Evaluation of EV Storage Buffer for Efficient Preservation of Engineered Extracellular Vesicles
Abstract
:1. Introduction
2. Results
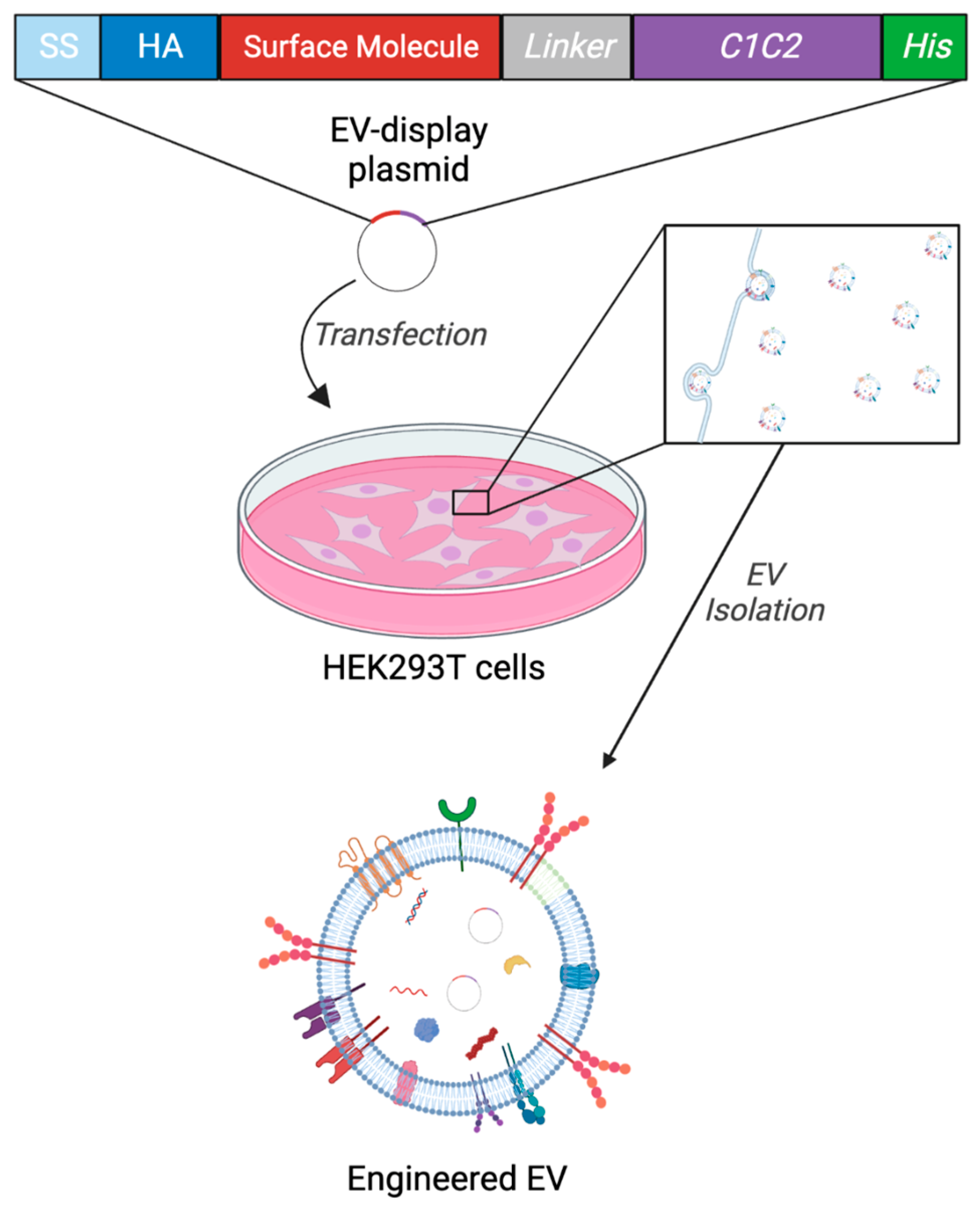
2.1. Experimental Design and Generation of Engineered EVs (eEVs) for Storage Test
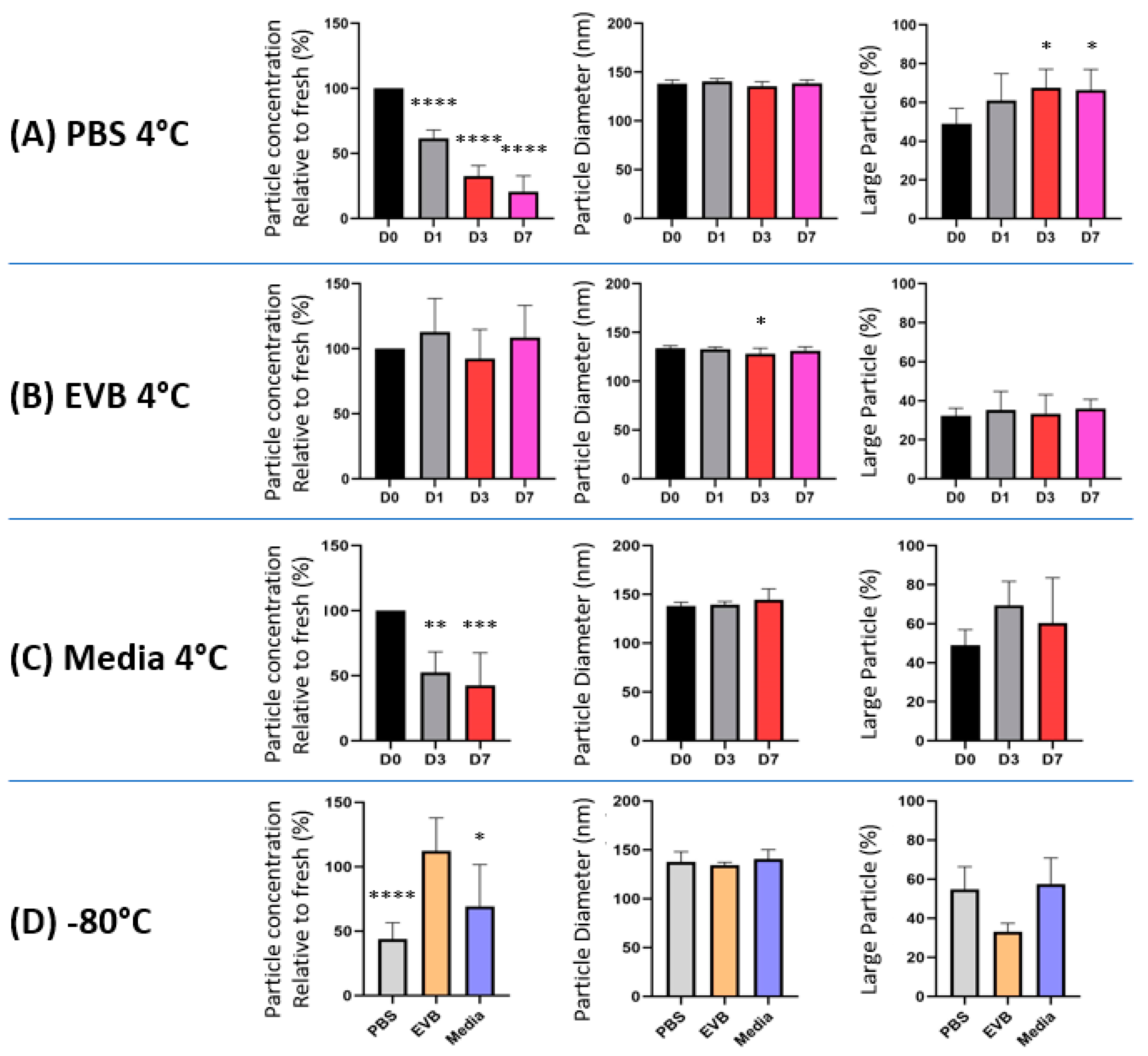
2.2. EV Storage Condition Influence Recovered Particle Numbers and Sizes
2.3. EV Storage Conditions Influence the Integrity of eEVs and Packaged DNA Contents
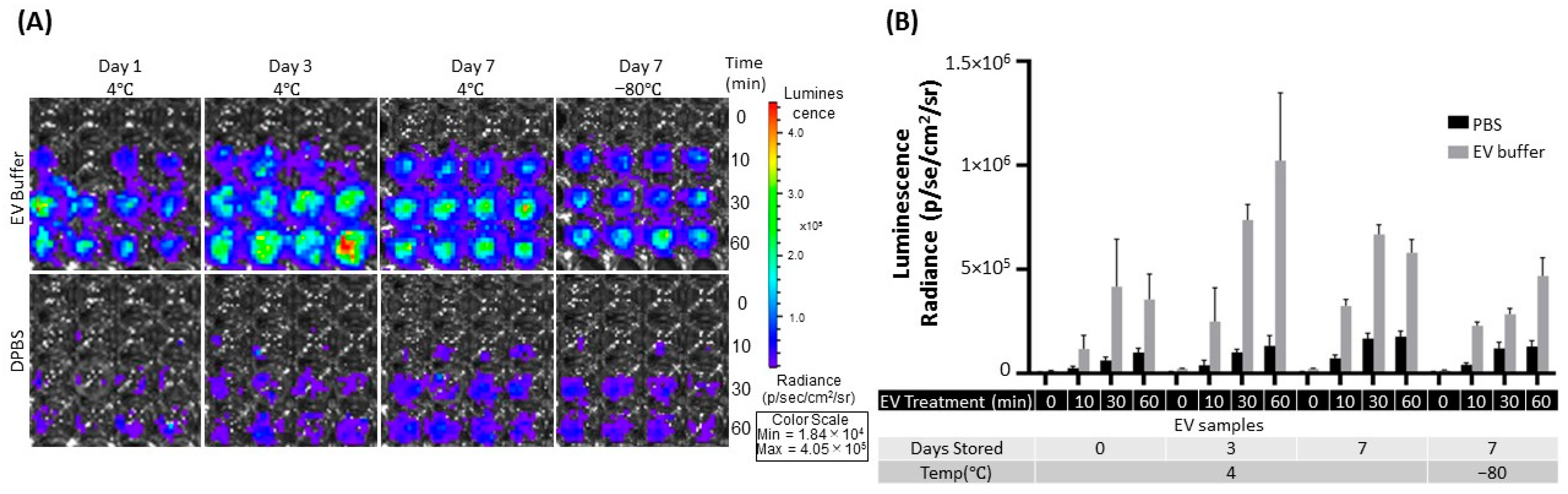
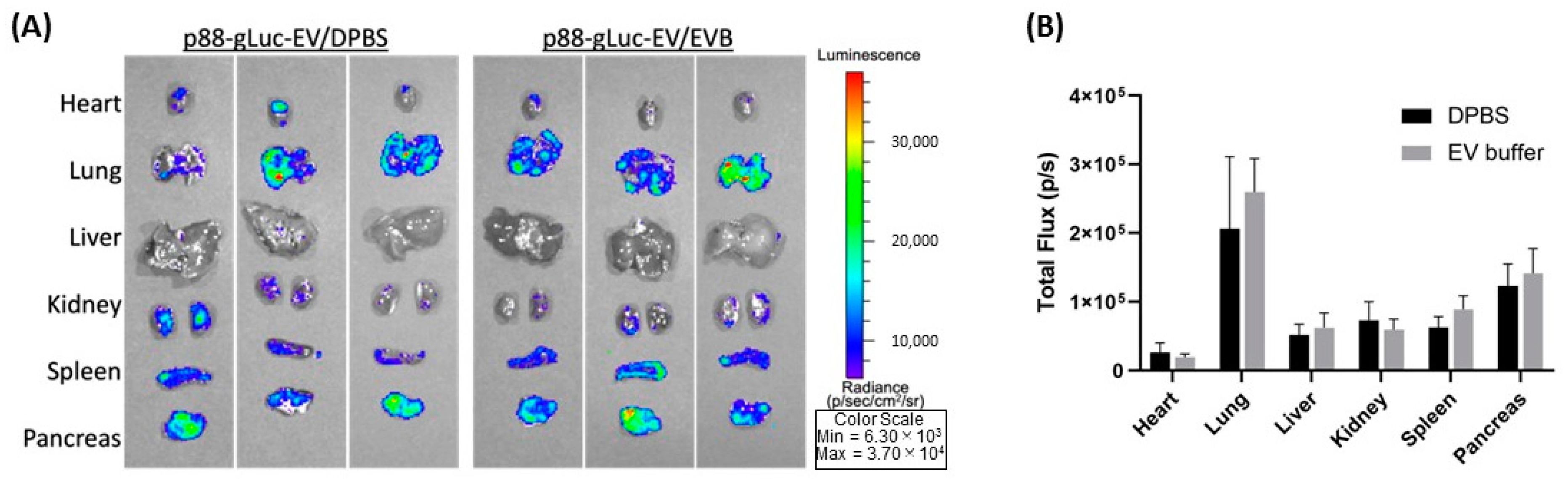
2.4. Effect on the eEV Functionality by EV Buffer Storage
3. Discussion
4. Materials and Methods
4.1. Cell Culture and Treatment
4.2. EV Isolation
4.3. Nanoparticle Tracking Analysis (NTA)
4.4. Immuno-Transmission Electron Microscopy (Immuno-TEM)
4.5. DNase I Treatment and Plasmid DNA Recovery from EVs
4.6. Quantitative Real-Time Polymerase Chain Reaction (qPCR)
4.7. In Vitro Bioluminescence Binding Assay
4.8. Animals and Ex Vivo Bioluminescence Biodistribution Assay
4.9. Data Analysis and Statistics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wolf, P. The nature and significance of platelet products in human plasma. Br. J. Haematol. 1967, 13, 269–288. [Google Scholar] [CrossRef] [PubMed]
- Yanez-Mo, M.; Siljander, P.R.; Andreu, Z.; Zavec, A.B.; Borras, F.E.; Buzas, E.I.; Buzas, K.; Casal, E.; Cappello, F.; Carvalho, J.; et al. Biological properties of extracellular vesicles and their physiological functions. J. Extracell. Vesicles 2015, 4, 27066. [Google Scholar] [CrossRef] [PubMed]
- Harding, C.; Heuser, J.; Stahl, P. Receptor-mediated endocytosis of transferrin and recycling of the transferrin receptor in rat reticulocytes. J. Cell Biol. 1983, 97, 329–339. [Google Scholar] [CrossRef]
- Pan, B.T.; Johnstone, R.M. Fate of the transferrin receptor during maturation of sheep reticulocytes in vitro: Selective externalization of the receptor. Cell 1983, 33, 967–978. [Google Scholar] [CrossRef]
- Robbins, P.D.; Morelli, A.E. Regulation of immune responses by extracellular vesicles. Nat. Rev. Immunol. 2014, 14, 195–208. [Google Scholar] [CrossRef]
- Thompson, A.G.; Gray, E.; Heman-Ackah, S.M.; Mager, I.; Talbot, K.; Andaloussi, S.E.; Wood, M.J.; Turner, M.R. Extracellular vesicles in neurodegenerative disease—Pathogenesis to biomarkers. Nat. Rev. Neurol. 2016, 12, 346–357. [Google Scholar] [CrossRef] [PubMed]
- Buzas, E.I.; Gyorgy, B.; Nagy, G.; Falus, A.; Gay, S. Emerging role of extracellular vesicles in inflammatory diseases. Nat. Rev. Rheumatol. 2014, 10, 356–364. [Google Scholar] [CrossRef] [PubMed]
- Jansen, F.; Nickenig, G.; Werner, N. Extracellular Vesicles in Cardiovascular Disease: Potential Applications in Diagnosis, Prognosis, and Epidemiology. Circ. Res. 2017, 120, 1649–1657. [Google Scholar] [CrossRef]
- Becker, A.; Thakur, B.K.; Weiss, J.M.; Kim, H.S.; Peinado, H.; Lyden, D. Extracellular Vesicles in Cancer: Cell-to-Cell Mediators of Metastasis. Cancer Cell 2016, 30, 836–848. [Google Scholar] [CrossRef]
- Schorey, J.S.; Harding, C.V. Extracellular vesicles and infectious diseases: New complexity to an old story. J. Clin. Investig. 2016, 126, 1181–1189. [Google Scholar] [CrossRef]
- Gandham, S.; Su, X.; Wood, J.; Nocera, A.L.; Alli, S.C.; Milane, L.; Zimmerman, A.; Amiji, M.; Ivanov, A.R. Technologies and Standardization in Research on Extracellular Vesicles. Trends Biotechnol. 2020, 38, 1066–1098. [Google Scholar] [CrossRef] [PubMed]
- Lotvall, J.; Hill, A.F.; Hochberg, F.; Buzas, E.I.; Di Vizio, D.; Gardiner, C.; Gho, Y.S.; Kurochkin, I.V.; Mathivanan, S.; Quesenberry, P.; et al. Minimal experimental requirements for definition of extracellular vesicles and their functions: A position statement from the International Society for Extracellular Vesicles. J. Extracell. Vesicles 2014, 3, 26913. [Google Scholar] [CrossRef]
- Thery, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef] [PubMed]
- Witwer, K.W.; Goberdhan, D.C.; O’Driscoll, L.; Thery, C.; Welsh, J.A.; Blenkiron, C.; Buzas, E.I.; Di Vizio, D.; Erdbrugger, U.; Falcon-Perez, J.M.; et al. Updating MISEV: Evolving the minimal requirements for studies of extracellular vesicles. J. Extracell. Vesicles 2021, 10, e12182. [Google Scholar] [CrossRef] [PubMed]
- Witwer, K.W.; Buzás, E.I.; Bemis, L.T.; Bora, A.; Lässer, C.; Lötvall, J.; Nolte-’t Hoen, E.N.; Piper, M.G.; Sivaraman, S.; Skog, J.; et al. Standardization of sample collection, isolation and analysis methods in extracellular vesicle research. J. Extracell. Vesicles 2013, 2, 20360. [Google Scholar] [CrossRef]
- Gelibter, S.; Marostica, G.; Mandelli, A.; Siciliani, S.; Podini, P.; Finardi, A.; Furlan, R. The impact of storage on extracellular vesicles: A systematic study. J. Extracell. Vesicles 2022, 11, e12162. [Google Scholar] [CrossRef]
- Royo, F.; Thery, C.; Falcon-Perez, J.M.; Nieuwland, R.; Witwer, K.W. Methods for Separation and Characterization of Extracellular Vesicles: Results of a Worldwide Survey Performed by the ISEV Rigor and Standardization Subcommittee. Cells 2020, 9, 1955. [Google Scholar] [CrossRef]
- van de Wakker, S.I.; van Oudheusden, J.; Mol, E.A.; Roefs, M.T.; Zheng, W.; Gorgens, A.; El Andaloussi, S.; Sluijter, J.P.G.; Vader, P. Influence of short term storage conditions, concentration methodsand excipients on extracellular vesicle recovery and function. Eur. J. Pharm. Biopharm. 2022, 170, 59–69. [Google Scholar] [CrossRef]
- Tegegn, T.Z.; De Paoli, S.H.; Orecna, M.; Elhelu, O.K.; Woodle, S.A.; Tarandovskiy, I.D.; Ovanesov, M.V.; Simak, J. Characterization of procoagulant extracellular vesicles and platelet membrane disintegration in DMSO-cryopreserved platelets. J. Extracell. Vesicles 2016, 5, 30422. [Google Scholar] [CrossRef]
- Bosch, S.; de Beaurepaire, L.; Allard, M.; Mosser, M.; Heichette, C.; Chretien, D.; Jegou, D.; Bach, J.M. Trehalose prevents aggregation of exosomes and cryodamage. Sci. Rep. 2016, 6, 36162. [Google Scholar] [CrossRef]
- Gorgens, A.; Corso, G.; Hagey, D.W.; Jawad Wiklander, R.; Gustafsson, M.O.; Felldin, U.; Lee, Y.; Bostancioglu, R.B.; Sork, H.; Liang, X.; et al. Identification of storage conditions stabilizing extracellular vesicles preparations. J. Extracell. Vesicles 2022, 11, e12238. [Google Scholar] [CrossRef] [PubMed]
- Belinskaia, D.A.; Voronina, P.A.; Goncharov, N.V. Integrative Role of Albumin: Evolutionary, Biochemical and Pathophysiological Aspects. J. Evol. Biochem. Physiol. 2021, 57, 1419–1448. [Google Scholar] [CrossRef]
- Maier, R.; Fries, M.R.; Buchholz, C.; Zhang, F.; Schreiber, F. Human versus Bovine Serum Albumin: A Subtle Difference in Hydrophobicity Leads to Large Differences in Bulk and Interface Behavior. Cryst. Growth Des. 2021, 21, 5451–5459. [Google Scholar] [CrossRef]
- Bujacz, A. Structures of bovine, equine and leporine serum albumin. Acta Crystallogr. Sect. D 2012, 68, 1278–1289. [Google Scholar] [CrossRef]
- Komuro, H.; Kawai-Harada, Y.; Aminova, S.; Pascual, N.; Malik, A.; Contag, C.H.; Harada, M. Engineering Extracellular Vesicles to Target Pancreatic Tissue In Vivo. Nanotheranostics 2021, 5, 378–390. [Google Scholar] [CrossRef] [PubMed]
- Komuro, H.; Aminova, S.; Lauro, K.; Woldring, D.; Harada, M. Design and Evaluation of Engineered Extracellular Vesicle (EV)-Based Targeting for EGFR-Overexpressing Tumor Cells Using Monobody Display. Bioengineering 2022, 9, 56. [Google Scholar] [CrossRef] [PubMed]
- Maroto, R.; Zhao, Y.; Jamaluddin, M.; Popov, V.L.; Wang, H.; Kalubowilage, M.; Zhang, Y.; Luisi, J.; Sun, H.; Culbertson, C.T.; et al. Effects of storage temperature on airway exosome integrity for diagnostic and functional analyses. J. Extracell. Vesicles 2017, 6, 1359478. [Google Scholar] [CrossRef]
- Zonneveld, M.I.; Brisson, A.R.; van Herwijnen, M.J.; Tan, S.; van de Lest, C.H.; Redegeld, F.A.; Garssen, J.; Wauben, M.H.; Nolte-’t Hoen, E.N. Recovery of extracellular vesicles from human breast milk is influenced by sample collection and vesicle isolation procedures. J. Extracell. Vesicles 2014, 3, 24215. [Google Scholar] [CrossRef]
- Nguyen, V.V.T.; Witwer, K.W.; Verhaar, M.C.; Strunk, D.; van Balkom, B.W.M. Functional assays to assess the therapeutic potential of extracellular vesicles. J. Extracell. Vesicles 2020, 10, e12033. [Google Scholar] [CrossRef]
- Trenkenschuh, E.; Richter, M.; Heinrich, E.; Koch, M.; Fuhrmann, G.; Friess, W. Enhancing the Stabilization Potential of Lyophilization for Extracellular Vesicles. Adv. Healthc. Mater. 2022, 11, e2100538. [Google Scholar] [CrossRef]
- Cheng, Y.; Zeng, Q.; Han, Q.; Xia, W. Effect of pH, temperature and freezing-thawing on quantity changes and cellular uptake of exosomes. Protein Cell 2019, 10, 295–299. [Google Scholar] [CrossRef]
- Park, S.J.; Jeon, H.; Yoo, S.M.; Lee, M.S. The effect of storage temperature on the biological activity of extracellular vesicles for the complement system. In Vitro Cell Dev. Biol. Anim. 2018, 54, 423–429. [Google Scholar] [CrossRef]
- Lorincz, A.M.; Timar, C.I.; Marosvari, K.A.; Veres, D.S.; Otrokocsi, L.; Kittel, A.; Ligeti, E. Effect of storage on physical and functional properties of extracellular vesicles derived from neutrophilic granulocytes. J. Extracell. Vesicles 2014, 3, 25465. [Google Scholar] [CrossRef]
- Evtushenko, E.G.; Bagrov, D.V.; Lazarev, V.N.; Livshits, M.A.; Khomyakova, E. Adsorption of extracellular vesicles onto the tube walls during storage in solution. PLoS ONE 2020, 15, e0243738. [Google Scholar] [CrossRef]
- McCann, J.; Sosa-Miranda, C.D.; Guo, H.; Reshke, R.; Savard, A.; Zardini Buzatto, A.; Taylor, J.A.; Li, L.; Gibbings, D.J. Contaminating transfection complexes can masquerade as small extracellular vesicles and impair their delivery of RNA. J. Extracell. Vesicles 2022, 11, e12220. [Google Scholar] [CrossRef]
- Wu, J.Y.; Li, Y.J.; Hu, X.B.; Huang, S.; Xiang, D.X. Preservation of small extracellular vesicles for functional analysis and therapeutic applications: A comparative evaluation of storage conditions. Drug Deliv. 2021, 28, 162–170. [Google Scholar] [CrossRef]
- Wang, L.; Li, Y.; Guo, R.; Li, S.; Chang, A.; Zhu, Z.; Tu, P. Optimized bioluminescence analysis of adenosine triphosphate (ATP) released by platelets and its application in the high throughput screening of platelet inhibitors. PLoS ONE 2019, 14, e0223096. [Google Scholar] [CrossRef]

| Temperature | Buffer | Fresh | Day 1 | Day 3 | Day 7 |
|---|---|---|---|---|---|
| 4 °C | PBS | + | + | + | + |
| EV Buffer | + | + | + | + | |
| Media (not isolated) | + | + | |||
| −80 °C | PBS | + | |||
| EV Buffer | + | ||||
| Media (not isolated) | + |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kawai-Harada, Y.; El Itawi, H.; Komuro, H.; Harada, M. Evaluation of EV Storage Buffer for Efficient Preservation of Engineered Extracellular Vesicles. Int. J. Mol. Sci. 2023, 24, 12841. https://doi.org/10.3390/ijms241612841
Kawai-Harada Y, El Itawi H, Komuro H, Harada M. Evaluation of EV Storage Buffer for Efficient Preservation of Engineered Extracellular Vesicles. International Journal of Molecular Sciences. 2023; 24(16):12841. https://doi.org/10.3390/ijms241612841
Chicago/Turabian StyleKawai-Harada, Yuki, Hanine El Itawi, Hiroaki Komuro, and Masako Harada. 2023. "Evaluation of EV Storage Buffer for Efficient Preservation of Engineered Extracellular Vesicles" International Journal of Molecular Sciences 24, no. 16: 12841. https://doi.org/10.3390/ijms241612841
APA StyleKawai-Harada, Y., El Itawi, H., Komuro, H., & Harada, M. (2023). Evaluation of EV Storage Buffer for Efficient Preservation of Engineered Extracellular Vesicles. International Journal of Molecular Sciences, 24(16), 12841. https://doi.org/10.3390/ijms241612841






