2016, 38, 177. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
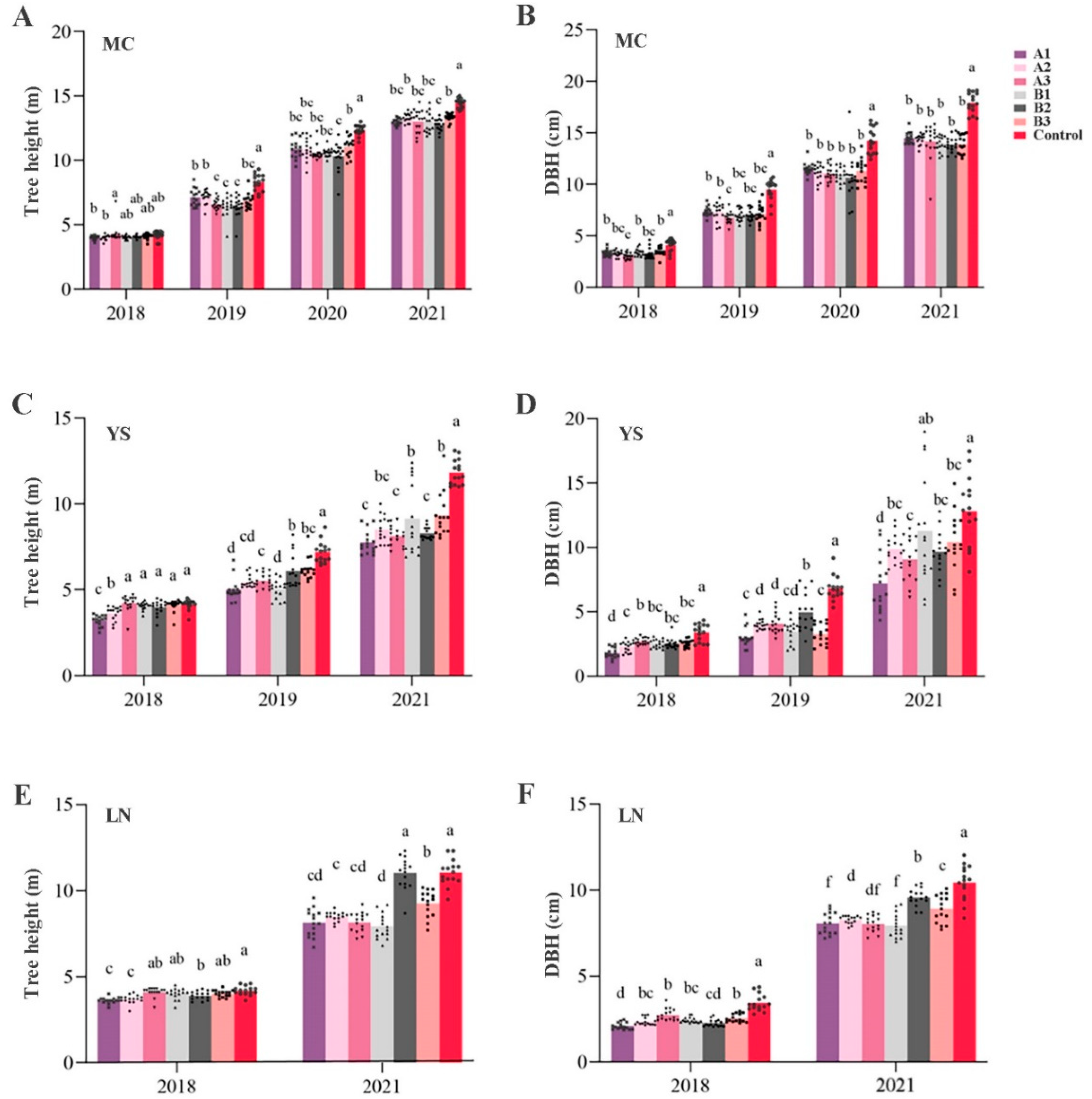

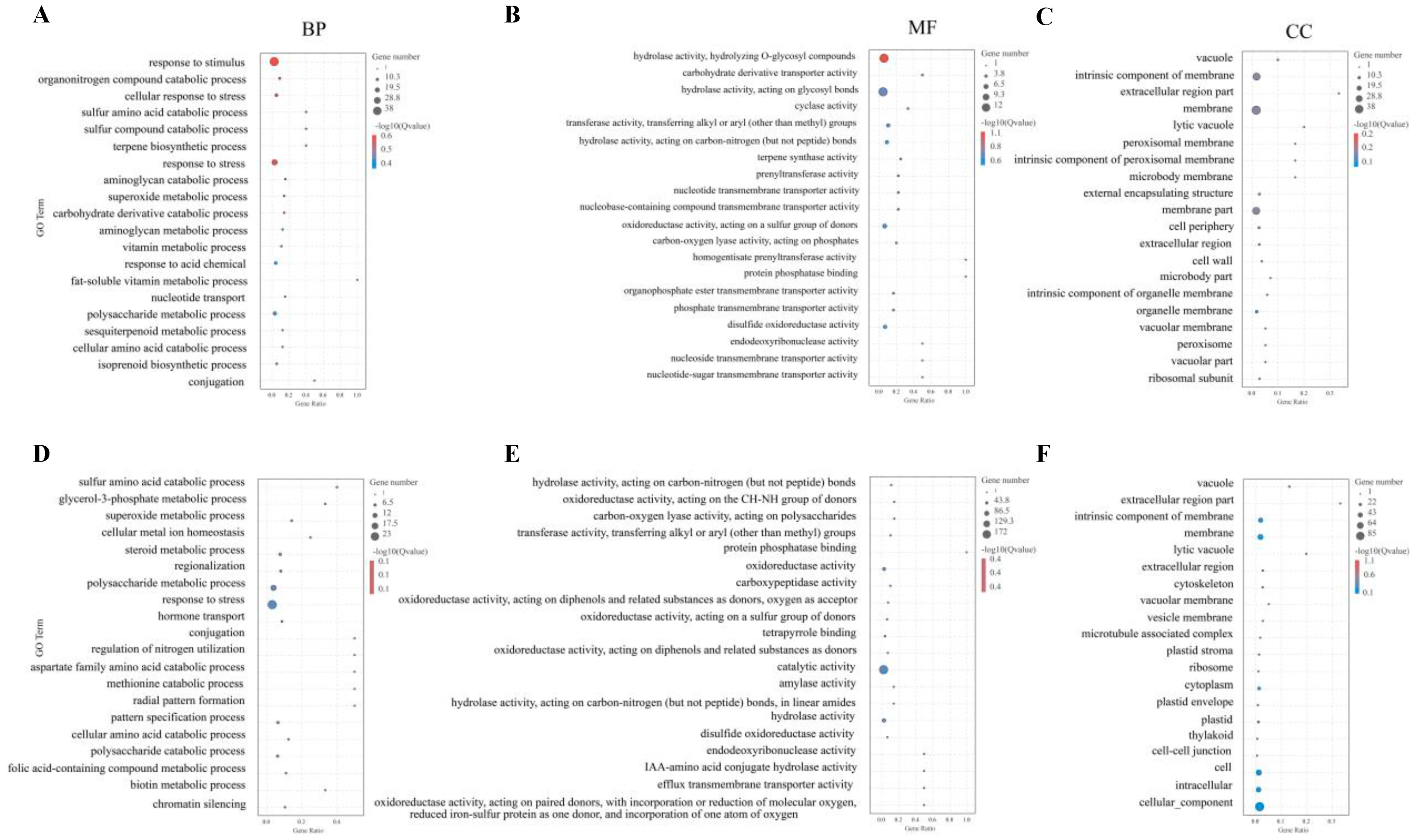
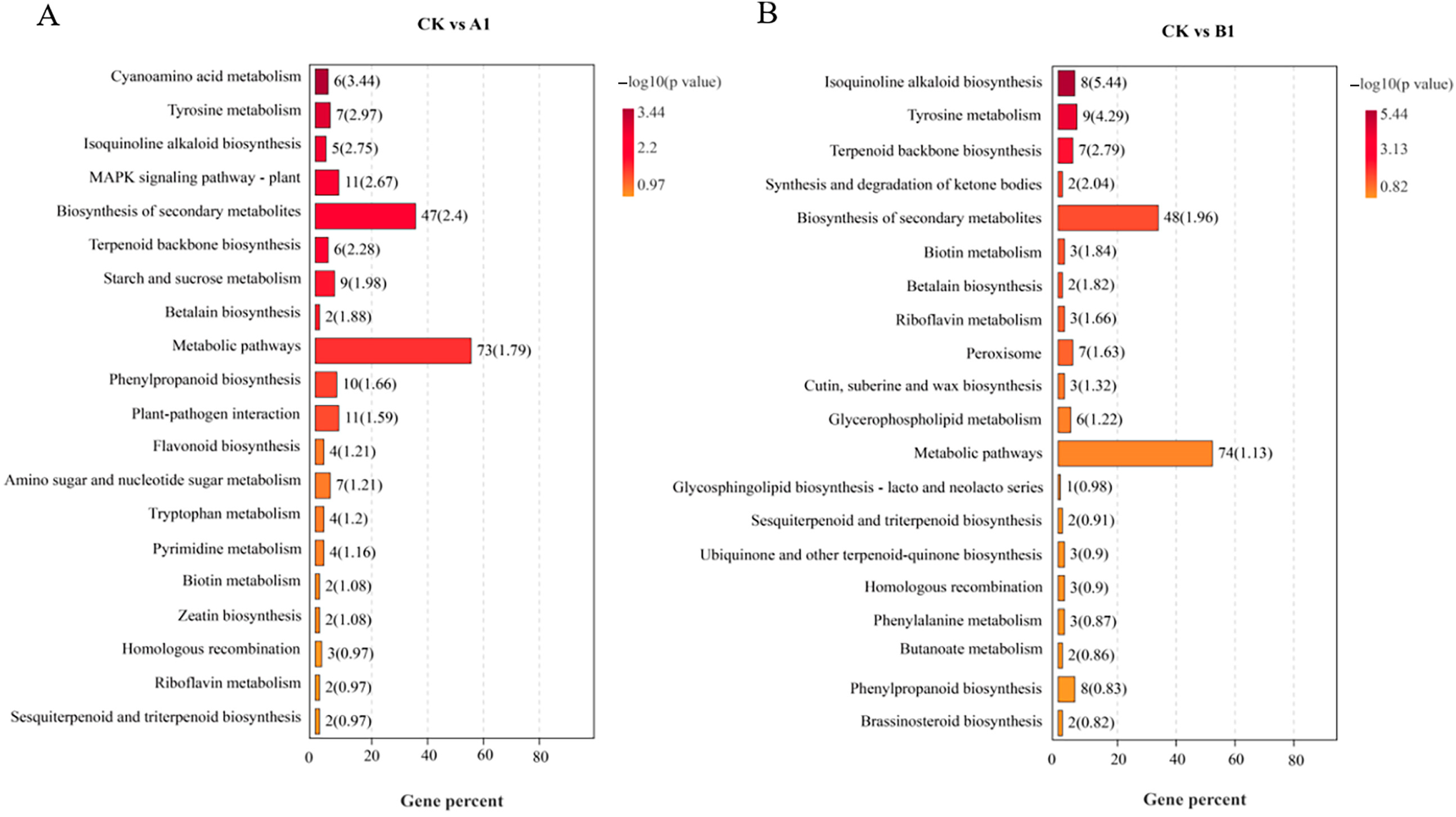

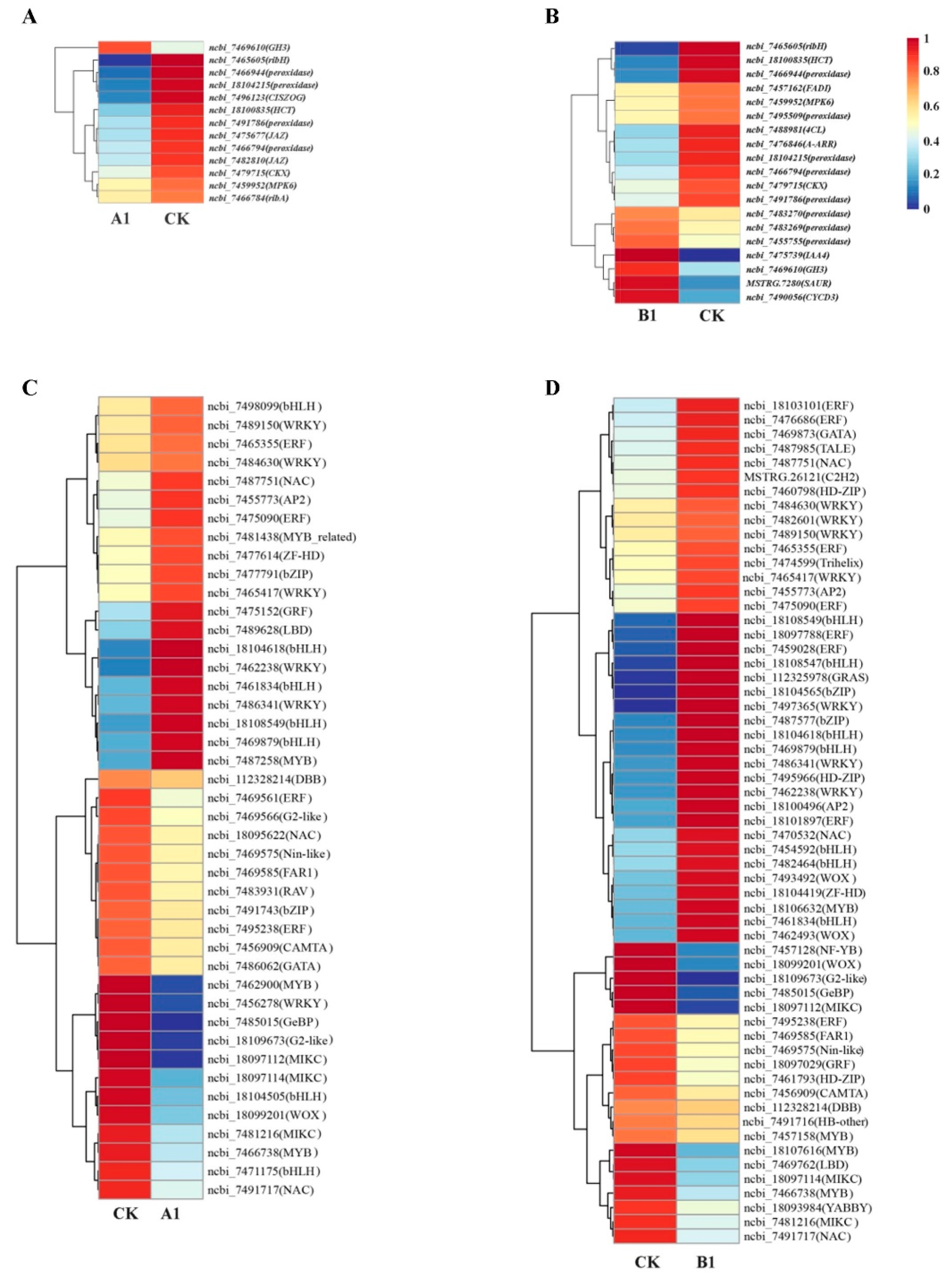

| Growth Trait | Type of Variation | Degree of Freedom | Sum of Squares | MEAN SQUARE | F | P |
|---|---|---|---|---|---|---|
| Tree height | Block | 6 | 0.4136 | 0.0689 | 0.34 | 0.9126 |
| Region | 2 | 251.855 | 125.9275 | 616.22 | 0.0001 | |
| Line | 6 | 55.7341 | 9.289 | 45.46 | 0.0001 | |
| Line × site | 12 | 24.447 | 2.0372 | 9.97 | 0.0001 | |
| Error | 36 | 7.3568 | 0.2044 | - | - | |
| Total | 62 | 339.8065 | - | - | - | |
| DBH | Block | 6 | 0.6221 | 0.1037 | 0.1562 | 0.9865 |
| Region | 2 | 289.2402 | 144.6201 | 217.8217 | 0.0001 | |
| Line | 6 | 171.7252 | 28.6209 | 43.1077 | 0.0001 | |
| Line × site | 12 | 60.1608 | 5.0134 | 7.551 | 0.0001 | |
| Error | 36 | 23.9018 | 0.6639 | - | - | |
| Total | 62 | 545.65 | - | - | - |
| Exogenous Genes | Chromosome | Transgenic Lines | Insertion Position | Insert Direction | Reads | Verified Subsequently |
|---|---|---|---|---|---|---|
| Cry1Ac-Cry3A-BADH | Chr09 | A1 | 4,383,212–4,383,306 | Reverse | 11 | - |
| Chr14 | A2 | 10,334,700–10,334,833 | Reverse | 4 | Yes (single-ended) | |
| Chr13 | A3 | 16,011,211–16,011,348 | Forward | 12 | Yes (single-ended) | |
| Cry1Ac-Cry3A-NTHK1 | Chr10 | B1 | 10,079,143–10,079,241 | Reverse | 6 | - |
| Chr14 | B2 | 1,944,472–1,944,710 | Forward | 12 | - | |
| Chr06 | B3 | 9,014,954–9,015,182 | Forward | 12 | Yes (single-ended) | |
| Chr18 | 15,956,346–1,595,632 | Reverse | 9 | Yes (single-ended) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Z.; Huang, Y.; Dong, Y.; Ren, Y.; Du, K.; Wang, J.; Yang, M. Effect of T-DNA Integration on Growth of Transgenic Populus × euramericana cv. Neva Underlying Field Stands. Int. J. Mol. Sci. 2023, 24, 12952. https://doi.org/10.3390/ijms241612952
Zhang Z, Huang Y, Dong Y, Ren Y, Du K, Wang J, Yang M. Effect of T-DNA Integration on Growth of Transgenic Populus × euramericana cv. Neva Underlying Field Stands. International Journal of Molecular Sciences. 2023; 24(16):12952. https://doi.org/10.3390/ijms241612952
Chicago/Turabian StyleZhang, Zijie, Yali Huang, Yan Dong, Yachao Ren, Kejiu Du, Jinmao Wang, and Minsheng Yang. 2023. "Effect of T-DNA Integration on Growth of Transgenic Populus × euramericana cv. Neva Underlying Field Stands" International Journal of Molecular Sciences 24, no. 16: 12952. https://doi.org/10.3390/ijms241612952
APA StyleZhang, Z., Huang, Y., Dong, Y., Ren, Y., Du, K., Wang, J., & Yang, M. (2023). Effect of T-DNA Integration on Growth of Transgenic Populus × euramericana cv. Neva Underlying Field Stands. International Journal of Molecular Sciences, 24(16), 12952. https://doi.org/10.3390/ijms241612952






