Abstract
CCR4-associated factor I (CAF1) is a deadenylase that plays a critical role in the initial step of mRNA degradation in most eukaryotic cells, and in plant growth and development. Knowledge of CAF1 proteins in woody plants remains limited. Wintersweet (Chimonanthus praecox) is a highly ornamental woody plant. In this study, CpCAF1 was isolated from wintersweet. CpCAF1 belongs to the DEDDh (Asp-Glu-Asp-Asp-His) subfamily of the DEDD (Asp-Glu-Asp-Asp) nuclease family. The amino acid sequence showed highest similarity to the homologous gene of Arabidopsis thaliana. In transgenic Arabidopsis overexpressing CpCAF1, the timing of bolting, formation of the first rosette, and other growth stages were earlier than those of the wild-type plants. Root, lateral branch, rosette leaf, and silique growth were positively correlated with CpCAF1 expression. FLOWERING LOCUS T (FT) and SUPPRESSOROF OVEREXPRESSION OF CO 1 (SOC1) gene expression was higher while EARLY FLOWERING3 (ELF3) and FLOWERING LOCUS C (FLC) gene expression of transgenic Arabidopsis was lower than the wild type grown for 4 weeks. Plant growth and flowering occurrences were earlier in transgenic Arabidopsis overexpressing CpCAF1 than in the wild-type plants. The abundance of the CpCAF1 transcript grew steadily, and significantly exceeded the initial level under 4 °C in wintersweet after initially decreasing. After low-temperature exposure, transgenic Arabidopsis had higher proline content and stronger superoxide dismutase activity than the wild type, and the malondialdehyde level in transgenic Arabidopsis was decreased significantly by 12 h and then increased in low temperature, whereas it was directly increased in the wild type. A higher potassium ion flux in the root was detected in transgenic plants than in the wild type with potassium deficiency. The CpCAF1 promoter was a constitutive promoter that contained multiple cis-acting regulatory elements. The DRE, LTR, and MYB elements, which play important roles in response to low temperature, were identified in the CpCAF1 promoter. These findings indicate that CpCAF1 is involved in flowering and low-temperature tolerance in wintersweet, and provide a basis for future genetic and breeding research on wintersweet.
1. Introduction
The CCR4-associated factor 1 (CAF1) protein is a deadenylase and an essential component of the CCR4–NOT complex, which plays a fundamental role in eukaryotic mRNA metabolism and has a multitude of different roles that impact eukaryotic gene expression [1]. Plants can develop healthily and respond more quickly to environmental changes if their mRNA metabolism affects the expression level of particular genes. The majority of previous studies on the functional roles of CAF1 are on yeasts and mammals [2,3,4,5]. The function of CAF1 has been studied in certain plants, such as Arabidopsis, pepper (Capsicum annuum), and rice (Oryza sativa), which has revealed that the gene is vital for plant growth and development, as well as response to biotic and abiotic stresses [6,7,8,9]. The Xanthomonas citri (Xc) effector protein PthA4 decreases CsCAF1 deadenylase activity in Xc-infected leaves and stabilizes CsLOB1 mRNA in Citrus sinensis [10]. The authors reported that PthA4 increases CsLOB1 transcription and translation to promote cell hypertrophy and hyperplasia in citrus by targeting the CCR4–NOT complex, implying that CsCAF1 limits cell growth in citrus. In Arabidopsis, hormones, cold damage, mechanical injury, and pathogen infection can cause the rapid expression of CAF1 genes [11]. In rice, the regulatory elements in the CAF1 promoter and variation in CAF1 expression caused by hormones and nitrogen deficiency have been studied. Only TaCAF1Ia, a novel non-typical member of the CAF1 subfamily that lacks the DEDD domain, may be a novel gene involved in another development. However, the suggested potential importance of typical CAF1 proteins in plant reproductive development has not been confirmed and their precise mechanism is currently unknown [12].
Low temperature influences plant growth, development, yield [13], and geographical distribution [14]. Frost and extreme low temperature in winter limit the sustainable growth of trees. Freezing damage is a persistent impediment to tree growth in a cold-winter climate. Many species have developed mechanisms to tolerate low-temperature conditions. Blooming in winter with strong fragrance, Chimonanthus praecox (wintersweet) is a deciduous shrub with high ornamental and economic value [15]. The reliable effects of low temperature on flower bud differentiation and blossoming is important for the regulation of flowering in wintersweet [16]. When the cumulative chilling requirement (CR) reached 570 chill units (CU), chilling caused an upregulation of ABA levels and a significant downregulation of SHORT VEGETATIVE PHASE (SVP) and FLOWERING LOCUS T (FT) homologs at the transcript level in opened and unopened flower buds (FBs, anther and ovary differentiation completed), which suggested that dormancy breaking of FBs could be regulated by the ABA-mediated SVP-FT module [17]. The molecular mechanisms of cold tolerance and flowering of wintersweet are currently under investigation. For example, the ectopic overexpression of CpNAC68, a transgene from wintersweet, in Arabidopsis enhanced the tolerance of transgenic plants to cold environments, yet had no effect on growth and development [18]. And, the overexpression of CpSIZ1, a SIZ/PIAS-Type SUMO E3 ligase from wintersweet, could delay flowering time and enhance cold tolerance in Arabidopsis [18]. Previous research has demonstrated that under CAF1 overexpression in Arabidopsis, rice, pepper, and other crops, the gene is actively expressed under low-temperature stress, effectively protecting the cell membranes and thus improving the cold tolerance of transgenic plants [19,20,21], although it has no effect on flower development. Therefore, it is probable that the biological traits caused by CAF1 in wintersweet are unique.
The aim of the present study was to explore the functions of CpCAF1 in wintersweet to gain insight into its regulatory mechanism. The full-length cDNA and upstream sequence of the CpCAF1 gene were cloned. The tolerance of transgenic Arabidopsis and the physiological effects of CpCAF1 expression in wintersweet were assessed. In addition, the upstream sequence promoter of CpCAF1 was cloned and analyzed. The results revealed that CpCAF1 plays an important role in low-temperature tolerance, plant growth, and flower development. The findings improve our understanding of how wintersweet is able to combat adverse environmental conditions and the mechanism of low-temperature-acclimation-induced low-temperature tolerance and flowering. Such knowledge is beneficial for breeding programs to enhance the ornamental value of wintersweet.
2. Results
2.1. Sequence Analysis of CpCAF1
The full-length cDNA of CpCAF1 (accession no: OP714123) was 1106 bp and contained an 807 bp long ORF encoding a protein of 269 amino acid residues with a calculated molecular mass of 30.37 kDa. The CpCAF1 protein contained a conserved domain (DEDD), including three aspartic acid (D), one glutamic acid (E), and several adjacent histidine residues belonging to the DEDDh subfamily of the DEDD RNase family.
It is speculated that the gene encodes a relatively stable hydrophobic protein, which is an important feature of biofilm proteins. The results of a neighbor-joining analysis (Figure 1A) showed that the CpCAF1 protein was highly similar to other CAF1 proteins. For instance, the CpCAF1 sequence showed 80–85% similarity with the CAF1 proteins of Arabidopsis thaliana, Nelumbo nucifera, Theobroma cacao, Vitis vinifera, Populus trichocarpa, Glycine max, Senna tora, and Corchorus olitorius. It indicated that CpCAF1 was more closely related to Arabidopsis thaliana CAF1 than to the other homologous proteins. Further homologous protein amino acid sequence alignment of CpCAF1 with 11 CAF1s of Arabidopsis marked a high degree of similarity (Figure 1B). The above conclusion was supported. No signal peptides were predicted in the sequence. The CpCAF1 is most likely to be found in the nucleus, according to the results of subcellular localization prediction. Confocal microscopic analysis showed that CpCAF1-mCherry had the fluorescence signals in both the nucleus and cytoplasm (Figure 2).
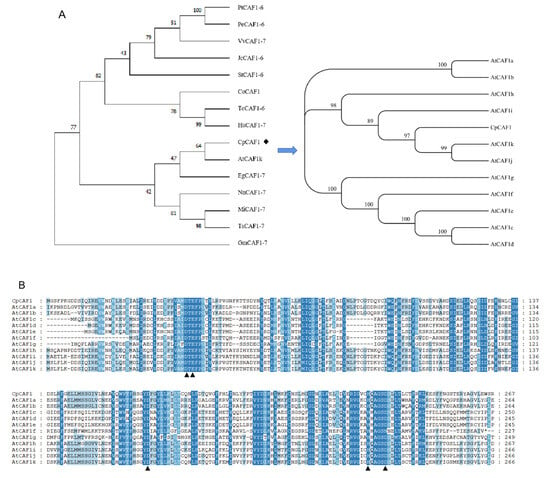
Figure 1.
Multiple sequence alignment and phylogenetic relationships of wintersweet CpCAF1. (A) Neighbor-joining tree for plant CAF1 proteins. The accession numbers from the GenBank of the sequences are as follows: Populus trichocarpa PtCAF1-6 (XP 024453230.1), Populus euphratica PeCAF1-6 (XP 011034532.1), Vitis vinifera VvCAF1-7 (XP 002272165.1), Jatropha curcas JcCAF1-6 (XP 012067515.1), Senna tora StCAF1-6 (KAF7814488.1), Corchorus olitorius CoCAF1 (OMO74191.1), Theobroma cacao TsCAF1-7 (XP 007023403.2), Herrania umbratica HuCAF1-7 (XP 021298364.1), Arabidopsis thaliana AtCAF1k (NP 565735.1), Elaeis guineensis EgCAF1-7 (XP 010929136.1), Nelumbo nucifera NnCAF1-7 (XP 010275748.1), Macadamia integrifolia MiCAF1-7 (XP 042480429.1), Telopea speciosissima TsCAF1-7 (XP 043695888.1), and Glycine max GmCAF1-7 (XP 003522433.1). (B) Multiple alignment of the deduced amino acid sequence of CpCAF1 and that of CAF1 proteins from Arabidopsis thaliana. Blue boxes indicate identical amino acids. The DEDDh catalytic site residues are indicated with arrowheads. AtCAF1a (AT3G44260.1), AtCAF1b (AT5G22250.1), AtCAF1c (AT1G27820.1), AtCAF1d (AT1G27890.1), AtCAF1e (AT1G61470.1), AtCAF1f (AT3G44240.1), AtCAF1g (AT1G06450.1), AtCAF1h (AT1G15920.1), AtCAF1i (AT5G10960.1), AtCAF1j (AT1G80780.3).
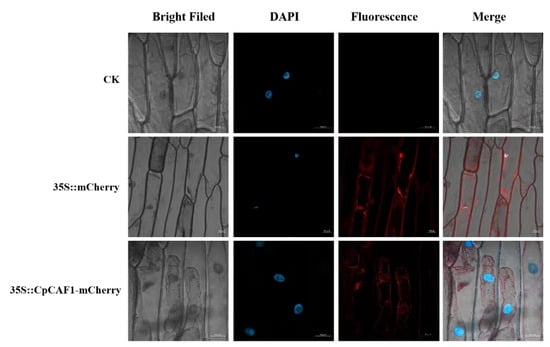
Figure 2.
Subcellular localization of 35S::CpCAF1-mCherry in onion epidermal cells. Transient expression of 35S::mCherry and 35S::CpCAF1-mCherry fusion constructs in onion epidermal cells. Cells were analyzed with confocal microscopy 36 h after bombardment. Images were taken under bright light vision (left) and fluorescence field vision (center). Merged images are shown in the right-hand panel. The blue fluorescence shows DAPI as a nuclear localization assay and the red fluorescence shows the subcellular location of the target protein. Bar = 50 μm.
2.2. CpCAF1 Response to Chilling and Flower Development in Wintersweet
A real-time quantitative PCR analysis revealed that CpCAF1 was expressed in all analyzed tissues, comprising roots, stems, cotyledons, young leaves, old leaves (mature leaves), outer petals, middle petals, inner petals, stamens, and pistils. However, the relative expression levels showed tissue-specific variation (Figure 3A). The highest expression level was detected in old leaves, followed by stamens, and expression was lowest in roots. The expression level in old leaves was more than 26 times that in roots. Similarly, during flower development, the expression level was lowest in the flower bud stage and peaked at flower senescence (Figure 3B). The expression level in the flower senescence stage was more than 4 times that in the flower bud stage.
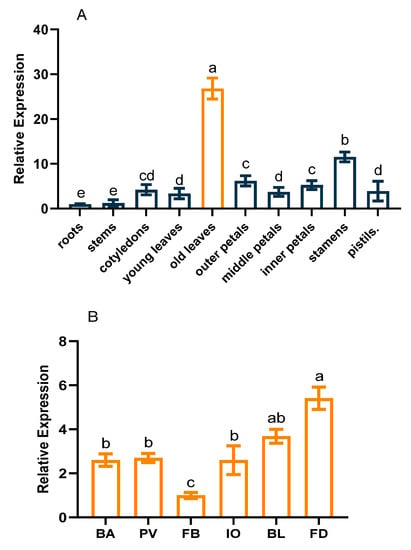
Figure 3.
CpCAF1 expression in different tissues and stages of floral development in wintersweet. (A) Relative expression of CpCAF1 in different tissues. (B) Relative expression of CpCAF1 at different stages of flower development (BA, flower buttress arises; PV, petals visible; FB, flower bud; IO, initial opening; BL, full bloom; FD, flower senescence). Each bar and error bar indicate the mean ± S.E. (N = 9). Results were analyzed using an analysis of variance, and a pairwise analysis of each event was performed using Duncan’s test for multiple comparison. The a–e represents the significance of differences analyzed applying Duncan’s multiple range test (α = 0.05).
The results suggested that CpCAF1 transcription was strongly induced by 4 °C low-temperature exposure. Compared with the control, low-temperature treatment initially reduced the abundance of CpCAF1 transcripts at 0.25 h, but thereafter the abundance gradually increased by 148% from 1 to 12 h and peaked at 12 h (Figure 4).
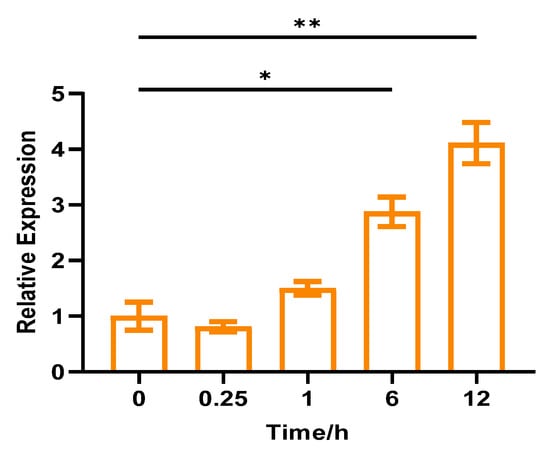
Figure 4.
Relative expression of CpCAF1 for low temperature (4 °C) in wintersweet. Each bar and error bar indicates the mean ± S.E. (N = 9). The significance of differences between means was assessed with one-way ANOVA using GraphPad Prism 9. * p < 0.05, ** p < 0.01.
2.3. Overexpression of CpCAF1 in Arabidopsis
Seven homozygous transgenic Arabidopsis lines were screened in the T4 generation. Two overexpression lines (L5 and L3) were selected for further study because they showed the highest and lowest levels of CpCAF1 expression, respectively, based on the qRT-PCR results (Figure 5).
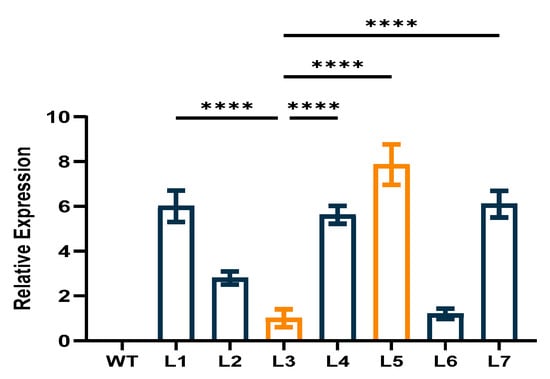
Figure 5.
Relative expression of CpCAF1 in leaves of transgenic Arabidopsis thaliana plants. WT, wild type (ecotype Col-0); L1–L7, CpCAF1-overexpressing lines. Each bar and error bar indicates the mean ± S.E. (N = 9). The significance of differences between L3 and the other group was assessed with one-way ANOVA using GraphPad Prism 9. **** p < 0.0001.
Among the phenotypic traits of L5 and L3, the timing of bolting, formation of the first rosette and stem leaves, the first inflorescence, the first flower, and the first silique were earlier than those of the wild-type plants (Table 1). Root growth, lateral branch growth, rosette leaf growth, and silique growth of the transgenic lines were positively correlated with CpCAF1 expression (Figure 6). It is noteworthy that, by the 30th day, the number of rosette leaves had reached 25 and 23 in L5 and L3, respectively, and flower opening had already occurred, whereas in the WT it had not (Figure 6C and Table 1).

Table 1.
Phenotypic traits in two T4 transgenic lines of Arabidopsis thaliana overexpressing CpCAF1 and the wild type.
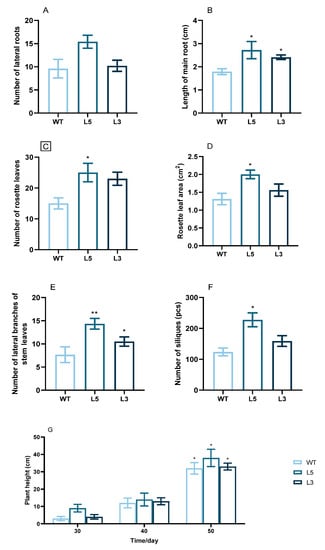
Figure 6.
Phenotypic traits of two T4 transgenic lines of Arabidopsis thaliana overexpressing CpCAF1. (A) Main root length after growth for 25 days. (B) Number of lateral roots after growth for 25 days. (C) Number of rosette leaves at 30 days. (D) Leaf area of rosette leaves at 30 days. (E) Lateral branch number of stem leaves at 30 days. (F) Number of siliques at 60 days. (G) Average plant height at different stages of growth. WT, wild type (ecotype Col-0); L3 and L5, CpCAF1-overexpressing lines. The mean of each column was compared with the mean of the WT, and each bar and error bar indicates the mean ± S.E. (N = 9). The significance of differences between the treatment and the control group was assessed with one-way ANOVA using GraphPad Prism 9. * p < 0.05, ** p < 0.01.
To investigate the effect of CpCAF1 on flowering time in transgenic Arabidopsis, plants grown for four weeks were sampled to detect the expression of genes related to flowering regulation. Plants were grown under short day (SD) conditions for 4 weeks. The expression levels of FT and SUPPRESSOROF OVEREXPRESSION OF CO 1 (SOC1) were increased significantly in L3 and L5, CpCAF1-overexpressing lines, whereas the expression levels of EARLY FLOWERING3 (ELF3) and FLOWERING LOCUS C (FLC) were decreased significantly in CpCAF1-overexpressing lines (Figure 7).
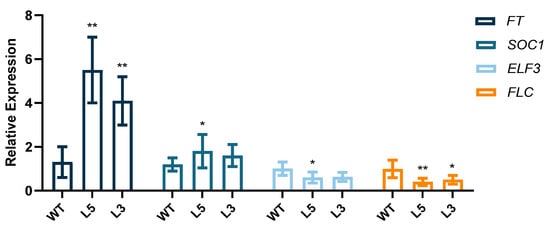
Figure 7.
Relative expression levels of FT, SOC1, ELF3, and FLC in WT, wild type (ecotype Col-0) and L3 and L5, CpCAF1-overexpressing lines grown for 4 weeks. Compare the mean of each column with the mean of WT and each bar and error bar indicates the mean ± S.E. (N = 9). The significance of differences between the treatment and the control group was assessed with one-way ANOVA using GraphPad Prism 9. * p < 0.05, ** p < 0.01.
Different levels of low-temperature damage were inflicted on wild-type and transgenic Arabidopsis after 12 h of low-temperature (−4 °C) exposure and 12 h of recovery thereafter (Figure 8). Wilting and yellowing were observed, but the wilting in transgenic lines was less severe than in Arabidopsis of the wild type. After 3 days of recovery, the survival rates of L5 were 72.4% and 68.2%, which were significantly much higher than the WT survival rate of 18.9%.
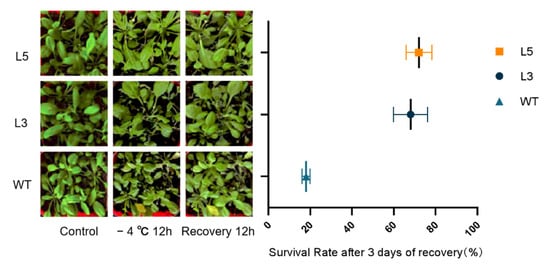
Figure 8.
Phenotype and survival rate of two T4 transgenic lines of Arabidopsis thaliana overexpressing CpCAF1 lines and wild type (WT) for low-temperature treatment and after recovery. Compare the mean of each plot with the mean of WT, and each bar and error bar indicates the mean ± S.E. (N = 9).
2.4. Biochemical Analysis of Transgenic Arabidopsis Overexpressing CpCAF1
Notable differences in proline content, MDA content, and SOD activity were observed between untreated transgenic Arabidopsis overexpressing CpCAF1 and wild-type plants for low-temperature treatment. The proline content of all plants gradually increased in response to 4 °C treatment, but that of transgenic Arabidopsis was significantly higher than that of the wild type (Figure 9A). At 48 h, the proline content of the strongly CpCAF1 overexpressing L5 line increased by 19 times, whereas that of the weakly CpCAF1 overexpressing L3 line increased by 17 times and the proline content of the wild type increased by 12.45 times. The proline content of L5 plants was 53.80% higher than the wild type and that of the L3 plants was 44.19% higher than the wild-type plants. The wild type suffered irreversible damage after 4 °C treatment as the MDA content increased. In the L5 and L3 lines, the MDA content decreased by 26.3% and 21.6% at 24 h and then increased at 48 h, and the plants suffered irreversible damage from low-temperature treatment (Figure 9B).
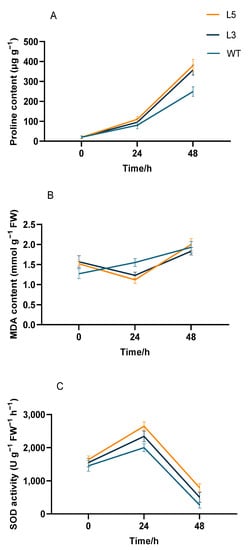
Figure 9.
Physiological indicators in two T4 transgenic lines of Arabidopsis thaliana overexpressing CpCAF1 under cold exposure. (A) Proline content, (B) malondialdehyde (MDA) content, and (C) superoxide dismutase (SOD) activity. WT, wild type (ecotype Col-0); L3 and L5, CpCAF1-overexpressing lines. Each bar and error bar indicates the mean ± S.E. (N = 9).
The SOD activity of all plants was increased in response to low-temperature treatment (Figure 9C). After 24 h of low-temperature treatment, the SOD activity of the L5 line was the highest and had increased by 67.0%. The SOD activity of the L3 line had increased by 51% and that of the wild type increased by 37.9%. The SOD activity of the L5 line was 32.5% higher than the wild type and that of the L3 was 17.0% higher than the wild type. After low-temperature treatment for 48 h, all plants showed decreased SOD activity, but the L5 and L3 transgenic Arabidopsis plants retained higher activity than the wild type. The potassium influx of the transgenic lines and wild type was induced to varying degrees with potassium deficiency. The potassium influx in mature root tissues of strongly CpCAF1 overexpressing L5 seedlings attained approximately 21 pmol cm−2 s−1, whereas that of weakly CpCAF1 overexpressing L3 seedlings was approximately 18 pmol cm−2 s−1, which were both higher than that of wild-type seedlings (Figure 10).
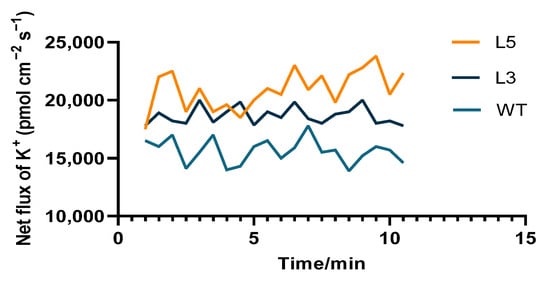
Figure 10.
Net flux of K+ in root tips of two T4 transgenic lines of Arabidopsis thaliana overexpressing CpCAF1 and the wild type. WT, wild type (ecotype Col-0); L3 and L5, CpCAF1-overexpressing lines (N = 9).
2.5. Cloning, Sequence Analysis, and Functional Characterization of the Promoter of CpCAF1
The 2133 bp of the 5′-upstream promoter region of CpCAF1 was cloned and designated CpCAF1pro. Sequence analysis showed that the TSS was located −225 bp upstream of the ATG and a map of the cis-acting regulatory elements in the promoter was constructed (Table 2). The AT1-motif, W-box, AuxRR-core, ABRE, SARE, TCA-motif, and other motifs play crucial roles in the responses to light, defense, auxin, abscisic acid (ABA), and salicylic acid. In addition, DRE (at position −456), LTR (at position −249), and MYB (at positions +151, −186, and −1231) domains, which are involved in the response to low-temperatures stress, were predicted.

Table 2.
Cis-acting regulatory elements predicted in the promoter region of CpCAF1.
To further explore the function of the CpCAF1 promoter, the transgenic plants were screened for secondary culture. Histochemical analyses of T2 transgenic Arabidopsis lines during the full developmental period indicated that the GUS gene driven by the CpCAF1 promoter was expressed in vegetative and reproductive organs (Figure 11). Quantitative fluorometric assays (Figure 12) were consistent with the results of the histochemical analysis, confirming that significant GUS activity was present in each of the major tissues and that the CpCAF1 promoter was a strongly constitutive promoter.
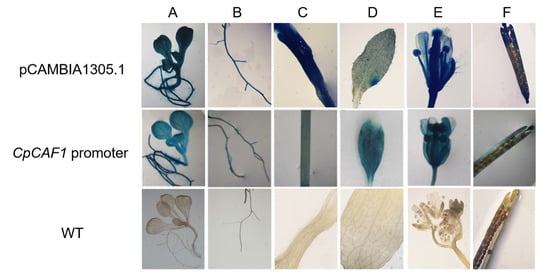
Figure 11.
Histochemical analysis of GUS activity driven by the CpCAF1 promoter in Arabidopsis thaliana. (A) Young seedling; (B) root; (C) stem; (D) leaf; (E) flower; (F) silique; CpCAF1 promoter, GUS activity driven by the CpCAF1 promoter in transgenic Arabidopsis; pCAMBIA1305.1, GUS activity driven by 35S of pCAMBIA1305.1 in transgenic Arabidopsis; WT, wild type (ecotype Col-0).
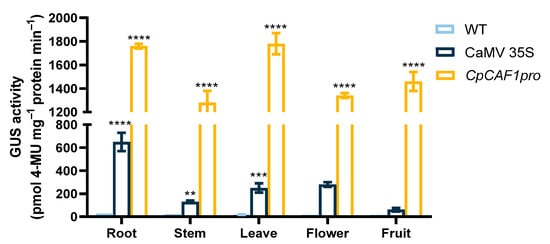
Figure 12.
GUS activity driven by the CpCAF1 or CaMV 35S promoters in Arabidopsis thaliana organs. Each bar and error bar indicates the mean ± S.E. (N = 9). The significance of differences between the CpCAF1 or CaMV 35S promoters and the wild type was assessed with two-way ANOVA using GraphPad Prism 9. ** p < 0.01, *** p < 0.001, **** p < 0.0001.
The results of serial deletion analysis in transgenic Arabidopsis (Figure 13) indicated that the promoter fragments CpCAF1pro and CpCAF1pro-P1/P2/P3 drove significantly higher GUS activity than the wild type. The CpCAF1pro and CpCAF1pro-P2 promoter fragment-driven GUS activity was maintained at high levels and both were significantly higher than that of the CpCAF1pro-P3 fragment. The CpCAF1pro-P1 promoter fragment drove significantly lower GUS activity than the CpCAF1pro promoter fragment.
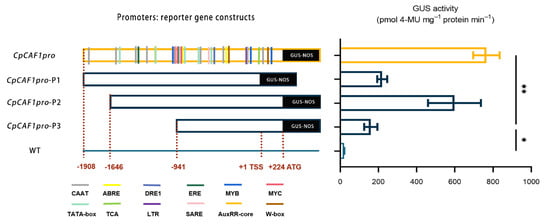
Figure 13.
Schematic representation of the CpCAF1 promoter and deletion fragment: reporter gene constructs and GUS activity in transgenic Arabidopsis thaliana. Each bar and error bar indicates the mean ± S.E. (N = 9). The significance of differences between the CpCAF1pro-P3 and the other group was assessed with one-way ANOVA using GraphPad Prism 9. * p < 0.05, ** p < 0.01.
3. Discussion
The CCR4–NOT complex in yeast and humans is composed of nine protein subunits and functions as a deadenylase, with CCR4 playing a prominent role and CAF1 filling in the gaps. A previous study discovered a novel pattern of interaction between OsCCR4 and OsCAF1 in the rice CCR4–NOT complex, and that OsCAF1 acts as a bridge between OsCCR4 and OsNOT1 in this complex [22]. CAF1 is involved in the deadenylation of mRNA in plants [23], and regulates gene expression and influences biological features [24]. The CAF1 gene is also essential for yeast and animal cells to grow and develop appropriately, although they harbor only one or two CAF1 homologs, in contrast to higher plants, which have the whole CAF1 gene family [25]. A total of 18 CAF1 homologs have been identified in rice [26], whereas 11 CAF1 orthologs have been identified in Arabidopsis. The presence of the CAF1 gene sequence in Vitis vinifera, Sorghum bicolor, Oryza sativa, and Arabidopsis was confirmed via a full-sequence genomic BLAST search of GenBank for certain plants [27]. Several CAF1 proteins are vital for plant growth and play essential roles in stress response [2,28,29,30].The potential function of CAF1 in flower development, however, has been reported previously only once: TaCAF1Ia1, a novel non-typical CAF1 lacking the DEDD domain, is specifically expressed during the early stages of anther development in wheat (Triticum aestivum). It is assumed that a distinct miR2275–CAF1 pathway evolved to regulate reproductive development in wheat [31]. However, the precise mechanism is currently not fully understood. The role of typical CAF1 proteins in reproductive development requires further investigation.
In the present study, CpCAF1, which was isolated from a wintersweet cDNA library, contained one ORF encoding a protein that shared high homology with the CAF1 of many eukaryotic organisms, especially Arabidopsis (Figure 1). Because the functional roles of CAF1 in wintersweet have not been studied previously, we explored the roles of CpCAF1 using wintersweet and the overexpression of CpCAF1 in transgenic Arabidopsis. The CpCAF1 promoter was successfully cloned and analyzed. The cis-acting regulatory elements of the CpCAF1 promoter were preliminarily investigated.
A qRT-PCR assay was used to examine the expression patterns of CpCAF1 in various organs and developmental stages of wintersweet (Figure 3A). The CpCAF1 gene was expressed in all analyzed tissues. However, the expression levels differed during flower development (Figure 3B). Between the flower senescence stage and the flower bud stage, the expression level of CpCAF1 showed an upward trend. It can be speculated that there may be unique traits and different mechanisms by which CpCAF1 was involved in floral organ development or delayed flower growth. This study provides a crucial foundation for investigating the important and uncommon potential roles of CAF1 in flower development. The present results also indicated that 4 °C treatment induced CpCAF1 transcription. The optimum temperature for flowering in wintersweet is a cooler temperature of 4 to 10 °C; 4 °C treatment initially decreased CpCAF1 transcript abundance, but subsequently the CpCAF1 transcript level climbed steadily and exceeded the original level (Figure 4). The expression levels of C-REPEAT BINDING FACTOR (CBF), whose ectopic expression can compensate for the absence of cold acclimation, and COLD-REGULATED (COR) genes in Arabidopsis tended to stabilize at 20 °C before cold treatment, while gene expression showed a fold change increase when treated at 4 °C within a day. The increased expression of these genes during cold acclimation increases freezing tolerance [32,33]. Therefore, based on the results of this study, we can speculate that CpCAF1 is implicated in chilling-response signaling pathways, which may be similar to many CAF1 genes induced by low-temperature treatment, such as AtCAF1 [34], DsCAF1 [35], OsCAF1B [36], and PtCAF1 [28].
In the T4 transgenic lines of Arabidopsis overexpressing CpCAF1, important growth stages such as the timing of bolting, first rosette leaf branch, and first flower appeared earlier than that of wild-type plants. Furthermore, the expression of the transgenic Arabidopsis FT and SOC1 gene was significantly much higher than that of the WT, whereas the expression of the ELF3 and FLC gene was significantly lower than that of the WT (Figure 7). The FT, expressed in leaves, functions as a florigen that is transported to the shoot apex to induce flowering. SOC1 was shown to directly bind to the FT promoter to activate FT transcription, thus forming a positive feedback, feed-forward loop at the leaf and growing point FT-SOC1 to ensure plant flowering induction [37]. FLC is a key gene that suppresses flowering in plants. It regulates flowering by repressing the flowering regulatory elements FT and SOC1 [38]. ELF3 is a negative regulator of flowering in Arabidopsis [39]. It indicates that CpCAF1 accelerated the transgenic Arabidopsis growth cycle, and notably also promoted earlier flowering in reproductive growth, compared with the wild type. It supports the results that suggest that GAF1 promotes the expression of FT and SOC1 by repressing genes encoding negative regulators of flowering including ELF3, a component of the Evening Complex (EC) in GA-controlled flowering [40]. Additionally, it is speculated that CAF1 may participate in the Arabidopsis flowering autonomous pathway by acting as a deadenylase, degrading the target gene’s (FLC) mRNA, lowering its abundance, reducing its negative regulatory impact on FT and SOC1, and encouraging flowering and the emergence of an early flowering phenotype. Another way CAF1 may function similarly to FY in Arabidopsis is by regulating the alternative polyadenylation (APA) of FLOWERING CONTROL LOCUS A (FCA) and FLC, two genes involved in blooming. Based on root length and other phenotypic indicators (Table 1, Figure 6), we concluded that transgenic Arabidopsis traits outperform wild-type Arabidopsis traits and the traits were proportional to gene expression level. Consistent with the aforementioned findings, CAF1 may play an important role in promoting plant growth.
To explore the changes in CpCAF1 expression in a low-temperature environment, we comparatively analyzed the lines that showed the highest and lowest levels of CpCAF1 expression (L5 and L3, respectively) for comparison in accordance with our previous research. Transgenic Arabidopsis showed strong low-temperature tolerance in the phenotype in a low-temperature environment (Figure 8). Proline accumulates in large amounts in plant tissues under certain stresses, protecting the cells against damage from reactive oxygen species [41], and is essential for osmoregulation [34,42]. The proline content of the high-CpCAF1-overexpressing L5 line showed the fastest and highest growth response to low-temperature treatment and improved the tolerance of cells to low-temperature stress (Figure 9A). Malondialdehyde is a cytotoxic product of lipid peroxidation that serves as a marker for free radical generation and tissue damage [43,44]. Under low-temperature stress, MDA, the product of membrane lipid peroxidation in plants, will accumulate in large quantities. Accumulation of MDA may cause some damage to the membrane and cells. With an increase in the duration of low-temperature treatment, the scavenging ability of the plant itself decreased and the MDA content increased. A slightly upward trend was observed for the MDA content in wild-type plants; however, the MDA content of transgenic Arabidopsis decreased initially and then increased, and the decrease in the high-CpCAF1-overexpressing L5 line was more obvious (Figure 9B). These results showed that the order of low-temperature tolerance of these three lines was L5 > L3 > wild type.
Superoxide dismutase is a metalloenzyme that catalyzes the dismutation of superoxide anions to oxygen and hydrogen peroxide. These responses serve as a defense strategy for the survival of an aerobic species [45]. The SOD activity in Arabidopsis leaves was increased by short-duration low-temperature treatment. The SOD activity of the L5 line increased the fastest and to the greatest extent, followed by the L3 line, and the difference in activity of the wild type was the smallest (Figure 9C). As low-temperature damage continues, SOD activity decreases due to the accumulation of superoxide anion free radicals and the broken balance of superoxide (O2·−) in the plant. In transgenic rice and pepper overexpressing CAF1, the gene is actively expressed in cold environments and the contents of proline and MDA, and SOD activity shows corresponding changes, thus effectively protecting the cell membranes and improving the cold tolerance of the transgenic plants [46]. The present results indicated that transgenic Arabidopsis plants were less damaged than wild-type or control plants, and thus they had superior cold tolerance, which was similar to the previous findings in rice and pepper. In wintersweet, CpNAC68 has a positive regulatory role in stress response processes. Under the stresses of heat, osmotic, and salt, the ectopic overexpression of CpNAC68 could decrease the relative electrolyte leakage and MDA content raised by transgenic Arabidopsis. The survival rate and chlorophyll content (SPAD values) were significantly greater than those of the WT plants [18,47]. According to our results, it is reasonable to speculate that CAF1 genes are involved in plant cold tolerance regulatory pathways by increasing the proline content, enhancing SOD activity, and reducing the MDA content.
Potassium is a macroelement essential for plant growth and development. A crucial role for sufficient K+ nutrition in plant tolerance to various abiotic and biotic stresses has been reported [48]. According to the currently available evidence, potassium supports plant survival in cold environments by decreasing reactive oxygen species production and increasing antioxidant concentrations [36,49]. We used non-destructive microscopy to evaluate the potassium ion flow rate of Arabidopsis roots for approximately 2 weeks to explore the mechanism of action of CpCAF1 on potassium ion absorption in Arabidopsis roots. In a low-potassium environment, the potassium influx of transgenic and wild-type seedlings was induced to varying degrees (Figure 10). The strongly CpCAF1 overexpressing L5 line showed the greatest induction, followed by the weakly CpCAF1 overexpressing L3 line, and induction was weakest in the wild type. Compared with the cold-sensitive tomato cultivar ‘S708′, the cold-tolerant tomato cultivar ‘T722’ exhibits a lower reduction in plant growth rate, whole-plant K+ content, and K+ net uptake [50]. Moreover, potassium functions in the plant signaling system to defend against low-temperature conditions and ensure an adequate potassium supply for the plant, whereas potassium deficiency can increase the susceptibility of plants to low temperatures [51]. The strongly CpCAF1 overexpressing L5 transgenic Arabidopsis line exhibited a significant potassium-deficiency-induced potassium influx and greater potassium ion flow into the cell even with potassium deficiency. Thus, it is reasonable to speculate that CpCAF1-overexpressing Arabidopsis has a better potassium deficiency tolerance and superior growth for low-temperature exposure than the wild type. This suggests that CpCAF1 is involved in the regulation of the potassium pathway to improve the low-temperature tolerance of plants.
In response to diverse environmental stimuli, plants have complex signal transduction networks and synthesize signal molecules for the defense response [46]. The CpCAF1 promoter sequence contained DRE, LTR, and MYB elements, which are involved in the low-temperature stress response, ABRE elements induced by drought, MYC elements that play a role in drought stress response, and ABA induction (Table 2). The promoter regions of the 19 PtCAF1 genes of Populus trichocarpa contain many cis-acting elements responsive to environmental stress [52]. The plant hormone ABA plays important roles in plant development and response to stresses [53,54,55]. The present results suggest that CpCAF1 plays a role in plant defense responses. A histochemical analysis of the construct containing the regulatory region of the CpCAF1 promoter revealed GUS expression in all tissues examined (Figure 10 and Figure 11). However, the GUS activity driven by the CpCAF1pro-P1 fragment was much lower than that driven by CpCAF1pro. The CpCAF1pro and CpCAF1pro-P2 promoter fragment-driven GUS activity was maintained at high levels and was significantly higher than that of the CpCAF1pro-P3 fragment (Figure 13). These results indicated that putative region-specific enhancers may exist within the promoter from −1621 to −941 bp and 0 to +224 bp. The high AT content (70.9%) in the CpCAF1 promoter sequence, which is generally accepted to be a characteristic of duplex DNA more prone to unwinding and improves the transcriptional efficiency of the gene [29], likely explains why the CpCAF1 promoter promotes GUS activity more significantly than the 35S promoter.
4. Materials and Methods
4.1. Plant Materials, Growth Conditions, and Low-Temperature Treatment
Adult individuals of wintersweet (C. praecox) were planted in a nursery at Southwest University (Chongqing, China). Arabidopsis thaliana ecotype Columbia (Col-0) was used for genetic transformation. All Arabidopsis plants were grown at 25 °C under a 16 h/8 h (light/dark) photoperiod and a light intensity of 55 µmol·m−2·s−1. Murashige and Skoog (MS) medium [56] supplemented with kanamycin (100 mg L−1) was used to screen for transformants. Wintersweet seeds were germinated in pots containing nutritional soil and seedlings at the four-leaf stage were used in the following treatments. For the low-temperature treatment, the seedlings were placed in a growth cabinet maintained at 4 °C for 0.25, 1, 6, or 12 h; refer to the research results of Li et al. [17]. The cotyledons, young leaves, roots, stem, and leaves were collected separately from plants at the four-leaf stage for RNA isolation. Flower buds and whole flowers were collected at different stages of floral development from the adult plants, following the method of Sui et al. [57] for RNA extraction and gene expression analysis.
4.2. Cloning of CpCAF1 and Bioinformatic Analysis
A potential target expressed sequence tag (EST) sequence was isolated from the wintersweet flower EST library using the 5-pTriplEx2 sequence primer (5′-TCCGAGATCTGGACGAGC-3′) [58]. The BlastN (nucleotide–nucleotide BLAST) and BlastX (translated query vs. protein database) tools accessible on the NCBI website (http://www.ncbi.nlm.nih.gov/ (accessed on 15 January 2020) and SeqMan version 11.1 (DNASTAR, Inc., Madison, WI, USA) were applied to analyze multiple sequence alignments and cluster analysis on the EST sequence. The full-length cDNA of CpCAF1 was cloned from the library with the primer pair T7 (5′-TAATACGACTCACTATAGGG-3′) and SP5 (5′-CTTCTGCTCTAAAAGCTGCG-3′). The sequence characteristics were analyzed with DNASTAR version 7.1 and DNAMAN version 6.03.99 bioinformatic software. The signal peptide and subcellular localization were predicted with the online software SignalP version 5.0 (http://www.cbs.dtu.dk/services/SignalP/ (accessed on 30 January 2020) and the TargetP 1.1 server (http://www.cbs.dtu.dk/services/TargetP/ (accessed on 30 January 2020), respectively. The alignment of the amino acid sequences of homologous proteins and the construction of a dendrogram using the neighbor-joining method were conducted with MEGA version 11 [59].
4.3. Subcellular Localization of CpCAF1
The coding sequence (CDS) of CpCAF1 without stop codon was inserted into SacI and BamHI of the modified pCAMBIA1300-mCherry vector to form a recombinant vector 35S::CpCAF1-mCherry. The epidermis of onion cells was cultured in MS media for 24 h in the dark, then agrobacterium GV3101 carrying 35S::CpCAF1-mCherry or 35S::mCherry were transferred into onion epidermal cells. After 36 h, mCherry fluorescent was observed using confocal laser microscopy (Olympus, Japan).
4.4. RNA Isolation and Quantitative Real-Time PCR Analysis of CpCAF1
Total RNA from all plant samples was isolated using the RNAprep Pure Kit (TIANGEN, Beijing, China). RNA sample OD260/OD280 and OD260/OD230 were determined with NanoDrop2000 (Thermo, Shanghai, China) and RNA quality was determined via agarose gel electrophoresis. The remnant genomic DNA from RNA was removed and the first-strand cDNA was synthesized using the All-in-One First-Strand Synthesis MasterMix (with dsDNase) (Yugong Biolabs, Jiangsu, China). The amount of RNA per PCR sample was 1 μg. Quantitative real-time PCR (qRT-PCR) was performed with the SsoFast™ EvaGreen SuperMix Kit (BioRad, Hercules, CA, USA). The primers used for real-time PCR were designed with Primer Premier 5.0. CpActin and were used as the internal reference genes. The reactions took place at a 20 μL volume and contained 4.0 μL 10 × each of forward and reverse primers, 2.0 μL 10× of cDNA, and 10.0 μL SsoFast™ EvaGreen® Supermix. The reactions were carried out using the following protocol: 95 °C for 30 s, followed by 40 cycles of 95 °C for 5 s and 56 °C for 30 s, 65 °C for 0.05 s, and 95 °C for 0.5 s. qRT-PCR equipment was c1000 touch thermal cycler combined with CFC96TM Real-Time system (BioRad, CA, USA). Relative gene expression levels were calculated using the 2−ΔΔCt method [60] with Bio-Rad Manager™ (Version 1.1) software. In addition, qRT-PCR was used to detect the CpCAF1 expression level in transgenic Arabidopsis, relative to the AtActin reference gene, using the above-mentioned method. All primers used in the study are listed in Table S1. Three biological replicates and three technical replicates were analyzed for each qRT-PCR reaction.
4.5. Construction of Plant Overexpression Vectors for CpCAF1 and Transformation of Arabidopsis
The open reading frame (ORF) of CpCAF1 was amplified with gene-specific primers (CpCAF1-F: 5′-GAATTCATGGGGAGTTTTCCCAAAGGTG-3′, CpCAF1-R: 5′-GGATCCTTACCACCGAGACCATTCCAAGACAC-3′) containing the restriction enzyme sites SacI and XbalI. The amplified fragments were inserted into the binary vector pCAMBIA2301, which contained the CaMV 35S promoter, to generate the recombinant vector designated pCAMBIA2301-CpCAF1. The recombinant vector was transformed into Agrobacterium tumefaciens strain GV3103 and the kanamycin-resistant lines were selected. Transformation of Arabidopsis with A. tumefaciens was performed using the floral-dip method [61]. The T1 transgenic lines were selected on MS medium supplemented with 100 mg L−1 kanamycin. The kanamycin-resistant seedlings were transplanted to soil and were grown to maturation. The T4 transgenic lines were selected using the same method. Genomic DNA was extracted from the leaves of putative transgenic Arabidopsis plants using the Plant Genomic DNA Kit (TIANGEN) in accordance with the manufacturer’s instructions. The PCR amplification was performed with the primers CpCAF1-F/R. The PCR procedure included an initial preheating step at 95 °C for 5 min, followed by 27 denaturation cycles run of 95 °C for 30 s, annealing at 56 °C for 30 s, and an extension at 72 °C for 1 min 40 s, followed by a final extension time at 72 °C for 10 min. The ensuing PCR products were detected with 1% gel electrophoresis to confirm CpCAF1’s insertion into the transgenic plants. Real-time quantitative PCR was used to verify the identity of the transgenic plants and classify the transgenic lines into different groups based on the CpCAF1 expression level.
4.6. Phenotypic Observation
To investigate whether CpCAF1 affects the nutritional and reproductive growth of transgenic plants, we observed and recorded data on phenotypic traits at different growth stages of transgenic Arabidopsis and compared their growth rate and growth status with those of wild-type Arabidopsis. The T4 generation Arabidopsis strains identified as positive for the CpCAF1 gene were subjected to expression analysis, and two transgenic strains (1 strongly and 1 slightly weakly expressed) were selected, and wild-type Arabidopsis (WT) was used as a control. In total, 90 plants of each transgenic strain and WT were selected as phenotypic observation record plants, and the phenotypes were observed and compared from seedling stage to scape-bearing stage to the decaying stage at regular intervals every day. In addition, the transgenic plants and WT were observed and photographed after 12 h of low-temperature (−4 °C) treatment and 12 h of resumption of normal culture (22 °C). Survival rates were then calculated after 3 days of recovery.
4.7. Measurement of Physiological Indicators
To assess the effect of CpCAF1 on the physiological indicators of low-temperature treatment, rosette leaves were sampled from the transgenic and wild-type plants (each sample comprised 0.1 g). The proline content was determined following the method of Bates et al. [62]. The malondialdehyde (MDA) content was estimated using the procedure described by Heath and Packer [27]. Superoxide dismutase (SOD) activity was assayed by measuring the ability to inhibit the photochemical reduction of nitroblue tetrazolium in accordance with the method of Beauchamp and Fridovich [19]. Each measurement was replicated three times.
For ion flux analyses, transgenic and wild-type Arabidopsis seedlings germinated over 2 weeks were measured using the non-invasive microtest system (NMT-YG-100, Younger, Amherst, MA, USA). The ion-selective electrodes were calibrated using the following solutions: Na+, 0.5 and 5.0 mmol L−1 NaCl; K+, 0.05 and 0.5 mmol L−1 KCl [21]. Taproot segments were immobilized on the bottom of a measuring dish and incubated in the measuring solution to equilibrate for 5 min. Steady fluxes of K+ in the meristematic zone of intact roots were measured and continuously recorded for 10 min.
4.8. Cloning and Analysis of the Upstream Sequence of CpCAF1
Genomic DNA was extracted from the leaves of young seedlings using the cetyltrimethylammonium bromide (CTAB) method [58]. Using the Chimonanthus salicifolius genome [31] assembly as the reference, the upstream sequence of CpCAF1 was amplified using the gene-specific primers CpCAF1-Pro-F (5′-GTGTAAGTGAAGAATTAAGAATTGTCG-3′) and CpCAF1-Pro-R (5′-CTTCTTCTCTTTTCCTTTTTTTTCCT-3′). The amplified fragment was designated CpCAF1pro. The purified product was cloned into the pMD19-T vector (TaKaRa Bio, Dalian, China), and then sequenced and analyzed. The prediction of cis-acting regulatory elements in the upstream sequence of CpCAF1 was performed using the PLACE (http://www.dna.affrc.go.jp/PLACE/signalscan.html (accessed on 10 January 2021) and PlantCARE (http://bioinformatics.psb.Ugent.be/webtools/plantcare.html (accessed on 10 January 2021) databases.
4.9. Construction and Transformation of CpCAF1 Promoter Deletions Fused to GUS
For analysis of the CpCAF1 promoter in transgenic tobacco plants, constructs containing the CpCAF1pro::GUS cassette were used. A 2133-bp promoter fragment (−1908 to +224 from the transcription start site; TSS), was amplified using specific primers (Table S2) containing the XbaI and NcoI restriction sites and cloned into the pCAMBIA1305.1 vector. Three truncated promoter fragments were generated and inserted into the vector using the same method: CpCAF1pro-P1 (−1980 to +2), CpCAF1pro-P2 (−1646 to +224), and CpCAF1pro-P3 (−941 to +224). The plasmids (CpCAF1pro/-P1/P2/P3::GUS) were transformed into Arabidopsis (Col-0) using the floral-dip method [19] mediated with Agrobacterium tumefaciens strain GV3101 cells. The seeds from T0 transgenic plants were selected on MS medium supplemented with 25 mg L−1 hygromycin B. Transgenic Arabidopsis T2 seeds were sown and the seedlings were cultured in MS liquid medium. Sampling was conducted after incubation for 6 h with three replicates per sample.
4.10. Histochemical and Fluorometric Analysis of GUS Activity
For histochemical and fluorometric assays of β-glucuronidase (GUS) activity, fresh samples (small plants or tubers) were treated with X-Gluc solution (Sigma-Aldrich, Shanghai, China). The GUS activity was quantified using 4-methylumbelliferyll-β-D-glucuronide (Sigma-Aldrich) as described previously [16]. The total protein activity was determined using Varioskan Flash (a full wavelength scanning multi-function reading device) in accordance with the method of Bradford [26].
4.11. Statistical Analyses
All statistical analyses were conducted with GraphPad Prism version 9.0.0 (GraphPad Software, Inc., San Diego, CA, USA).
5. Conclusions
In conclusion, the present findings showed that wintersweet CpCAF1 accelerates each growth stage in Arabidopsis and specifically implied that the gene plays a role in promoting flowering through gibberellin or the autonomous pathway. Furthermore, CpCAF1 enhances the low-temperature tolerance of Chimonanthus praecox by increasing the proline content, decreasing the MDA content, enhancing SOD activity, and increasing potassium ion flux, which supports the hypothesis that CpCAF1 is a cold-related transcription factor with a positive regulatory effect in low-temperature tolerance enhancement. However, its specific regulatory mechanism still needs further study. In addition, the CpCAF1 promoter is predicted to be a constitutive promoter containing elements involved in low-temperature stress response, which further supports the above conclusions. Given that CpCAF1 is indicated to play important roles in flower development and low-temperature tolerance, the next logical step is to investigate the regulatory mechanism of the gene in promoting plant growth and earlier flowering and flower senescence, as well as the structural function of proteins encoded by CpCAF1 and the potential regulators of CpCAF1 in low-temperature tolerance.
Supplementary Materials
The supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms241612945/s1.
Author Contributions
M.L. and S.S. conceived the research and designed the experiments; S.Z. performed all experiments of CpCAF1 including the cloning, production of transgenic materials, QRT-PCR analysis, and measurement of physiological indicators; B.W. contributed to the upstream sequence of CpCAF1 experiments; Y.L. analyzed the data and wrote the manuscript; M.X. assisted with writing the manuscript; J.M. supervised the experiments, manuscript revision, and approved the submitted version. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the National Natural Science Foundation of China (grant nos. 32271937 and 31370698).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available in the article.
Acknowledgments
We thank Southwest University for their support with the sample collection.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Behm-Ansmant, I.; Rehwinkel, J.; Doerks, T.; Stark, A.; Bork, P.; Izaurralde, E. MRNA degradation by miRNAs and GW182 requires both CCR4: NOT deadenylase and DCP1: DCP2 decapping complexes. Gene Dev. 2006, 20, 1885–1898. [Google Scholar] [CrossRef] [PubMed]
- Balu, B.; Maher, S.P.; Pance, A.; Chauhan, C.; Naumov, A.V.; Andrews, R.M.; Ellis, P.D.; Khan, S.M.; Lin, J.W.; Janse, C.J.; et al. CCR4-Associated Factor 1 Coordinates the Expression of Plasmodium falciparum Egress and Invasion Proteins. Eukaryot. Cell 2011, 10, 1257–1263. [Google Scholar] [CrossRef] [PubMed]
- Molin, L.; Puisieux, A. C-elegans homologue of the Caf1 gene, which encodes a subunit of the CCR4-NOT complex, is essential for embryonic and larval development and for meiotic progression. Gene 2005, 358, 73–81. [Google Scholar] [CrossRef]
- Kopylov, P.K.; Platonov, M.E.; Ablamunits, V.G.; Kombarova, T.I.; Ivanov, S.A.; Kadnikova, L.A.; Somov, A.N.; Dentovskaya, S.V.; Uversky, V.N.; Anisimov, A.P. Yersinia pestis Caf1 Protein: Effect of Sequence Polymorphism on Intrinsic Disorder Propensity, Serological Cross-Reactivity and Cross-Protectivity of Isoforms. PLoS ONE 2016, 11, e0162308. [Google Scholar] [CrossRef]
- Niinuma, S.; Fukaya, T.; Tomari, Y. CCR4 and CAF1 deadenylases have an intrinsic activity to remove the post-poly(A) sequence. RNA 2016, 22, 1550–1559. [Google Scholar] [CrossRef] [PubMed]
- Chou, W.L.; Huang, L.F.; Fang, J.C.; Yeh, C.H.; Hong, C.Y.; Wu, S.J.; Lu, C.A. Divergence of the expression and subcellular localization of CCR4-associated factor 1 (CAF1) deadenylase proteins in Oryza sativa. Plant Mol. Biol. 2014, 85, 443–458. [Google Scholar] [CrossRef]
- Fang, J.C.; Tsai, Y.C.; Chou, W.L.; Liu, H.Y.; Chang, C.C.; Wu, S.J.; Lu, C.A. A CCR4-associated factor 1, OsCAF1B, confers tolerance of low-temperature stress to rice seedlings. Plant Mol. Biol. 2021, 105, 177–192. [Google Scholar] [CrossRef]
- Sarowar, S.; Oh, H.W.; Cho, H.S.; Baek, K.H.; Seong, E.S.; Joung, Y.H.; Choi, G.J.; Lee, S.; Choi, D. Capsicum annuum CCR4-associated factor CaCAF1 is necessary for plant development and defence response. Plant J. 2007, 51, 792–802. [Google Scholar] [CrossRef]
- Walley, J.W.; Kelley, D.R.; Nestorova, G.; Hirschberg, D.L.; Dehesh, K. Arabidopsis Deadenylases AtCAF1a and AtCAF1b Play Overlapping and Distinct Roles in Mediating Environmental Stress Responses. Plant Physiol. 2010, 152, 866–875. [Google Scholar] [CrossRef]
- Shimo, H.M.; Terassi, C.; Silva, C.C.L.; Zanella, J.D.; Mercaldi, G.F.; Rocco, S.A.; Benedetti, C.E. Role of the Citrus sinensis RNA deadenylase CsCAF1 in citrus canker resistance. Mol. Plant Pathol. 2019, 20, 1105–1118. [Google Scholar] [CrossRef]
- Liang, W.X.; Li, C.B.; Liu, F.; Jiang, H.L.; Li, S.Y.; Sun, J.Q.; Wu, X.Y.; Li, C.Y. The Arabidopsis homologs of CCR4-associated factor 1 show mRNA deadenylation activity and play a role in plant defence responses. Cell Res. 2009, 19, 307–316. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Song, R.; Wang, Y.; Wang, X.; Peng, J.; Nevo, E.; Ren, X.; Sun, D. New insights into the evolution of CAF1 family and utilization of TaCAF1Ia1 specificity to reveal the origin of the maternal progenitor for common wheat. J. Adv. Res. 2022, 42, 135–148. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.Y.; Nawaz, M.A.; Wei, N.N.; Cheng, F.; Bie, Z.L. Suboptimal Temperature Acclimation Enhances Chilling Tolerance by Improving Photosynthetic Adaptability and Osmoregulation Ability in Watermelon. Hortic. Plant J. 2020, 6, 49–60. [Google Scholar] [CrossRef]
- Wu, Y.Y.; Wu, S.H.; Wang, X.Q.; Mao, T.Y.; Bao, M.Z.; Zhang, J.W.; Zhang, J. Genome-wide identification and characterization of the bHLH gene family in an ornamental woody plant Prunus mume. Hortic. Plant J. 2022, 8, 531–544. [Google Scholar] [CrossRef]
- Liu, D.F.; Sui, S.Z.; Ma, J.; Li, Z.N.; Guo, Y.L.; Luo, D.P.; Yang, J.F.; Li, M.Y. Transcriptomic Analysis of Flower Development in Wintersweet (Chimonanthus praecox). PLoS ONE 2014, 9, e86976. [Google Scholar] [CrossRef]
- Jefferson, R.A.; Kavanagh, T.A.; Bevan, M.W. Gus Fusions—Beta-Glucuronidase as a Sensitive and Versatile Gene Fusion Marker in Higher-Plants. Embo J. 1987, 6, 3901–3907. [Google Scholar] [CrossRef]
- Li, Z.N.; Liu, N.; Zhang, W.; Wu, C.Y.; Jiang, Y.J.; Ma, J.; Li, M.Y.; Sui, S.Z. Integrated transcriptome and proteome analysis provides insight into chilling-induced dormancy breaking in Chimonanthus praecox. Hortic. Res. 2020, 7, 198. [Google Scholar] [CrossRef]
- Lin, J.; Liu, D.F.; Wang, X.; Ahmed, S.; Li, M.Y.; Kovinich, N.; Sui, S.Z. Transgene CpNAC68 from Wintersweet (Chimonanthus praecox) Improves Arabidopsis Survival of Multiple Abiotic Stresses. Plants 2021, 10, 1403. [Google Scholar] [CrossRef]
- Beauchamp, C.; Fridovich, I. Superoxide dismutase: Improved assays and an assay applicable to acrylamide gels. Anal. Biochem. 1971, 44, 276–287. [Google Scholar] [CrossRef]
- Ferrari, S.; Galletti, R.; Denoux, C.; De Lorenzo, G.; Ausubel, F.M.; Dewdney, J. Resistance to Botrytis cinerea induced in Arabidopsis by elicitors is independent of salicylic acid, ethylene, or jasmonate signaling but requires PHYTOALEXIN DEFICIENT3. Plant Physiol. 2007, 144, 367–379. [Google Scholar] [CrossRef]
- Sun, J.; Chen, S.; Dai, S.; Wang, R.; Li, N.; Shen, X.; Zhou, X.; Lu, C.; Zheng, X.; Hu, Z.; et al. NaCl-induced alternations of cellular and tissue ion fluxes in roots of salt-resistant and salt-sensitive poplar species. Plant Physiol. 2009, 149, 1141–1153. [Google Scholar] [CrossRef] [PubMed]
- Chou, W.L.; Chung, Y.L.; Fang, J.C.; Lu, C.A. Novel interaction between CCR4 and CAF1 in rice CCR4-NOT deadenylase complex. Plant Mol. Biol. 2017, 93, 79–96. [Google Scholar] [CrossRef] [PubMed]
- Collart, M.A. Global control of gene expression in yeast by the Ccr4-Not complex. Gene 2003, 313, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Ramnarain, D.B.; Chiang, Y.C.; Ding, L.H.; McMahon, J.S.; Denis, C.L. Genome wide expression analysis of the CCR4-NOT complex indicates that it consists of three modules with the NOT module controlling SAGA-responsive genes. Mol. Genet. Genom. 2008, 279, 323–337. [Google Scholar] [CrossRef]
- Schwede, A.; Manful, T.; Jha, B.A.; Helbig, C.; Bercovich, N.; Stewart, M.; Clayton, C. The role of deadenylation in the degradation of unstable mRNAs in trypanosomes. Nucleic Acids Res. 2009, 37, 5511–5528. [Google Scholar] [CrossRef]
- Bradford, M.M. Rapid and Sensitive Method for Quantitation of Microgram Quantities of Protein Utilizing Principle of Protein-Dye Binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Heath, R.L.; Packer, L. Photoperoxidation in Isolated Chloroplasts. I. Kinetics and Stoichiometry of Fatty Acid Peroxidation. Arch. Biochem. Biophys. 1968, 125, 189–198. [Google Scholar] [CrossRef]
- Wang, P.; Li, L.L.; Wei, H.; Sun, W.B.; Zhou, P.J.; Zhu, S.; Li, D.W.; Zhuge, Q. Genome-Wide and Comprehensive Analysis of the Multiple Stress-Related CAF1 (CCR4-Associated Factor 1) Family and Its Expression in Poplar. Plants 2021, 10, 981. [Google Scholar] [CrossRef]
- Stalberg, K.; Ellerstom, M.; Ezcurra, I.; Ablov, S.; Rask, L. Disruption of an overlapping E-box/ABRE motif abolished high transcription of the napA storage-protein promoter in transgenic Brassica napus seeds. Planta 1996, 199, 515–519. [Google Scholar] [CrossRef]
- Walley, J.W.; Kelley, D.R.; Savchenko, T.; Dehesh, K. Investigating the function of CAF1 deadenylases during plant stress responses. Plant Signal Behav. 2010, 5, 802–805. [Google Scholar] [CrossRef][Green Version]
- Lv, Q.D.; Qiu, J.; Liu, J.; Li, Z.; Zhang, W.T.; Wang, Q.; Fang, J.; Pan, J.J.; Chen, Z.D.; Cheng, W.L.; et al. The Chimonanthus salicifolius genome provides insight into magnoliid evolution and flavonoid biosynthesis. Plant J. 2020, 103, 1910–1923. [Google Scholar] [CrossRef] [PubMed]
- Knight, M.R.; Knight, H. Low-temperature perception leading to gene expression and cold tolerance in higher plants. New Phytol. 2012, 195, 737–751. [Google Scholar] [CrossRef] [PubMed]
- Thomashow, M.F. Molecular Basis of Plant Cold Acclimation: Insights Gained from Studying the CBF Cold Response Pathway. Plant Physiol. 2010, 154, 571–577. [Google Scholar] [CrossRef] [PubMed]
- Laliberte, G.; Hellebust, J.A. Regulation of Proline Content of Chlorella autotrophica in Response to Changes in Salinity. Can. J. Bot. 1989, 67, 1959–1965. [Google Scholar] [CrossRef]
- Chen, X.J.; Zhang, X.H.; Hu, L.D.; Zhang, J.Q.; Jiang, Y.; Yang, Y.; Yan, Y.B. DsCaf1 is involved in environmental stress response of Dunaliella salina. Int. J. Biol. Macromol. 2016, 82, 369–374. [Google Scholar] [CrossRef]
- Cakmak, I. The role of potassium in alleviating detrimental effects of abiotic stresses in plants. J. Plant Nutr. Soil Sc. 2005, 168, 521–530. [Google Scholar] [CrossRef]
- Kou, K.; Yang, H.; Li, H.; Fang, C.; Chen, L.; Yue, L.; Nan, H.; Kong, L.; Li, X.; Wang, F.; et al. A functionally divergent SOC1 homolog improves soybean yield and latitudinal adaptation. Curr. Biol. 2022, 32, 1728–1742.e6. [Google Scholar] [CrossRef]
- Crevillen, P.; Dean, C. Regulation of the floral repressor gene FLC: The complexity of transcription in a chromatin context. Curr. Opin. Plant Biol. 2011, 14, 38–44. [Google Scholar] [CrossRef]
- Hartmann, U.; Hohmann, S.; Nettesheim, K.; Wisman, E.; Saedler, H.; Huijser, P. Molecular cloning of SVP: A negative regulator of the floral transition in Arabidopsis. Plant J. 2000, 21, 351–360. [Google Scholar] [CrossRef]
- Fukazawa, J.; Miyamoto, C.; Ando, H.; Mori, K.; Takahashi, Y. DELLA-GAF1 complex is involved in tissue-specific expression and gibberellin feedback regulation of GA20ox1 in Arabidopsis. Plant Mol. Biol. 2021, 107, 147–158. [Google Scholar] [CrossRef]
- Choudhary, M.; Jetley, U.K.; Khan, M.A.; Zutshi, S.; Fatma, T. Effect of heavy metal stress on proline, malondialdehyde, and superoxide dismutase activity in the cyanobacterium Spirulina platensis-S5. Ecotoxicol. Environ. Saf. 2007, 66, 204–209. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, I.; Hellebust, J.A. The Relationship between Inorganic Nitrogen-Metabolism and Proline Accumulation in Osmoregulatory Responses of 2 Euryhaline Microalgae. Plant Physiol. 1988, 88, 348–354. [Google Scholar] [CrossRef] [PubMed]
- Seto, H.; Ohkubo, T. Determination of a Guanosine Malonaldehyde Adduct in Urine by High-Performance Liquid-Chromatography with a Thiobarbituric Acid Reaction Detector. J. Chromatogr. Biomed. 1991, 570, 301–307. [Google Scholar] [CrossRef] [PubMed]
- Ohkawa, H.; Ohishi, N.; Yagi, K. Assay for Lipid Peroxides in Animal-Tissues by Thiobarbituric Acid Reaction. Anal. Biochem. 1979, 95, 351–358. [Google Scholar] [CrossRef]
- Beyer, W.; Imlay, J.; Fridovich, I. Superoxide Dismutases. Prog. Nucleic Acid Res. Mol. Biol. 1991, 40, 221–253. [Google Scholar] [PubMed]
- Yang, Y.; Lee, J.H.; Poindexter, M.R.; Shao, Y.H.; Liu, W.S.; Lenaghan, S.C.; Ahkami, A.H.; Blumwald, E.; Stewart, C.N. Rational design and testing of abiotic stress-inducible synthetic promoters from poplar cis-regulatory elements. Plant Biotechnol. J. 2021, 19, 1354–1369. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.K. Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 2002, 53, 247–273. [Google Scholar] [CrossRef]
- Wang, M.; Zheng, Q.S.; Shen, Q.R.; Guo, S.W. The Critical Role of Potassium in Plant Stress Response. Int. J. Mol. Sci. 2013, 14, 7370–7390. [Google Scholar] [CrossRef]
- Devi, B.S.R.; Kim, Y.J.; Selvi, S.K.; Gayathri, S.; Altanzul, K.; Parvin, S.; Yang, D.U.; Lee, O.R.; Lee, S.; Yang, D.C. Influence of potassium nitrate on antioxidant level and secondary metabolite genes under cold stress in Panax ginseng. Russ. J. Plant Physiol. 2012, 59, 318–325. [Google Scholar] [CrossRef]
- Gao, H.; Yang, W.J.; Li, C.X.; Zhou, X.G.; Gao, D.M.; Rahman, M.K.U.; Li, N.H.; Wu, F.Z. Gene Expression and K+ Uptake of Two Tomato Cultivars in Response to Sub-Optimal Temperature. Plants 2020, 9, 65. [Google Scholar] [CrossRef]
- Hasanuzzaman, M.; Bhuyan, M.H.M.B.; Nahar, K.; Hossain, M.S.; Al Mahmud, J.; Hossen, M.S.; Masud, A.A.C.; Moumita; Fujita, M. Potassium: A Vital Regulator of Plant Responses and Tolerance to Abiotic Stresses. Agronomy 2018, 8, 31. [Google Scholar] [CrossRef]
- Li, S.; Lin, Y.C.J.; Wang, P.Y.; Zhang, B.F.; Li, M.; Chen, S.; Shi, R.; Tunlaya-Anukit, S.; Liu, X.Y.; Wang, Z.F.; et al. The AREB1 Transcription Factor Influences Histone Acetylation to Regulate Drought Responses and Tolerance in Populus trichocarpa. Plant Cell 2019, 31, 663–686. [Google Scholar] [CrossRef] [PubMed]
- Li, S.J.; Yang, N.; Chen, L.Q. Paraffin section observation of flower bud differentiation of Chimonanthus praecox in Kunming and comparison of the differentiation processes in different regions, China. Hortic. Plant J. 2022, 8, 221–229. [Google Scholar] [CrossRef]
- Mornya, P.M.P.; Cheng, F.Y. Effect of Combined Chilling and GA(3) Treatment on Bud Abortion in Forced ‘Luoyanghong’ Tree Peony (Paeonia suffruticosa Andr.). Hortic. Plant J. 2018, 4, 250–256. [Google Scholar] [CrossRef]
- Tao, H.X.; Sun, H.Q.; Wang, Y.F.; Wang, X.; Guo, Y.P. Effects of water stress on quality and sugar metabolism in ‘Gala’ apple fruit. Hortic. Plant J. 2023, 9, 60–72. [Google Scholar] [CrossRef]
- Murashige, T. Citation Classic—Revised Medium for Rapid Growth and Bioassays with Tobacco Tissue-Cultures. Curr. Contents 1978, 10, 473–497. [Google Scholar] [CrossRef]
- Sui, S.Z.; Luo, J.H.; Ma, J.; Zhu, Q.L.; Lei, X.H.; Li, M.Y. Generation and Analysis of Expressed Sequence Tags from Chimonanthus praecox (Wintersweet) Flowers for Discovering Stress-Responsive and Floral Development-Related Genes. Comp. Funct. Genom. 2012, 2012, 134596. [Google Scholar] [CrossRef]
- Michiels, A.; Van den Ende, W.; Tucker, M.; Van Riet, L.; Van Laere, A. Extraction of high-quality genomic DNA from latex-containing plants. Anal. Biochem. 2003, 315, 85–89. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11 Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [PubMed]
- Bates, L.S.; Waldren, R.P.; Teare, I.D. Rapid Determination of Free Proline for Water-Stress Studies. Plant Soil 1973, 39, 205–207. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).