Expression Profile of New Marker Genes Involved in Differentiation of Human Wharton’s Jelly-Derived Mesenchymal Stem Cells into Chondrocytes, Osteoblasts, Adipocytes and Neural-like Cells
Abstract
:1. Introduction
2. Results
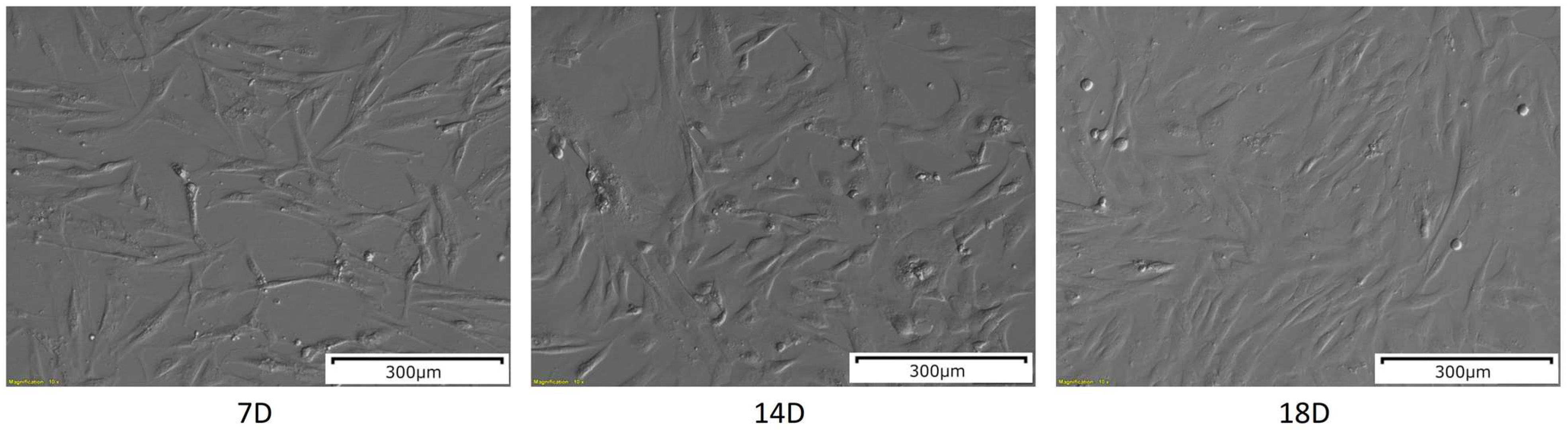
2.1. Morphological Analysis
2.2. Evaluation of WJ-MSCs Differentiation
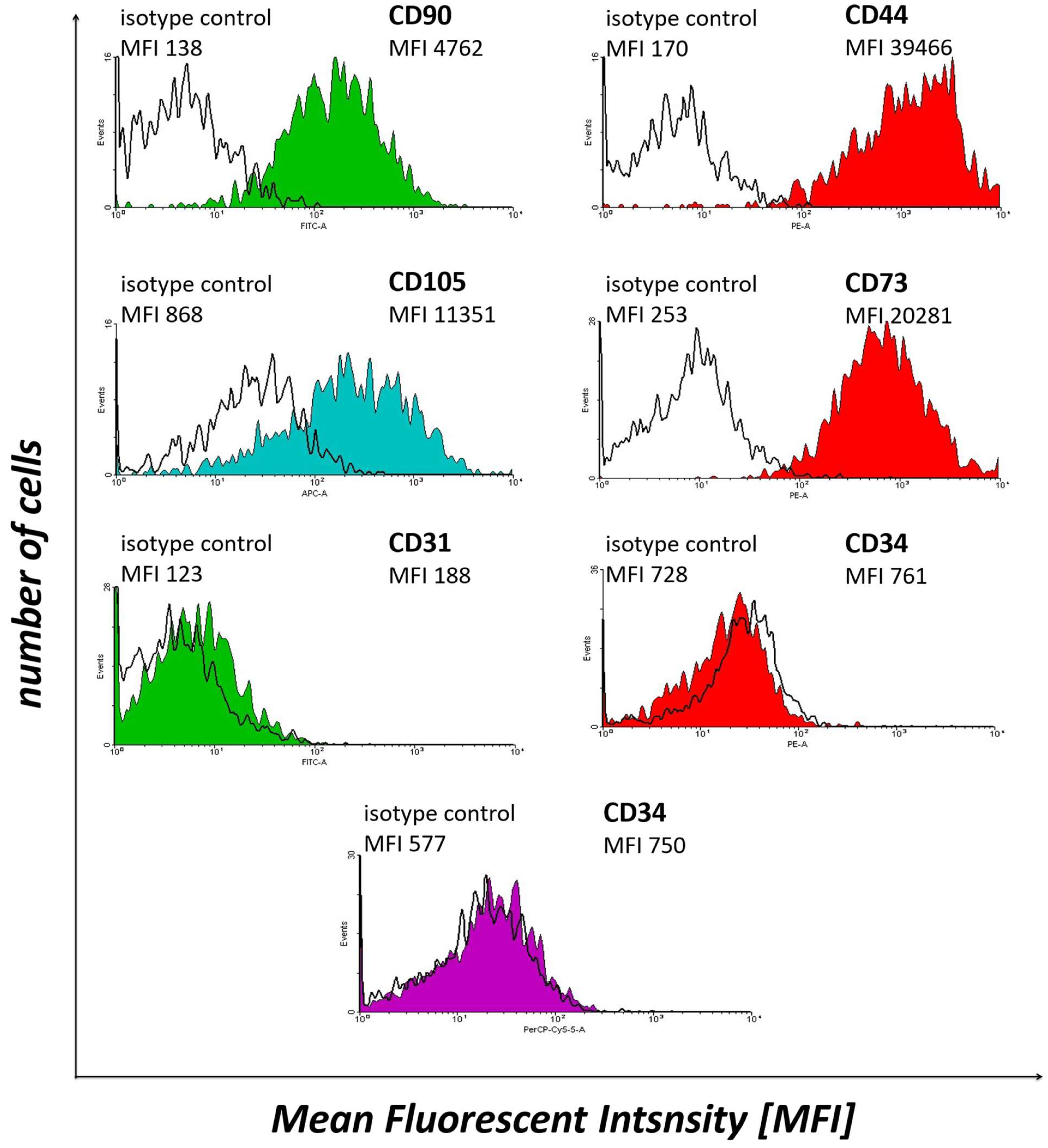
2.3. Flow Cytometry Analysis
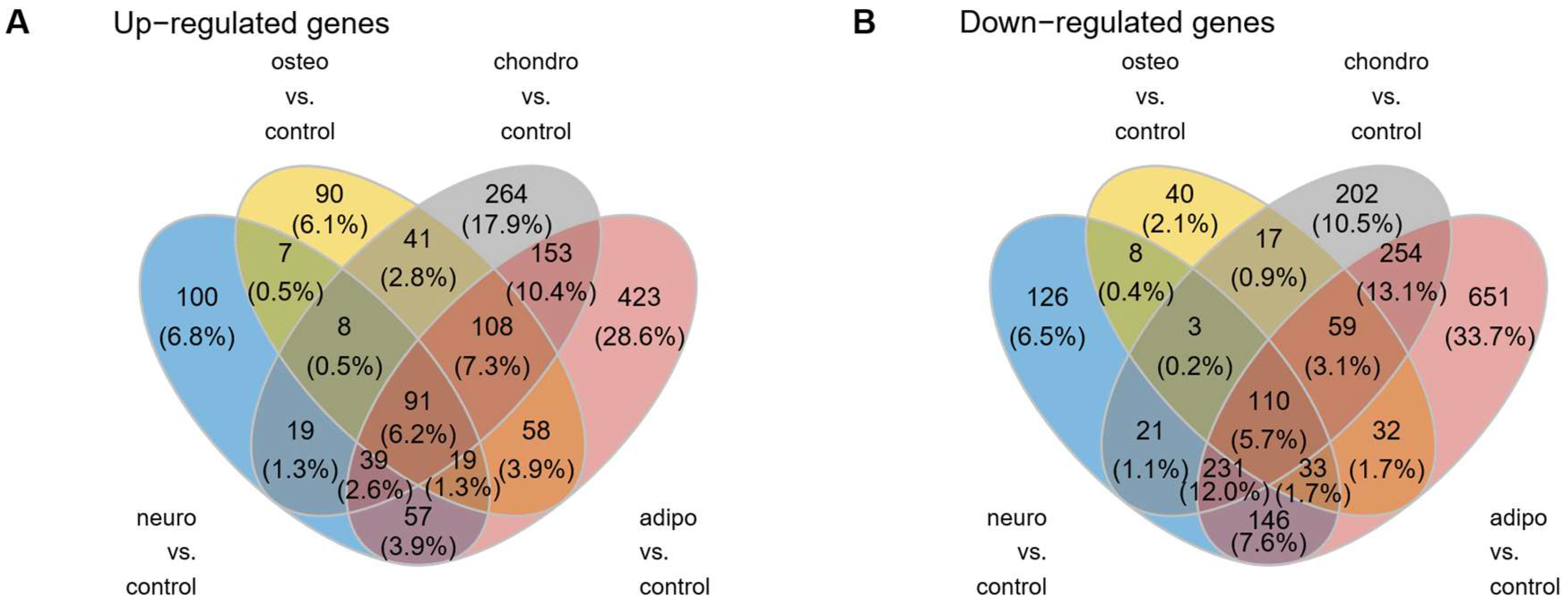
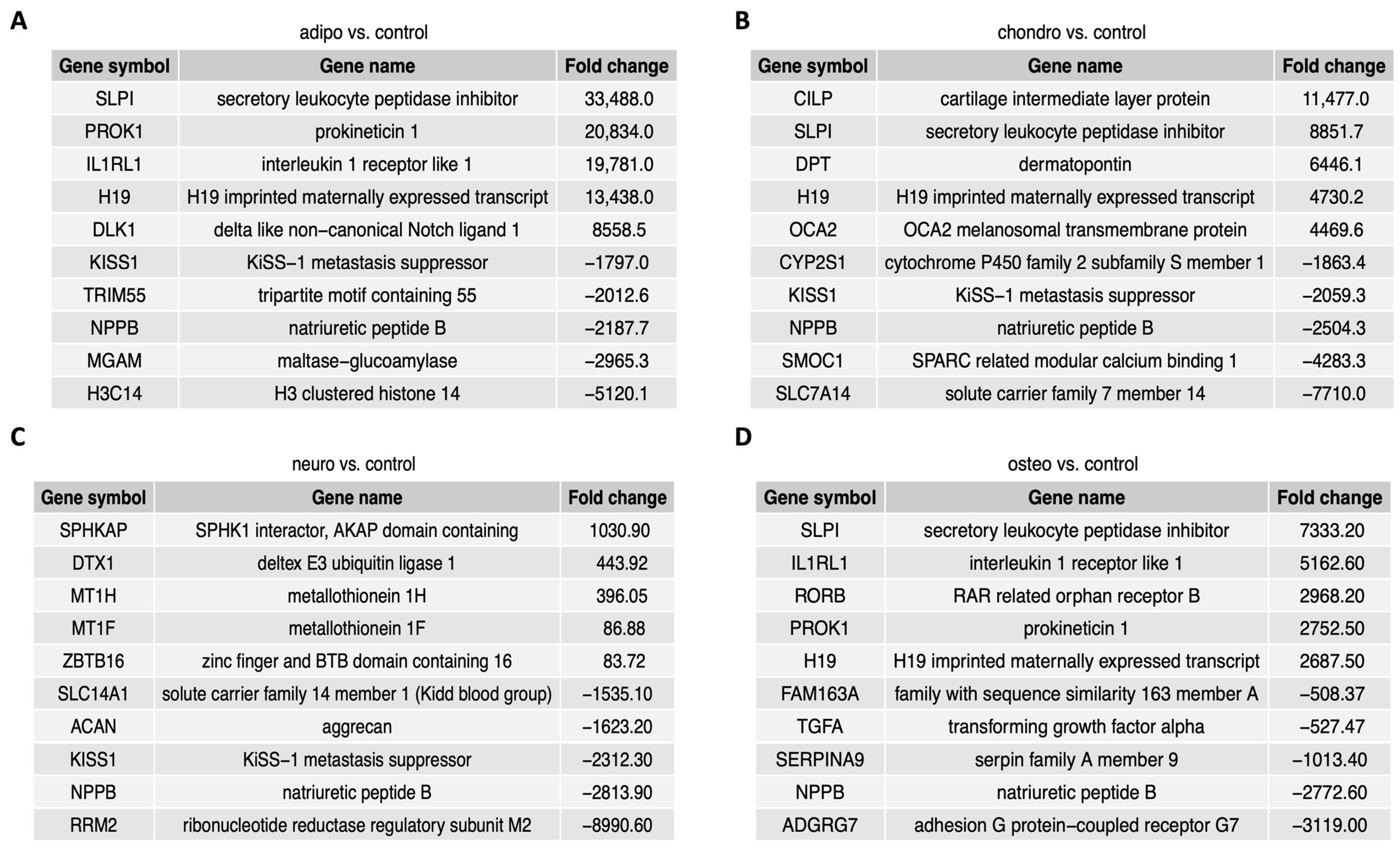
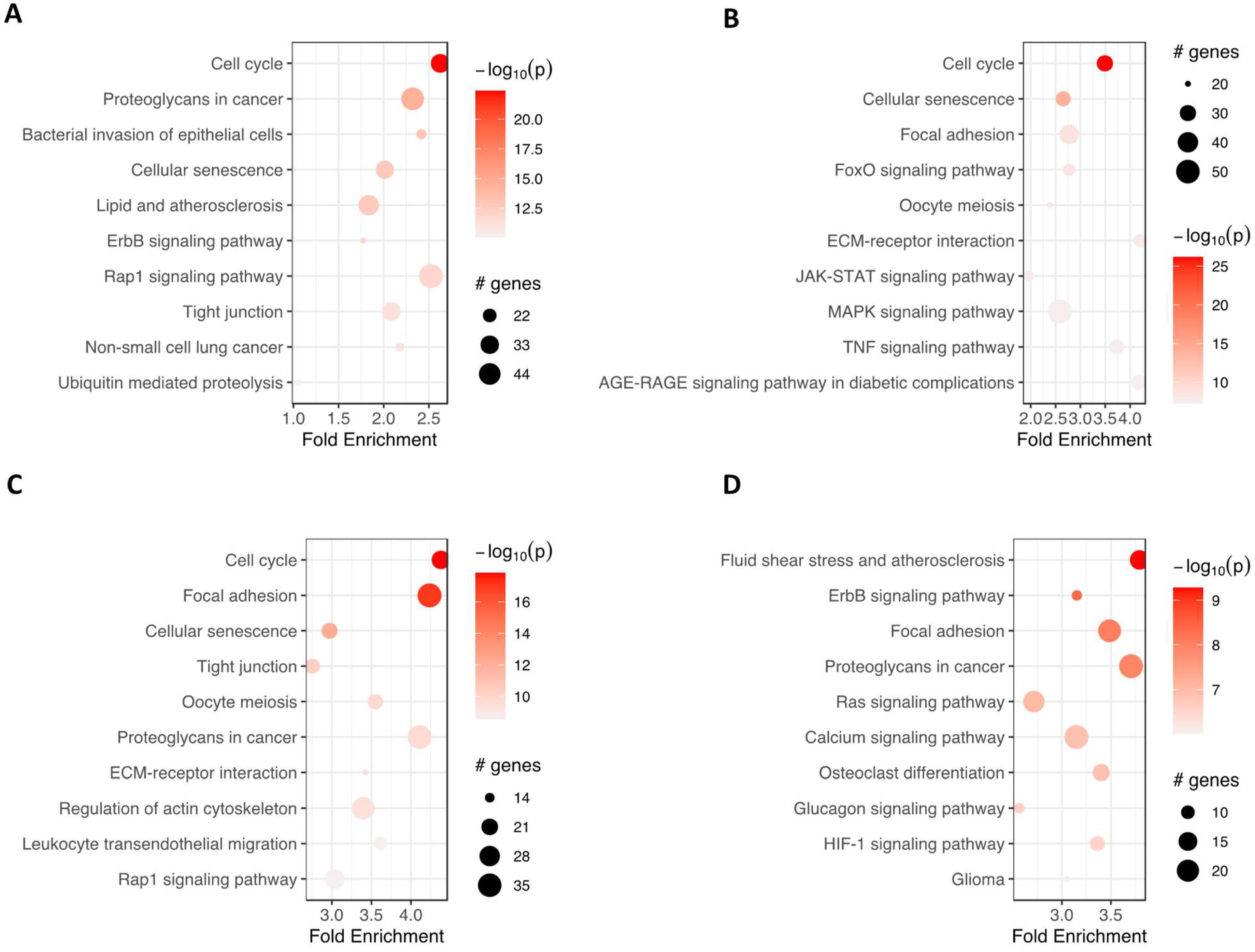
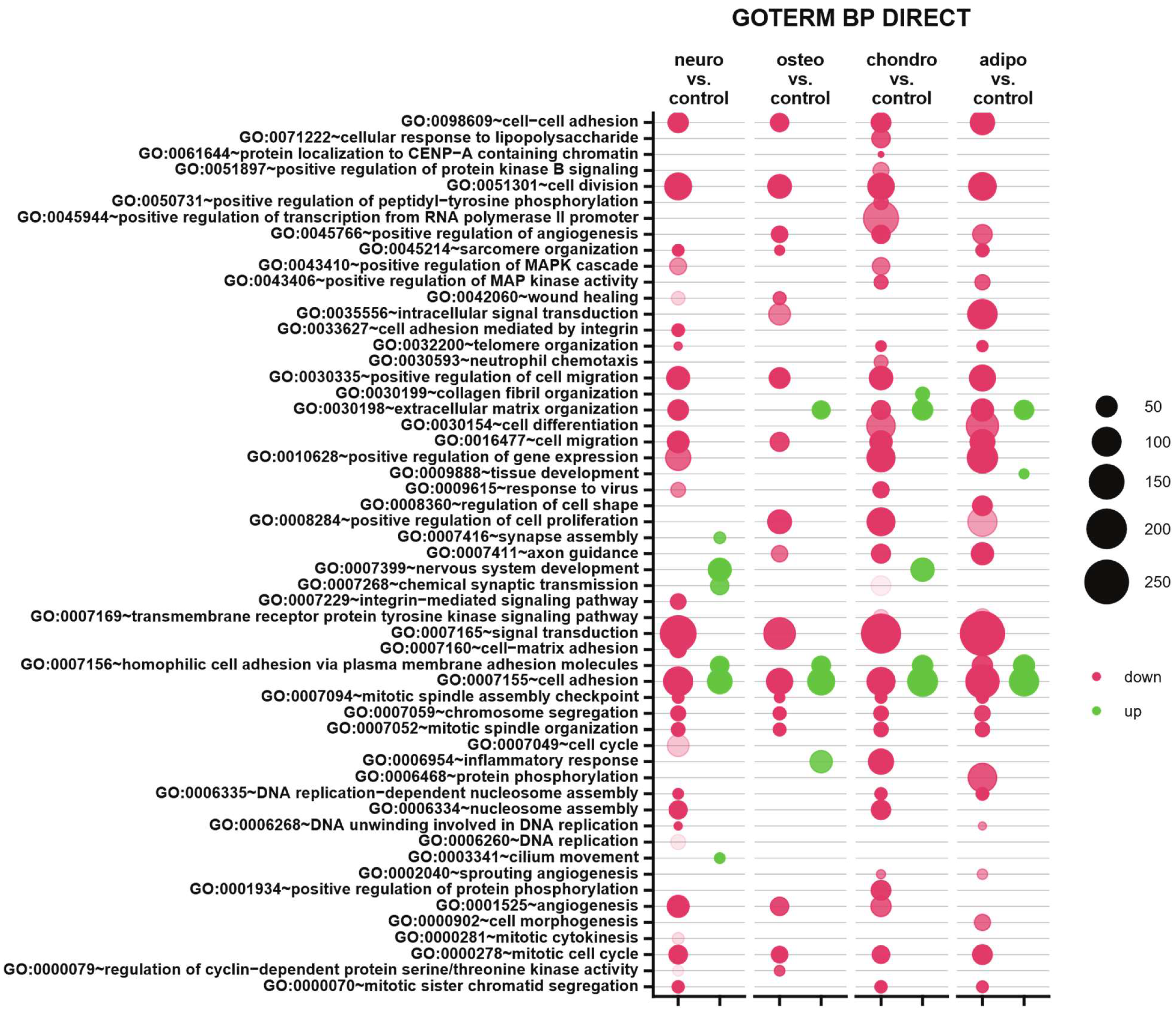
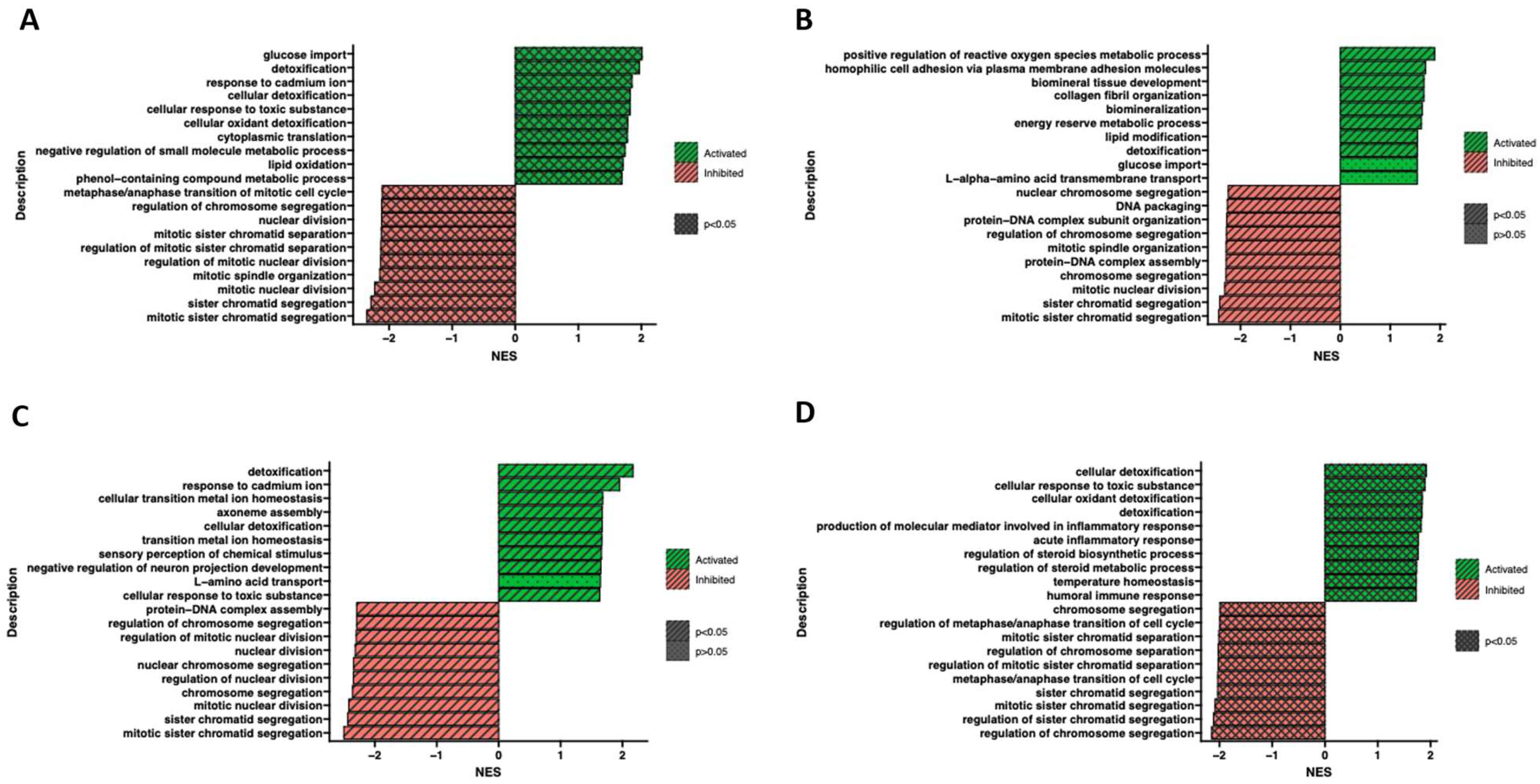
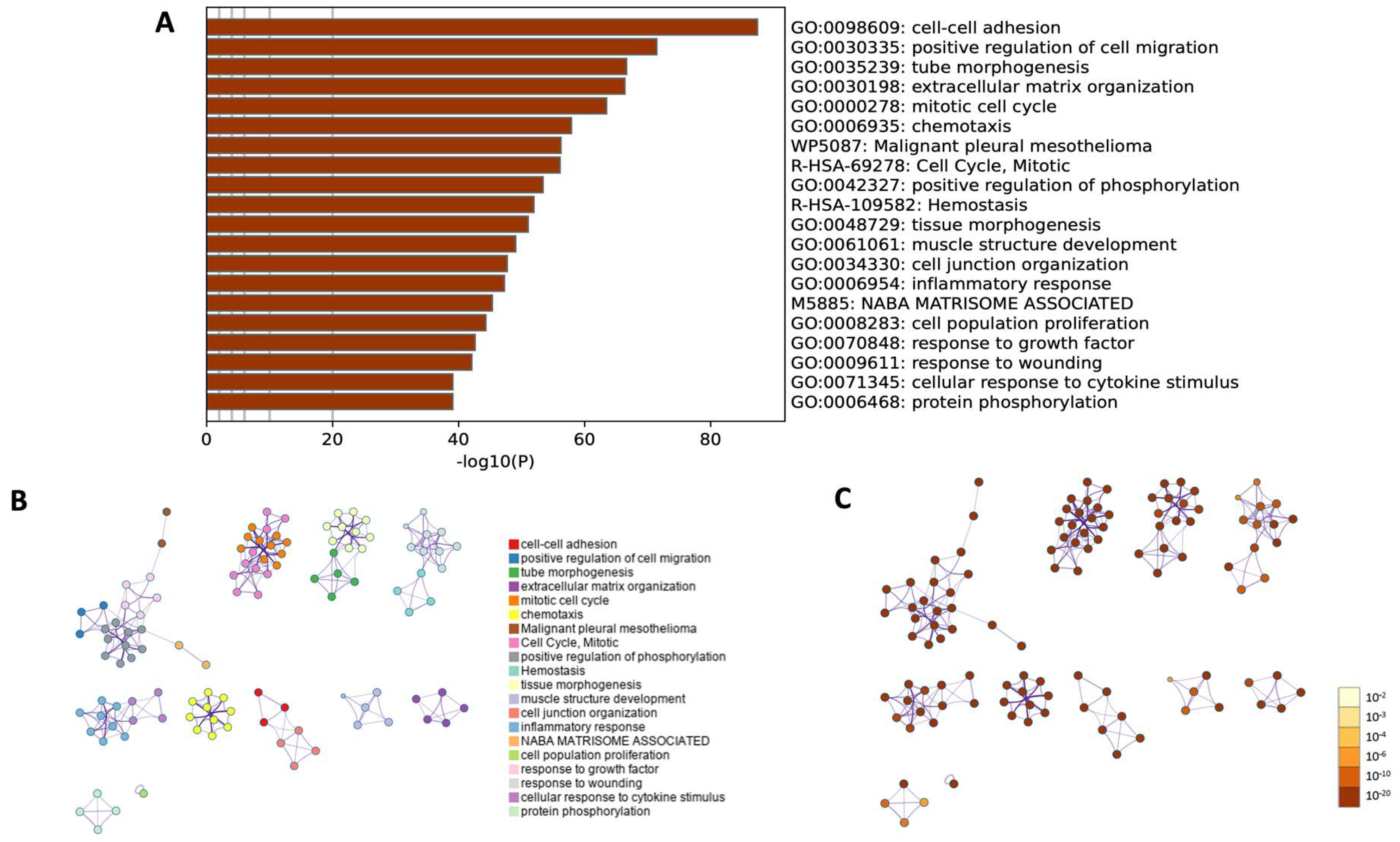
2.4. RNA-seq Analysis
3. Discussion
4. Materials and Methods
4.1. Material Collection
4.2. Isolation of Wharton’s Jelly-Derived Mesenchymal Stem Cells
4.3. In Vitro Cell Culture
4.4. Flow Cytometry Analysis
4.5. Multilineage Differentiation
4.5.1. Osteogenic Differentiation
4.5.2. Neurogenic Differentiation
4.5.3. Chondrogenic Differentiation
4.5.4. Adipogenic Differentiation
4.6. RNA Isolation
4.7. RNA-seq
4.8. Bioinformatical and Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Arutyunyan, I.; Elchaninov, A.; Makarov, A.; Fatkhudinov, T. Umbilical Cord as Prospective Source for Mesenchymal Stem Cell-Based Therapy. Stem Cells Int. 2016, 2016, 6901286. [Google Scholar] [CrossRef]
- Dominici, M.; Le Blanc, K.; Mueller, I.; Slaper-Cortenbach, I.; Marini, F.C.; Krause, D.S.; Deans, R.J.; Keating, A.; Prockop, D.J.; Horwitz, E.M. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy 2006, 8, 315–317. [Google Scholar] [CrossRef]
- Rucinski, M.; Zok, A.; Guidolin, D.; de Caro, R.; Malendowicz, L.K. Expression of precerebellins in cultured rat calvaria osteoblast-like cells. Int. J. Mol. Med. 2008, 22, 553–558. [Google Scholar] [CrossRef] [PubMed]
- Lyons, F.G.; Mattei, T.A. Sources, Identification, and Clinical Implications of Heterogeneity in Human Umbilical Cord Stem Cells. Adv. Exp. Med. Biol. 2019, 1169, 243–256. [Google Scholar] [CrossRef]
- Subramanian, A.; Fong, C.Y.; Biswas, A.; Bongso, A. Comparative characterization of cells from the various compartments of the human umbilical cord shows that the Wharton’s jelly compartment provides the best source of clinically utilizable mesenchymal stem cells. PLoS ONE 2015, 10, e0127992. [Google Scholar] [CrossRef] [PubMed]
- Martin-Rendon, E.; Sweeney, D.; Lu, F.; Girdlestone, J.; Navarrete, C.; Watt, S.M. 5-Azacytidine-treated human mesenchymal stem/progenitor cells derived from umbilical cord, cord blood and bone marrow do not generate cardiomyocytes in vitro at high frequencies. Vox Sang. 2008, 95, 137–148. [Google Scholar] [CrossRef]
- Tong, C.K.; Vellasamy, S.; Chong Tan, B.; Abdullah, M.; Vidyadaran, S.; Fong Seow, H.; Ramasamy, R. Generation of mesenchymal stem cell from human umbilical cord tissue using a combination enzymatic and mechanical disassociation method. Cell Biol. Int. 2011, 35, 221–226. [Google Scholar] [CrossRef]
- Karahuseyinoglu, S.; Cinar, O.; Kilic, E.; Kara, F.; Akay, G.G.; Demiralp, D.Ö.; Tukun, A.; Uckan, D.; Can, A. Biology of Stem Cells in Human Umbilical Cord Stroma: In Situ and In Vitro Surveys. Stem Cells 2007, 25, 319–331. [Google Scholar] [CrossRef] [PubMed]
- Weiss, M.L.; Anderson, C.; Medicetty, S.; Seshareddy, K.B.; Weiss, R.J.; VanderWerff, I.; Troyer, D.; McIntosh, K.R. Immune Properties of Human Umbilical Cord Wharton’s Jelly-Derived Cells. Stem Cells 2008, 26, 2865–2874. [Google Scholar] [CrossRef]
- da Rosa, N.N.; Appel, J.M.; Irioda, A.C.; Mogharbel, B.F.; de Oliveira, N.B.; Perussolo, M.C.; Stricker, P.E.F.; Rosa-Fernandes, L.; Marinho, C.R.F.; de Carvalho, K.A.T. Three-Dimensional Bioprinting of an In Vitro Lung Model. Int. J. Mol. Sci. 2023, 24, 5852. [Google Scholar] [CrossRef]
- Zhang, W.; Sun, T.; Zhang, J.; Hu, X.; Yang, M.; Han, L.; Xu, G.; Zhao, Y.; Li, Z. Construction of artificial periosteum with methacrylamide gelatin hydrogel-wharton’s jelly based on stem cell recruitment and its application in bone tissue engineering. Mater. Today Bio 2023, 18, 100528. [Google Scholar] [CrossRef] [PubMed]
- Mansour, R.N.; Hasanzadeh, E.; Abasi, M.; Gholipourmalekabadi, M.; Mellati, A.; Enderami, S.E. The Effect of Fetal Bovine Acellular Dermal Matrix Seeded with Wharton’s Jelly Mesenchymal Stem Cells for Healing Full-Thickness Skin Wounds. Genes 2023, 14, 909. [Google Scholar] [CrossRef] [PubMed]
- Lizarazo-Fonseca, L.; Correa-Araujo, L.; Prieto-Abello, L.; Camacho-Rodríguez, B.; Silva-Cote, I. In vitro and in vivo evaluation of electrospun poly (ε-caprolactone)/collagen scaffolds and Wharton’s jelly mesenchymal stromal cells (hWJ-MSCs) constructs as potential alternative for skin tissue engineering. Regen. Ther. 2023, 24, 11–24. [Google Scholar] [CrossRef] [PubMed]
- Taherpour, A.; Hosseini-khah, Z.; Zargari, M.; Ehsan Enderami, S. The microenvironment of silk/gelatin nanofibrous scaffold improves proliferation and differentiation of Wharton’s jelly-derived mesenchymal cells into islet-like cells. Gene 2022, 833, 146586. [Google Scholar] [CrossRef]
- Doğan, A. Embryonic Stem Cells in Development and Regenerative Medicine. In Cell Biology and Translational Medicine, Volume 1; Advances in Experimental Medicine and Biology; Springer: Cham, Switzerland, 2018; Volume 1079, pp. 1–15. [Google Scholar] [CrossRef]
- Cabrera-Pérez, R.; Monguió-Tortajada, M.; Gámez-Valero, A.; Rojas-Márquez, R.; Borràs, F.E.; Roura, S.; Vives, J. Osteogenic commitment of Wharton’s jelly mesenchymal stromal cells: Mechanisms and implications for bioprocess development and clinical application. Stem Cell Res. Ther. 2019, 10, 356. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.-S.; Hung, S.-C.; Peng, S.-T.; Huang, C.-C.; Wei, H.-M.; Guo, Y.-J.; Fu, Y.-S.; Lai, M.-C.; Chen, C.-C. Mesenchymal Stem Cells in the Wharton’s Jelly of the Human Umbilical Cord. Stem Cells 2004, 22, 1330–1337. [Google Scholar] [CrossRef]
- Ansari, A.S.; Yazid, M.D.; Sainik, N.Q.A.V.; Razali, R.A.; Saim, A.B.; Idrus, R.B.H. Osteogenic Induction of Wharton’s Jelly-Derived Mesenchymal Stem Cell for Bone Regeneration: A Systematic Review. Stem Cells Int. 2018, 2018, 2406462. [Google Scholar] [CrossRef]
- Stefańska, K.; Nemcova, L.; Blatkiewicz, M.; Pieńkowski, W.; Ruciński, M.; Zabel, M.; Mozdziak, P.; Podhorska-Okołów, M.; Dzięgiel, P.; Kempisty, B. Apoptosis Related Human Wharton’s Jelly-Derived Stem Cells Differentiation into Osteoblasts, Chondrocytes, Adipocytes and Neural-like Cells—Complete Transcriptomic Assays. Int. J. Mol. Sci. 2023, 24, 10023. [Google Scholar] [CrossRef]
- Stelcer, E.; Komarowska, H.; Jopek, K.; Żok, A.; Iżycki, D.; Malińska, A.; Szczepaniak, B.; Komekbai, Z.; Karczewski, M.; Wierzbicki, T.; et al. Biological response of adrenal carcinoma and melanoma cells to mitotane treatment. Oncol. Lett. 2022, 23, 120. [Google Scholar] [CrossRef]
- Budna, J.; Chachuła, A.; Kaźmierczak, D.; Rybska, M.; Ciesiółka, S.; Bryja, A.; Kranc, W.; Borys, S.; Zok, A.; Bukowska, D.; et al. Morphogenesis-related gene-expression profile in porcine oocytes before and after in vitro maturation. Zygote 2017, 25, 331–340. [Google Scholar] [CrossRef]
- Golkar-Narenji, A.; Antosik, P.; Nolin, S.; Rucinski, M.; Jopek, K.; Zok, A.; Sobolewski, J.; Jankowski, M.; Zdun, M.; Bukowska, D.; et al. Gene Ontology Groups and Signaling Pathways Regulating the Process of Avian Satellite Cell Differentiation. Genes 2022, 13, 242. [Google Scholar] [CrossRef]
- Potier, E.; Ferreira, E.; Meunier, A.; Sedel, L.; Logeart-Avramoglou, D.; Petite, H. Prolonged hypoxia concomitant with serum deprivation induces massive human mesenchymal stem cell death. Tissue Eng. 2007, 13, 1325–1331. [Google Scholar] [CrossRef]
- Binder, B.Y.K.; Genetos, D.C.; Leach, J.K. Lysophosphatidic Acid Protects Human Mesenchymal Stromal Cells from Differentiation-Dependent Vulnerability to Apoptosis. Tissue Eng. Part A 2014, 20, 1156. [Google Scholar] [CrossRef]
- Pesarini, J.R.; de Oliveira, E.J.T.; Pessatto, L.R.; Rabacow, A.P.M.; Camassola, M.; dos Santos, B.P.; de Barros, M.E.; Cantero, W.d.B.; Antoniolli-Silva, A.C.M.B.; Oliveira, R.J. Calcitriol combined with calcium chloride causes apoptosis in undifferentiated adipose tissue-derived human mesenchymal stem cells, but this effect decreases during adipogenic differentiation. Biomed. Pharmacother. 2018, 108, 914–924. [Google Scholar] [CrossRef] [PubMed]
- Oliver, L.; Hue, E.; Séry, Q.; Lafargue, A.; Pecqueur, C.; Paris, F.; Vallette, F.M. Differentiation-related response to DNA breaks in human mesenchymal stem cells. Stem Cells 2013, 31, 800–807. [Google Scholar] [CrossRef]
- Manna, C.; Das, K.; Mandal, D.; Banerjee, D.; Mukherjee, J.; Ganguly, I.; Naskar, S.; Bag, S. Canine umbilical cord tissue derived mesenchymal stem cells naturally express mRNAs of some antimicrobial peptides. Vet. Res. Commun. 2023; in press. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Shao, Y.; Xie, D.; Pan, J.; Chen, H.; Yao, J.; Liang, J.; Ke, H.; Cai, D.; Zeng, C. Effect of secretory leucocyte protease inhibitor on early tendon-to-bone healing after anterior cruciate ligament reconstruction in a rat model. Bone Jt. Res. 2022, 11, 503. [Google Scholar] [CrossRef]
- Choi, B.D.; Lee, S.Y.; Jeong, S.J.; Lim, D.S.; Cha, H.J.; Chung, W.G.; Jeong, M.J. Secretory leukocyte protease inhibitor promotes differentiation and mineralization of MC3T3-E1 preosteoblasts on a titanium surface. Mol. Med. Rep. 2016, 14, 1241–1246. [Google Scholar] [CrossRef] [PubMed]
- Ohlsson, S.; Tufvesson, B.; Polling, Å.; Ohlsson, K. Distribution of the secretory leucocyte proteinase inhibitor in human articular cartilage. Biol. Chem. 1997, 378, 1055–1058. [Google Scholar] [CrossRef] [PubMed]
- Jacoby, A.S.; Melrose, J.; Robinson, B.G.; Hyland, V.J.; Ghosh, P. Secretory leucocyte proteinase inhibitor is produced by human articular cartilage chondrocytes and intervertebral disc fibrochondrocytes. Eur. J. Biochem. 1993, 218, 951–957. [Google Scholar] [CrossRef]
- Adapala, V.J.; Buhman, K.K.; Ajuwon, K.M. Novel anti-inflammatory role of SLPI in adipose tissue and its regulation by high fat diet. J. Inflamm. 2011, 8, 5. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Q.Q.; Wang, X.; Li, Y.F.; Peng, L.J.; Jiang, Z.S. Secretory leukocyte protease inhibitor promising protective roles in obesity-associated atherosclerosis. Exp. Biol. Med. 2017, 242, 250. [Google Scholar] [CrossRef]
- Liang, W.C.; Fu, W.M.; Wang, Y.B.; Sun, Y.X.; Xu, L.L.; Wong, C.W.; Chan, K.M.; Li, G.; Waye, M.M.Y.; Zhang, J.F. H19 activates Wnt signaling and promotes osteoblast differentiation by functioning as a competing endogenous RNA. Sci. Rep. 2016, 6, 20121. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Gan, S.; Su, C.; Zheng, Z.; Liao, Y.; Shao, J.; Zhu, Z.; Chen, W. Screening and preliminary identification of long non-coding RNAs critical for osteogenic differentiation of human umbilical cord mesenchymal stem cells. Bioengineered 2022, 13, 6880. [Google Scholar] [CrossRef] [PubMed]
- Cao, B.; Dai, X. Platelet lysate induces chondrogenic differentiation of umbilical cord-derived mesenchymal stem cells by regulating the lncRNA H19/miR-29b-3p/SOX9 axis. FEBS Open Bio 2020, 10, 2656. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Zheng, Y.; Jin, C.; Li, X.; Jia, L.; Li, W. Long Non-coding RNA H19 Inhibits Adipocyte Differentiation of Bone Marrow Mesenchymal Stem Cells through Epigenetic Modulation of Histone Deacetylases. Sci. Rep. 2016, 6, 28897. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Wu, Y.; Yang, H.; Hong, P.; Fang, X.; Hu, Y. H19/miR-30a/C8orf4 axis modulates the adipogenic differentiation process in human adipose tissue-derived mesenchymal stem cells. J. Cell. Physiol. 2019, 234, 20925–20934. [Google Scholar] [CrossRef]
- Saidi, S.; Bouri, F.; Lencel, P.; Duplomb, L.; Baud’huin, M.; Delplace, S.; Leterme, D.; Miellot, F.; Heymann, D.; Hardouin, P.; et al. IL-33 is expressed in human osteoblasts, but has no direct effect on bone remodeling. Cytokine 2011, 53, 347–354. [Google Scholar] [CrossRef]
- Schulze, J.; Bickert, T.; Beil, F.T.; Zaiss, M.M.; Albers, J.; Wintges, K.; Streichert, T.; Klaetschke, K.; Keller, J.; Hissnauer, T.N.; et al. Interleukin-33 is expressed in differentiated osteoblasts and blocks osteoclast formation from bone marrow precursor cells. J. Bone Miner. Res. 2011, 26, 704–717. [Google Scholar] [CrossRef]
- Wood, I.S.; Wang, B.; Trayhurn, P. IL-33, a recently identified interleukin-1 gene family member, is expressed in human adipocytes. Biochem. Biophys. Res. Commun. 2009, 384, 105–109. [Google Scholar] [CrossRef]
- Ngan, E.S.W.; Lee, K.Y.; Sit, F.Y.L.; Poon, H.C.; Chan, J.K.Y.; Sham, M.H.; Lui, V.C.H.; Tam, P.K.H. Prokineticin-1 modulates proliferation and differentiation of enteric neural crest cells. Biochim. Biophys. Acta 2007, 1773, 536–545. [Google Scholar] [CrossRef] [PubMed]
- da Silva, C.; Durandt, C.; Kallmeyer, K.; Ambele, M.A.; Pepper, M.S. The Role of Pref-1 during Adipogenic Differentiation: An Overview of Suggested Mechanisms. Int. J. Mol. Sci. 2020, 21, 4104. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Hirai, M.; Cantero, S.; Ciubotariu, R.; Dobrila, L.; Hirsh, A.; Igura, K.; Satoh, H.; Yokomi, I.; Nishimura, T.; et al. Isolation and characterization of mesenchymal stem cells from human umbilical cord blood: Reevaluation of critical factors for successful isolation and high ability to proliferate and differentiate to chondrocytes as compared to mesenchymal stem cells from bone marrow and adipose tissue. J. Cell. Biochem. 2011, 112, 1206–1218. [Google Scholar] [CrossRef] [PubMed]
- Karagianni, M.; Brinkmann, I.; Kinzebach, S.; Grassl, M.; Weiss, C.; Bugert, P.; Bieback, K. A comparative analysis of the adipogenic potential in human mesenchymal stromal cells from cord blood and other sources. Cytotherapy 2013, 15, 76–88. [Google Scholar] [CrossRef]
- Morganstein, D.L.; Wu, P.; Mane, M.R.; Fisk, N.M.; White, R.; Parker, M.G. Human fetal mesenchymal stem cells differentiate into brown and white adipocytes, and reveal a role for ERRα in human UCP1 expression. Cell Res. 2010, 20, 434. [Google Scholar] [CrossRef]
- Farr, J.N.; Weivoda, M.M.; Nicks, K.M.; Fraser, D.G.; Negley, B.A.; Onken, J.L.; Thicke, B.S.; Ruan, M.; Liu, H.; Forrest, D.; et al. Osteoprotection Through the Deletion of the Transcription Factor Rorβ in Mice. J. Bone Miner. Res. 2018, 33, 720. [Google Scholar] [CrossRef]
- Roforth, M.M.; Khosla, S.; Monroe, D.G. Identification of Rorβ targets in cultured osteoblasts and in human bone. Biochem. Biophys. Res. Commun. 2013, 440, 768–773. [Google Scholar] [CrossRef] [PubMed]
- Lorenzo, P.; Bayliss, M.T.; Heinegårdt, D. A novel cartilage protein (CILP) present in the mid-zone of human articular cartilage increases with age. J. Biol. Chem. 1998, 273, 23463–23468. [Google Scholar] [CrossRef]
- Derfoul, A.; Perkins, G.L.; Hall, D.J.; Tuan, R.S. Glucocorticoids Promote Chondrogenic Differentiation of Adult Human Mesenchymal Stem Cells by Enhancing Expression of Cartilage Extracellular Matrix Genes. Stem Cells 2006, 24, 1487–1495. [Google Scholar] [CrossRef] [PubMed]
- Vidács, D.L.; Veréb, Z.; Bozó, R.; Flink, L.B.; Polyánka, H.; Németh, I.B.; Póliska, S.; Papp, B.T.; Manczinger, M.; Gáspár, R.; et al. Phenotypic plasticity of melanocytes derived from human adult skin. Pigment Cell Melanoma Res. 2022, 35, 38–51. [Google Scholar] [CrossRef]
- Lecorguillé, M.; McAuliffe, F.M.; Twomey, P.J.; Viljoen, K.; Mehegan, J.; Kelleher, C.C.; Suderman, M.; Phillips, C.M. Maternal Glycaemic and Insulinemic Status and Newborn DNA Methylation: Findings in Women with Overweight and Obesity. J. Clin. Endocrinol. Metab. 2022, 108, 85–98. [Google Scholar] [CrossRef]
- Lacana, E.; Maceyka, M.; Milstien, S.; Spiegel, S. Cloning and characterization of a protein kinase A anchoring protein (AKAP)-related protein that interacts with and regulates sphingosine kinase 1 activity. J. Biol. Chem. 2002, 277, 32947–32953. [Google Scholar] [CrossRef]
- Jiang, Z.J.; Gong, L.W. The SphK1/S1P Axis Regulates Synaptic Vesicle Endocytosis via TRPC5 Channels. J. Neurosci. 2023, 43, 3807–3824. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.Y.; Hu, Q.D.; Tekaya, M.; Shimoda, Y.; Ang, B.T.; Nie, D.Y.; Sun, L.; Hu, W.P.; Karsak, M.; Duka, T.; et al. NB-3/Notch1 pathway via Deltex1 promotes neural progenitor cell differentiation into oligodendrocytes. J. Biol. Chem. 2004, 279, 25858–25865. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.-C.; Huang, Y.-C.; Yeh, T.-H.; Shih, H.-Y.; Lin, C.-Y.; Lin, S.-J.; Chiu, C.-C.; Huang, C.-W.; Jiang, Y.-J. Deltex1 is inhibited by the Notch–Hairy/E(Spl) signaling pathway and induces neuronal and glial differentiation. Neural Dev. 2015, 10, 28. [Google Scholar] [CrossRef] [PubMed]
- Michael, G.J.; Esmailzadeh, S.; Moran, L.B.; Christian, L.; Pearce, R.K.B.; Graeber, M.B. Up-regulation of metallothionein gene expression in Parkinsonian astrocytes. Neurogenetics 2011, 12, 295–305. [Google Scholar] [CrossRef] [PubMed]
- Sobieszczuk, D.F.; Poliakov, A.; Xu, Q.; Wilkinson, D.G. A feedback loop mediated by degradation of an inhibitor is required to initiate neuronal differentiation. Genes Dev. 2010, 24, 206. [Google Scholar] [CrossRef]
- Zhu, X.; Wang, Z.; Sun, Y.E.; Liu, Y.; Wu, Z.; Ma, B.; Cheng, L. Neuroprotective Effects of Human Umbilical Cord-Derived Mesenchymal Stem Cells From Different Donors on Spinal Cord Injury in Mice. Front. Cell. Neurosci. 2021, 15, 768711. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Yan, L. Draw Venn Diagram by “ggplot2” [R Package ggvenn Version 0.1.10]. 2023. Available online: https://cran.r-project.org/web/packages/ggvenn/ggvenn.pdf (accessed on 28 May 2023).
- Dennis, G.; Sherman, B.T.; Hosack, D.A.; Yang, J.; Gao, W.; Lane, H.C.; Lempicki, R.A. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003, 4, R60. [Google Scholar] [CrossRef]
- Fresno, C.; Fernández, E.A. RDAVIDWebService: A versatile R interface to DAVID. Bioinformatics 2013, 29, 2810–2811. [Google Scholar] [CrossRef]
- Benjamini, Y.; Cohen, R. Weighted false discovery rate controlling procedures for clinical trials. Biostatistics 2017, 18, 91–104. [Google Scholar] [CrossRef]
- Goesmann, A.; Haubrock, M.; Meyer, F.; Kalinowski, J.; Giegerich, R. PathFinder: Reconstruction and dynamic visualization of metabolic pathways. Bioinformatics 2002, 18, 124–129. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef]
- Bader, G.D.; Hogue, C.W.V. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinform. 2003, 4, 2. [Google Scholar] [CrossRef]


| Antibody | Fluorochrome | Isotype | Clone | Producer |
|---|---|---|---|---|
| CD90 | FITC | IgG1 | REA897 | Miltenyi Biotec (Bergisch Gladbach, Germany) |
| CD105 | APC | IgG1 | REA794 | Miltenyi Biotec |
| CD73 | PE | IgG1 | REA804 | Miltenyi Biotec |
| CD44 | PE | IgG1k | DB105 | Miltenyi Biotec |
| CD45 | PerCP | IgG2ak | 5B1 | Miltenyi Biotec |
| CD34 | PE | IgG2ak | AC136 | Miltenyi Biotec |
| Isotype control IgG1k | PE | IgG1k | IS5-21F5 | Miltenyi Biotec |
| Isotype control IgG1 | FITC | IgG1k | IS5-21F5 | Miltenyi Biotec |
| Isotype control IgG2a | PerCP | IgG2a | S43.10 | Miltenyi Biotec |
| Isotype control IgG2a | PE | IgG2a | S43.10 | Miltenyi Biotec |
| REA Control (S) | APC | IgG1 | REA293 | Miltenyi Biotec |
| REA Control (S) | PE | IgG1 | REA293 | Miltenyi Biotec |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stefańska, K.; Nemcova, L.; Blatkiewicz, M.; Żok, A.; Kaczmarek, M.; Pieńkowski, W.; Mozdziak, P.; Piotrowska-Kempisty, H.; Kempisty, B. Expression Profile of New Marker Genes Involved in Differentiation of Human Wharton’s Jelly-Derived Mesenchymal Stem Cells into Chondrocytes, Osteoblasts, Adipocytes and Neural-like Cells. Int. J. Mol. Sci. 2023, 24, 12939. https://doi.org/10.3390/ijms241612939
Stefańska K, Nemcova L, Blatkiewicz M, Żok A, Kaczmarek M, Pieńkowski W, Mozdziak P, Piotrowska-Kempisty H, Kempisty B. Expression Profile of New Marker Genes Involved in Differentiation of Human Wharton’s Jelly-Derived Mesenchymal Stem Cells into Chondrocytes, Osteoblasts, Adipocytes and Neural-like Cells. International Journal of Molecular Sciences. 2023; 24(16):12939. https://doi.org/10.3390/ijms241612939
Chicago/Turabian StyleStefańska, Katarzyna, Lucie Nemcova, Małgorzata Blatkiewicz, Agnieszka Żok, Mariusz Kaczmarek, Wojciech Pieńkowski, Paul Mozdziak, Hanna Piotrowska-Kempisty, and Bartosz Kempisty. 2023. "Expression Profile of New Marker Genes Involved in Differentiation of Human Wharton’s Jelly-Derived Mesenchymal Stem Cells into Chondrocytes, Osteoblasts, Adipocytes and Neural-like Cells" International Journal of Molecular Sciences 24, no. 16: 12939. https://doi.org/10.3390/ijms241612939
APA StyleStefańska, K., Nemcova, L., Blatkiewicz, M., Żok, A., Kaczmarek, M., Pieńkowski, W., Mozdziak, P., Piotrowska-Kempisty, H., & Kempisty, B. (2023). Expression Profile of New Marker Genes Involved in Differentiation of Human Wharton’s Jelly-Derived Mesenchymal Stem Cells into Chondrocytes, Osteoblasts, Adipocytes and Neural-like Cells. International Journal of Molecular Sciences, 24(16), 12939. https://doi.org/10.3390/ijms241612939










