S100A4 Promotes BCG-Induced Pyroptosis of Macrophages by Activating the NF-κB/NLRP3 Inflammasome Signaling Pathway
Abstract
1. Introduction
2. Results
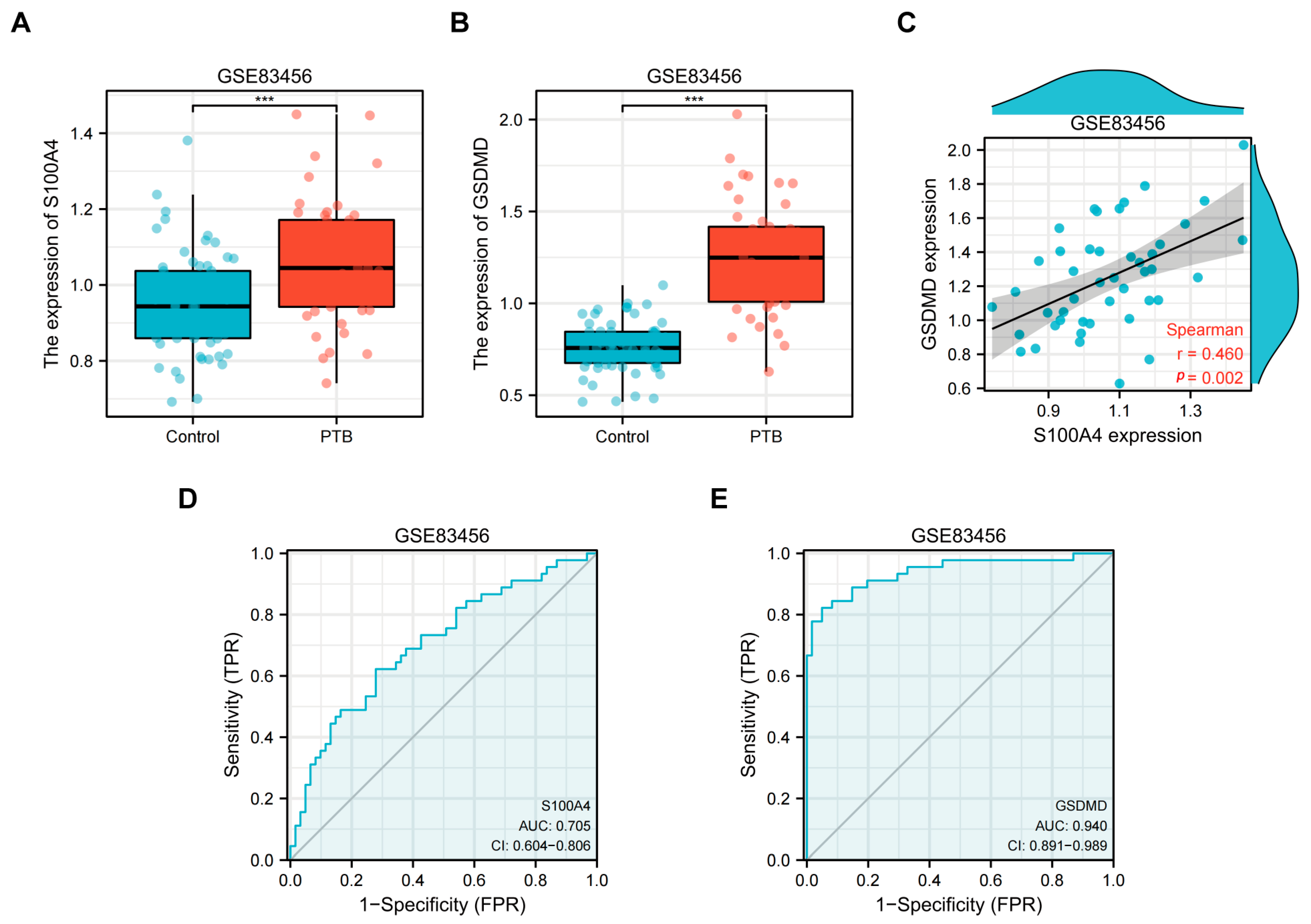
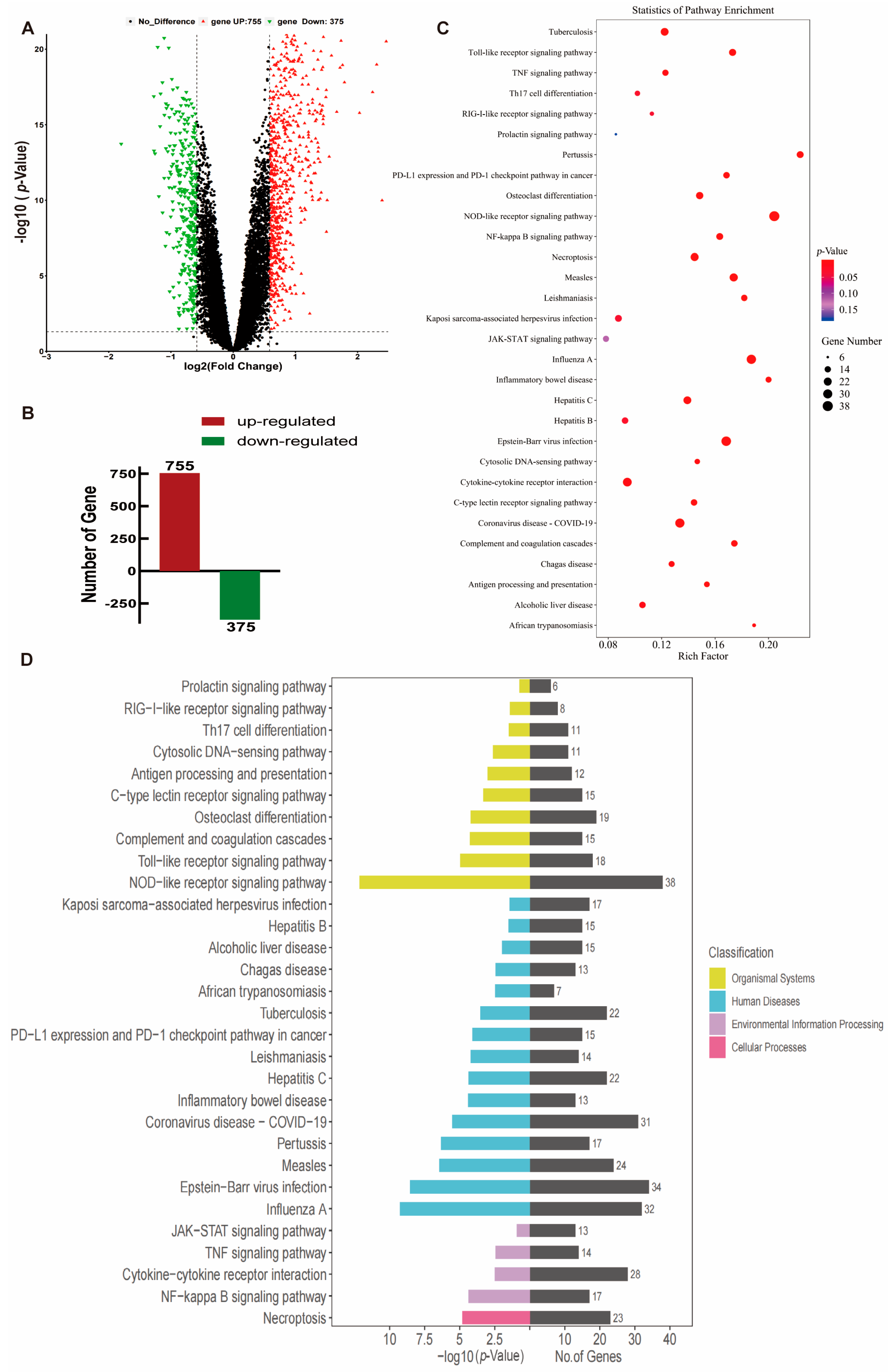
2.1. Transcriptome Analysis of Peripheral Blood from TB Patients
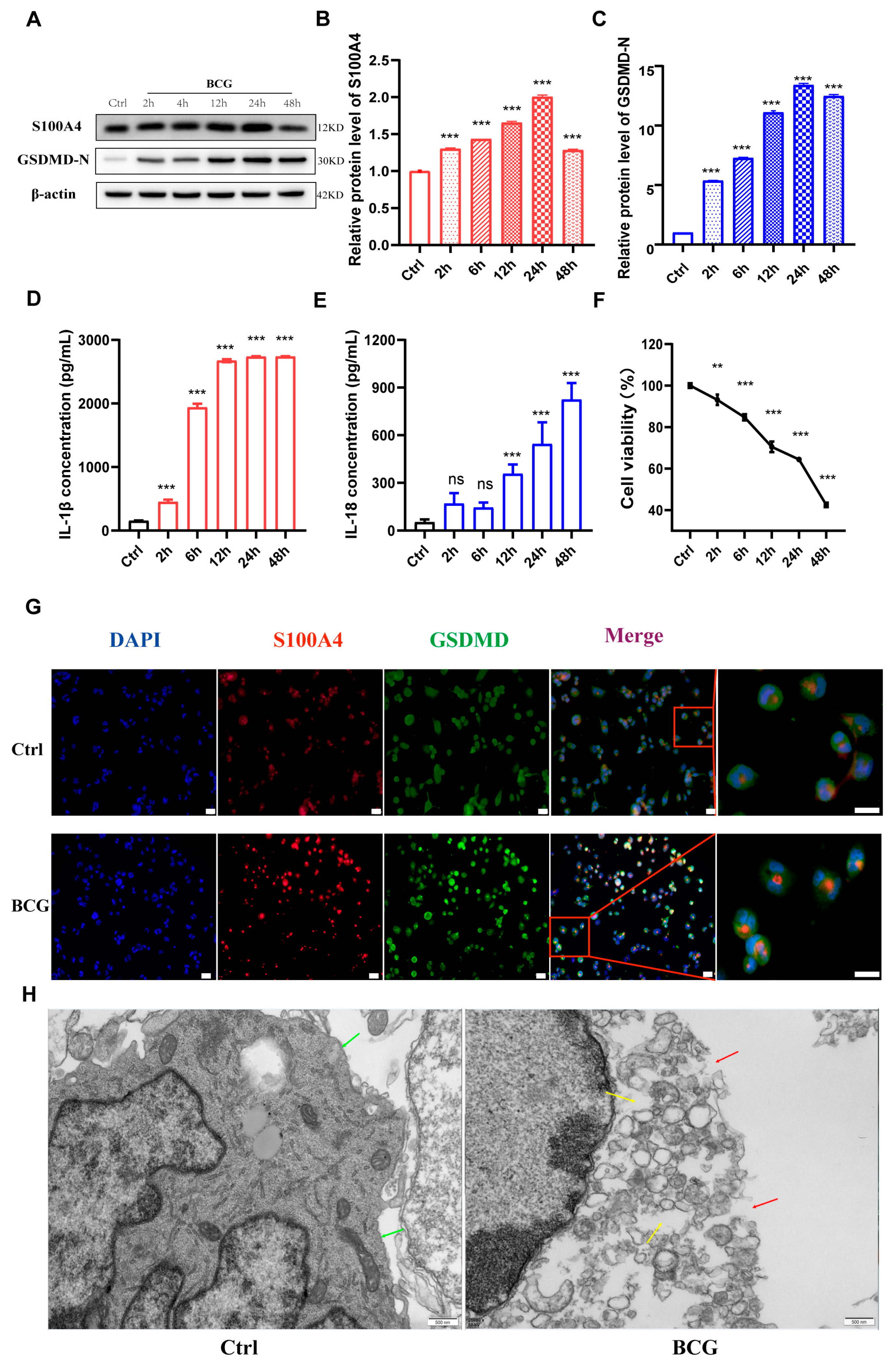
2.2. Infection of THP-1 Macrophage with BCG Up-Regulate S100A4 and Induce Pyroptosis
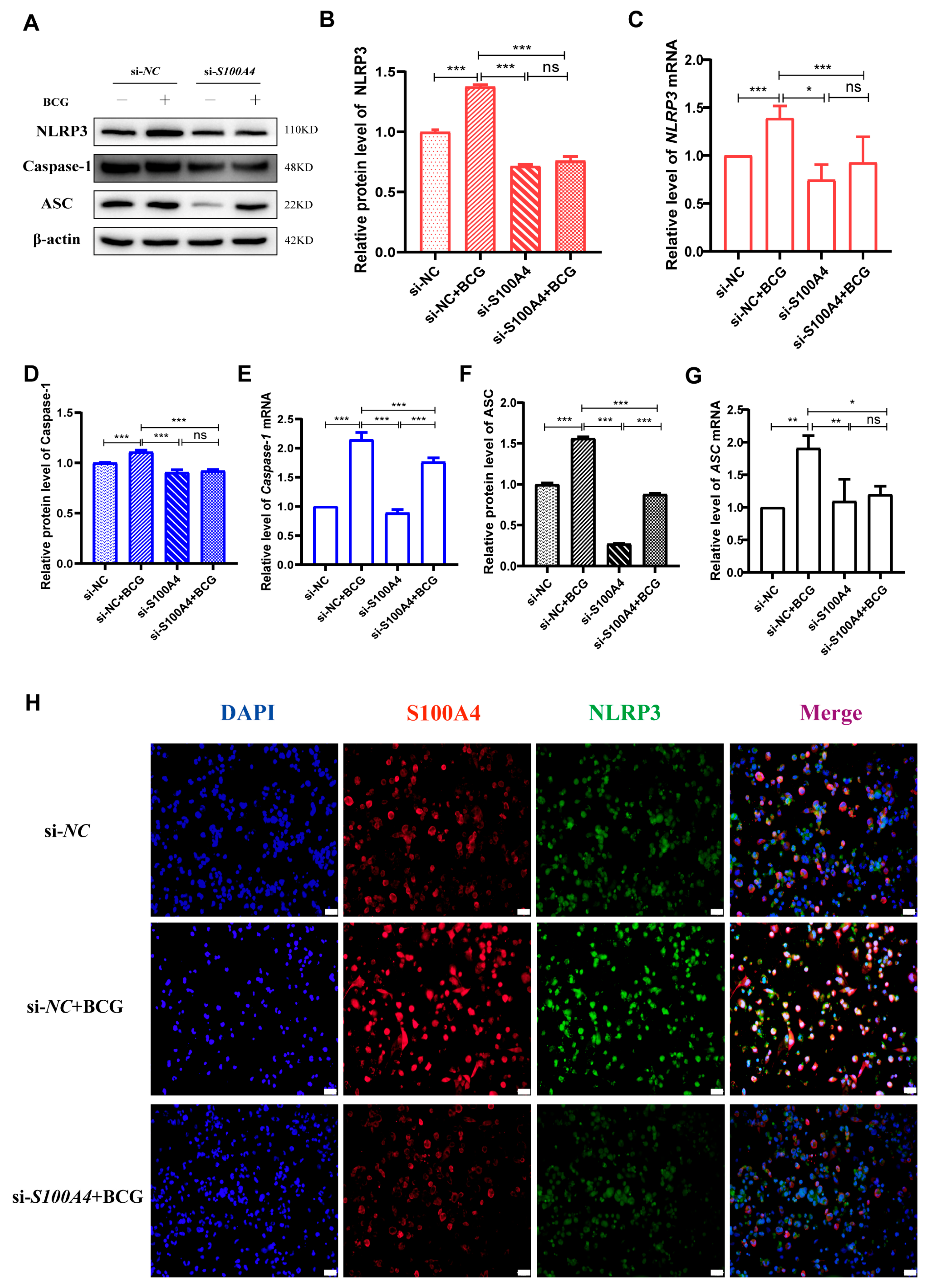
2.3. si-S100A4 Inhibited Pyroptosis in BCG-Infected THP-1 Macrophage
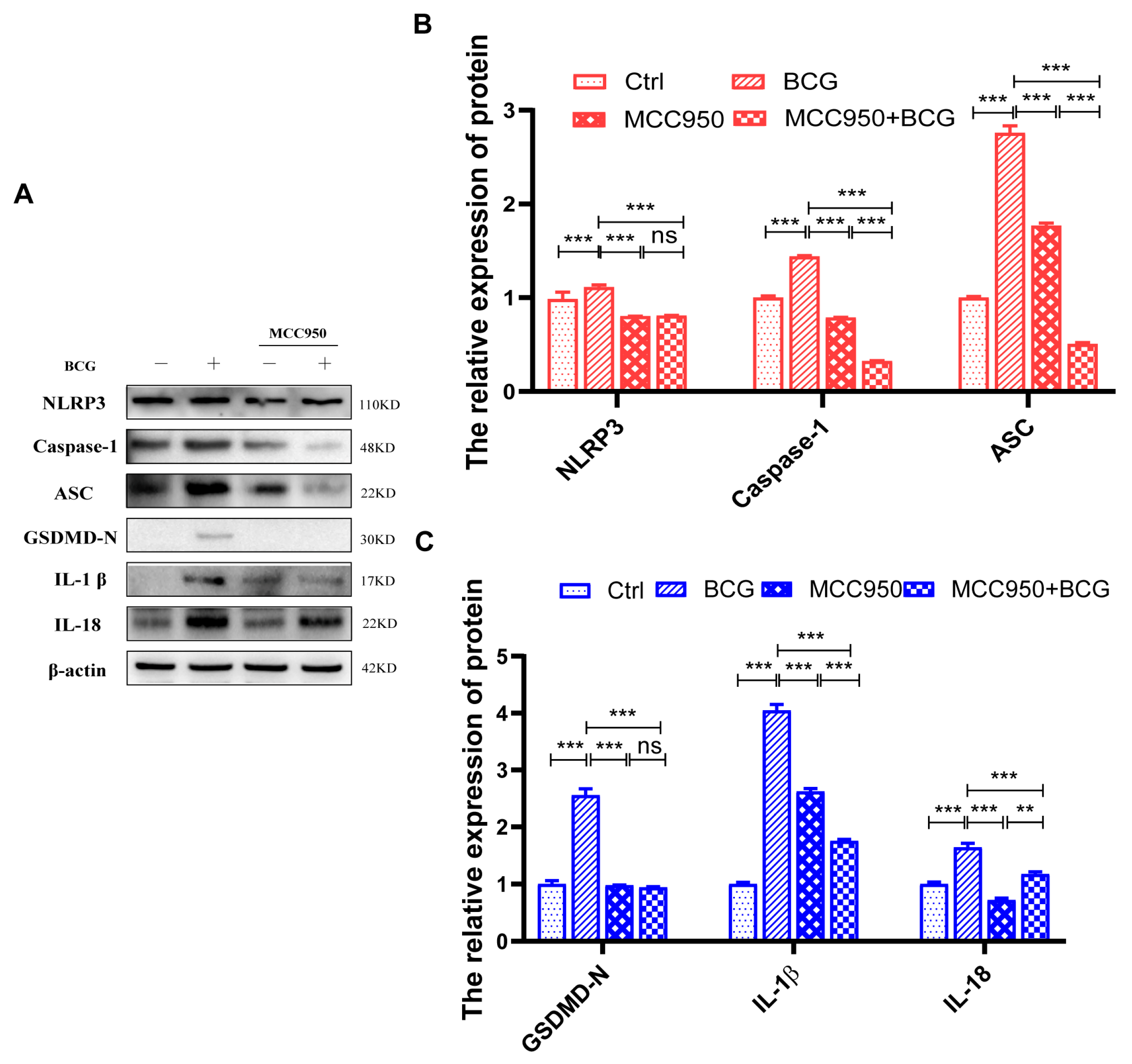
2.4. si-S100A4 Inhibited the Activation of NLRP3 Inflammasome in BCG-Infected THP-1 Macrophage
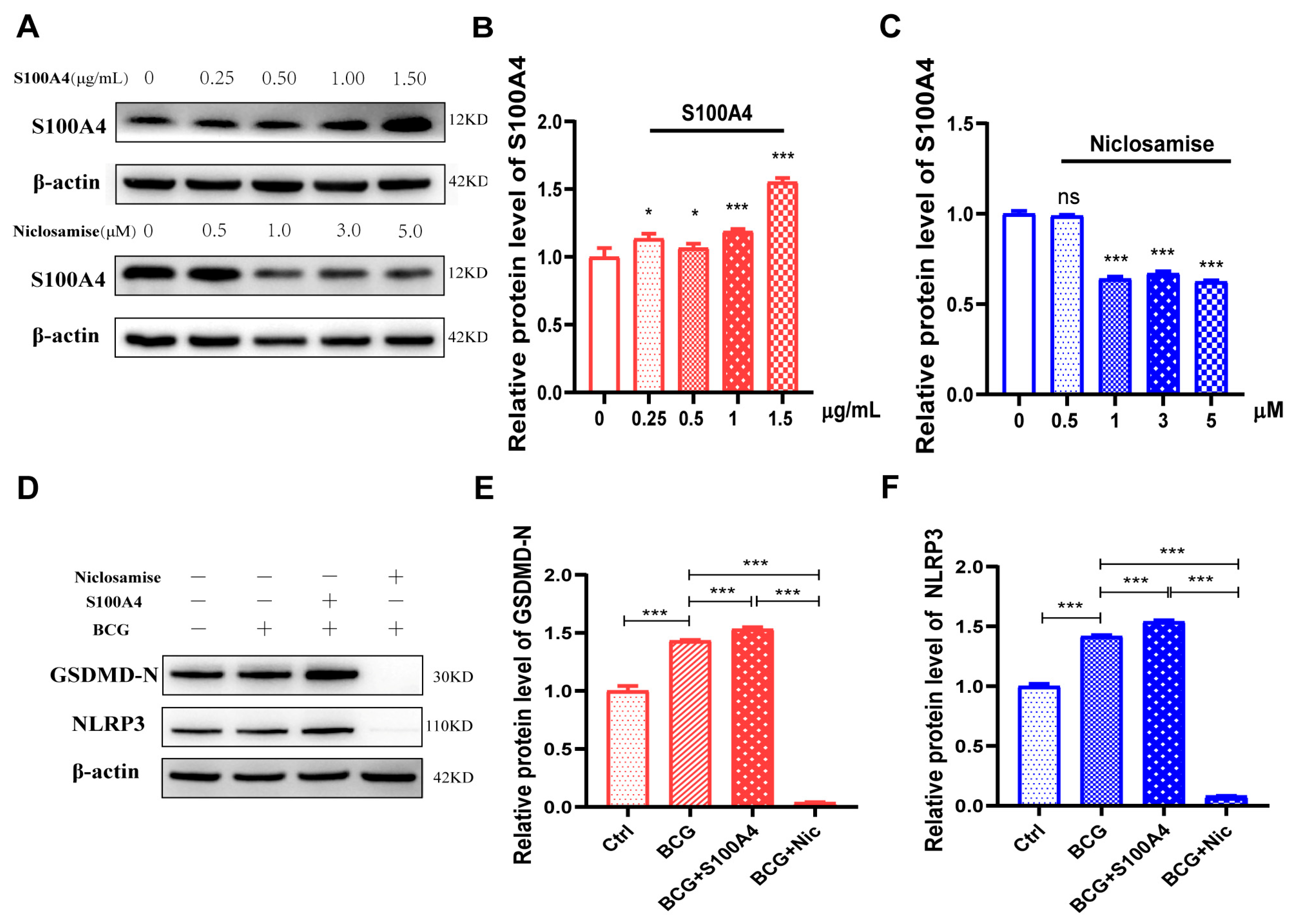
2.5. Exogenous S100A4 Up-Regulated GSDMD-N and NLRP3 Protein Expression, While Niclosamide Had the Opposite Effects in BCG-Infected THP-1 Macrophage
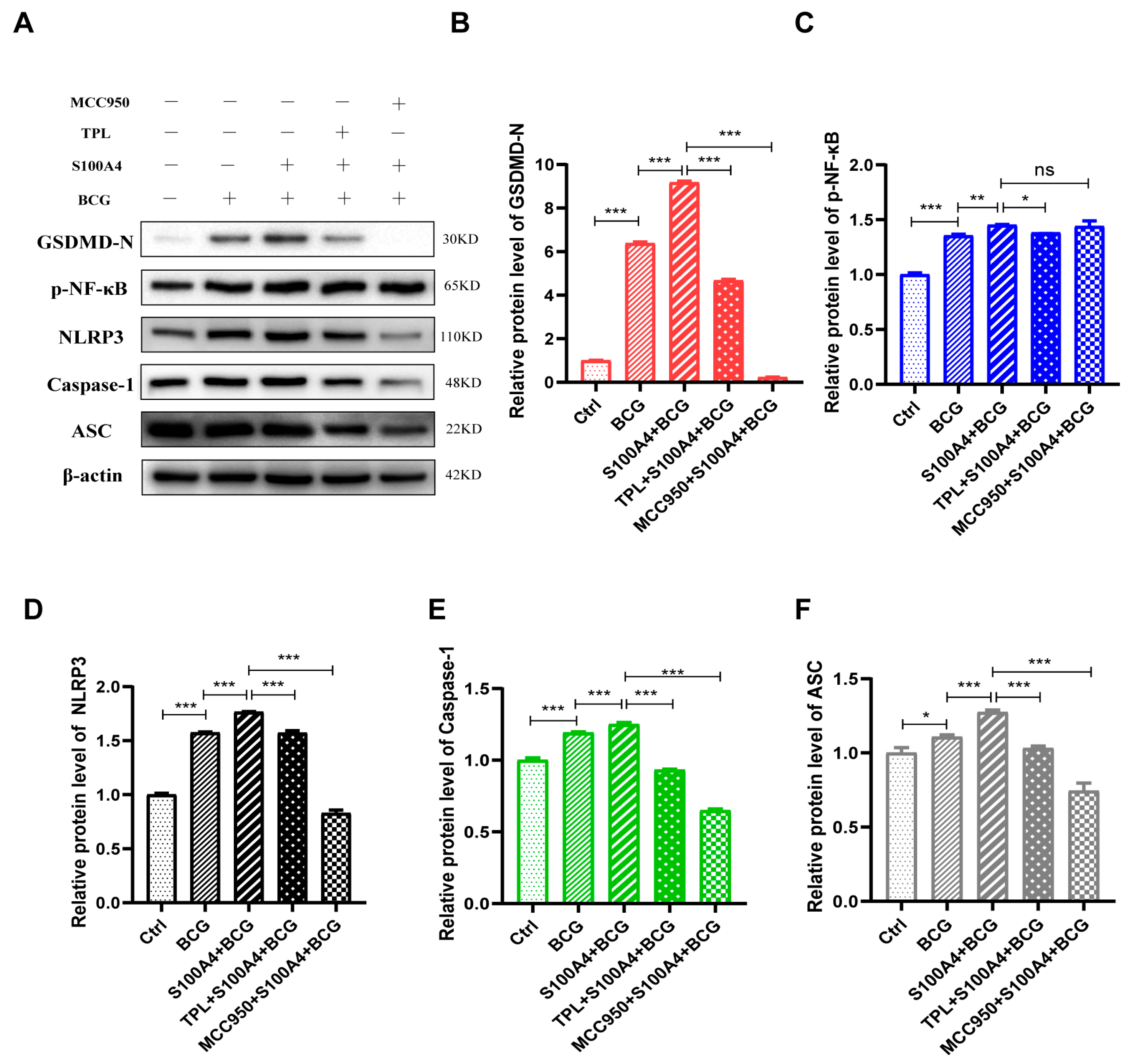
2.6. S100A4 Up-Regulates Pyroptosis of BCG-Infected THP-1 Macrophage by Activating the NF-κB/NLRP3 Inflammasome Signaling Pathway
3. Discussion
4. Materials and Methods
4.1. Bacterial Culture
4.2. Cell Culture and Infection
4.3. Small Interfering RNA Transfection
4.4. Chemicals and Inhibitors Treatment
4.5. Western Blot
4.6. Quantitative Reverse-Transcription PCR (qRT-PCR)
4.7. Immunofluorescent Staining
4.8. Transmission Electron Microscopy
4.9. Enzyme-Linked Immunosorbent Assay (ELISA)
4.10. CCK-8 Assay
4.11. GEO Data Analysis
4.12. Statistical Analysis
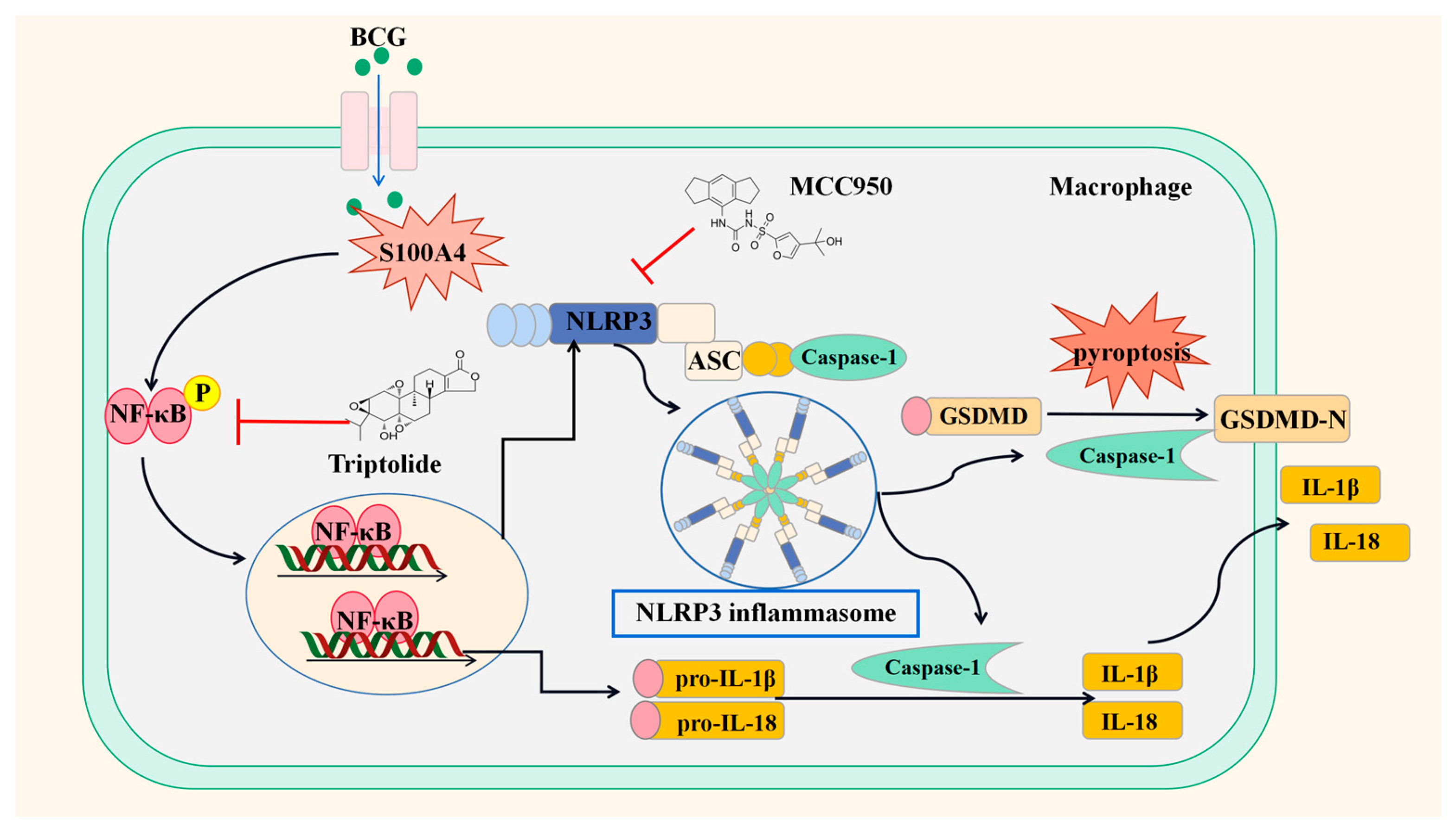
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Huang, L.; Nazarova, E.V.; Russell, D.G. Mycobacterium tuberculosis: Bacterial Fitness within the Host Macrophage. Microbiol. Spectr. 2019, 7, 127–138. [Google Scholar]
- McQuaid, C.F.; McCreesh, N.; Read, J.M.; Sumner, T.; Houben, R.; White, R.G.; Harris, R.C. The potential impact of COVID-19-related disruption on tuberculosis burden. Eur. Respir. J. 2020, 56, 2001718. [Google Scholar] [CrossRef] [PubMed]
- Dheda, K.; Perumal, T.; Moultrie, H.; Perumal, R.; Esmail, A.; Scott, A.J.; Udwadia, Z.; Chang, K.C.; Peter, J.; Pooran, A.; et al. The intersecting pandemics of tuberculosis and COVID-19: Population-level and patient-level impact, clinical presentation, and corrective interventions. Lancet Respir. Med. 2022, 10, 603–622. [Google Scholar] [CrossRef]
- Schito, M.; Migliori, G.B.; Fletcher, H.A.; McNerney, R.; Centis, R.; D’Ambrosio, L.; Bates, M.; Kibiki, G.; Kapata, N.; Corrah, T.; et al. Perspectives on Advances in Tuberculosis Diagnostics, Drugs, and Vaccines. Clin. Infect. Dis. 2015, 61 (Suppl. 3), S102–S118. [Google Scholar] [CrossRef]
- Shi, L.; Jiang, Q.; Bushkin, Y.; Subbian, S.; Tyagi, S. Biphasic Dynamics of Macrophage Immunometabolism during Mycobacterium tuberculosis Infection. Mbio 2019, 10, e02550-18. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Xue, D.; Song, F.; Liu, X.; Li, W.; Wang, Y. DUSP5 (dual-specificity protein phosphatase 5) suppresses BCG-induced autophagy via ERK 1/2 signaling pathway. Mol. Immunol. 2020, 126, 101–109. [Google Scholar] [CrossRef]
- Zheng, X.; Li, M.; Chen, Q.; Ma, B.; Nie, X.; Liu, Y.; Yang, Y.; Xu, J.; Wang, Y. Ror2-mediated cholesterol accumulation regulates autophagic activity within BCG-infected macrophages. Microb. Pathog. 2022, 167, 105564. [Google Scholar] [CrossRef]
- Chen, Q.; Zheng, X.; Li, Y.; Ma, B.; Nie, X.; Li, M.; Liu, Y.; Xu, J.; Yang, Y. Wnt5a regulates autophagy in Bacille Calmette-Guérin (BCG)-Infected pulmonary epithelial cells. Microb. Pathog. 2022, 173, 105826. [Google Scholar] [CrossRef]
- Ma, C.; Wu, X.; Zhang, X.; Liu, X.; Deng, G. Heme oxygenase-1 modulates ferroptosis by fine-tuning levels of intracellular iron and reactive oxygen species of macrophages in response to Bacillus Calmette-Guerin infection. Front. Cell. Infect. Microbiol. 2022, 12, 1004148. [Google Scholar] [CrossRef]
- Fu, Y.; Shen, J.; Liu, F.; Zhang, H.; Zheng, Y.; Jiang, X. Andrographolide Suppresses Pyroptosis in Mycobacterium tuberculosis-Infected Macrophages via the microRNA-155/Nrf2 Axis. Oxid. Med. Cell. Longev. 2022, 2022, 1885066. [Google Scholar] [CrossRef]
- Li, Y.; Fu, Y.; Sun, J.; Shen, J.; Liu, F.; Ning, B.; Lu, Z.; Wei, L.; Jiang, X. Tanshinone IIA alleviates NLRP3 inflammasome-mediated pyroptosis in Mycobacterium tuberculosis-(H37Ra-) infected macrophages by inhibiting endoplasmic reticulum stress. J. Ethnopharmacol. 2022, 282, 114595. [Google Scholar] [CrossRef] [PubMed]
- Qian, J.; Hu, Y.; Zhang, X.; Chi, M.; Xu, S.; Wang, H.; Zhang, X. Mycobacterium tuberculosis PE_PGRS19 Induces Pyroptosis through a Non-Classical Caspase-11/GSDMD Pathway in Macrophages. Microorganisms 2022, 10, 2473. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Xia, S.; Zhang, Z.; Wu, H.; Lieberman, J. Channelling inflammation: Gasdermins in physiology and disease. Nat. Rev. Drug. Discov. 2021, 20, 384–405. [Google Scholar] [CrossRef]
- Pereira, M.; Tourlomousis, P.; Wright, J.; Monie, P.T.; Bryant, C.E. CARD9 negatively regulates NLRP3-induced IL-1β production on Salmonella infection of macrophages. Nat. Commun. 2016, 7, 12874. [Google Scholar] [CrossRef]
- Broz, P.; Dixit, V.M. Inflammasomes: Mechanism of assembly, regulation and signalling. Nat. Rev. Immunol. 2016, 16, 407–420. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Ma, W.; Gao, W.; Xing, Y.; Chen, L.; Xia, Z.; Zhang, Z.; Dai, Z. Propofol directly induces caspase-1-dependent macrophage pyroptosis through the NLRP3-ASC inflammasome. Cell Death Dis. 2019, 10, 542. [Google Scholar] [CrossRef] [PubMed]
- Ma, M.Q.; Zheng, S.S.; Chen, H.L.; Xu, H.B.; Zhang, D.L.; Zhang, Y.A.; Xiang, S.Y.; Cheng, B.H.; Jin, S.W.; Fu, P.H. Protectin Conjugates in Tissue Regeneration 1 Inhibits Macrophage Pyroptosis by Restricting NLRP3 Inflammasome Assembly to Mitigate Sepsis via the cAMP-PKA Pathway. Lab. Investig. 2023, 103, 100028. [Google Scholar] [CrossRef]
- Mulvihill, E.; Sborgi, L.; Mari, S.A.; Pfreundschuh, M.; Hiller, S.; Müller, D.J. Mechanism of membrane pore formation by human gasdermin-D. EMBO J. 2018, 37, e98321. [Google Scholar] [CrossRef]
- Donato, R.; Cannon, B.R.; Sorci, G.; Riuzzi, F.; Hsu, K.; Weber, D.J.; Geczy, C.L. Functions of S100 proteins. Curr. Mol. Med. 2013, 13, 24–57. [Google Scholar] [CrossRef]
- Bertheloot, D.; Latz, E. HMGB1, IL-1α, IL-33 and S100 proteins: Dual-function alarmins. Cell Mol. Immunol. 2017, 14, 43–64. [Google Scholar] [CrossRef]
- Bresnick, A.R.; Weber, D.J.; Zimmer, D.B. S100 proteins in cancer. Nat. Rev. Cancer 2015, 15, 96–109. [Google Scholar] [CrossRef]
- Fei, F.; Qu, J.; Li, C.; Wang, X.; Li, Y.; Zhang, S. Role of metastasis-induced protein S100A4 in human non-tumor pathophysiologies. Cell Biosci. 2017, 7, 64. [Google Scholar] [CrossRef]
- Boye, K.; Maelandsmo, G.M. S100A4 and metastasis: A small actor playing many roles. Am. J. Pathol. 2010, 176, 528–535. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Zhang, H.; Li, Y.; Zhang, Y.; Bian, Y.; Zeng, Y.; Yao, X.; Wan, J.; Chen, X.; Li, J.; et al. S100A4 enhances protumor macrophage polarization by control of PPAR-γ-dependent induction of fatty acid oxidation. J. Immunother. Cancer 2021, 9, e002548. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Wu, Y.H.; Liu, S.Q.; Sheng, Z.Y.; Zhen, Z.D.; Gao, R.Q.; Cui, X.Y.; Fan, D.Y.; Qin, Z.H.; Zheng, A.H.; et al. S100A4+ macrophages facilitate zika virus invasion and persistence in the seminiferous tubules via interferon-gamma mediation. PLoS Pathog. 2020, 16, e1009019. [Google Scholar] [CrossRef]
- Yadav, A.; Vallabu, S.; Kumar, D.; Ding, G.; Charney, D.N.; Chander, P.N.; Singhal, P.C. HIVAN phenotype: Consequence of epithelial mesenchymal transdifferentiation. Am. J. Physiol. Renal. Physiol. 2010, 298, F734–F744. [Google Scholar] [CrossRef]
- Zhu, K.; Huang, W.; Wang, W.; Liao, L.; Li, S.; Yang, S.; Xu, J.; Li, L.; Meng, M.; Xie, Y.; et al. Up-regulation of S100A4 expression by HBx protein promotes proliferation of hepatocellular carcinoma cells and its correlation with clinical survival. Gene 2020, 749, 144679. [Google Scholar] [CrossRef] [PubMed]
- Blankley, S.; Graham, C.M.; Turner, J.; Berry, M.P.; Bloom, C.I.; Xu, Z.; Pascual, V.; Banchereau, J.; Chaussabel, D.; Breen, R.; et al. The Transcriptional Signature of Active Tuberculosis Reflects Symptom Status in Extra-Pulmonary and Pulmonary Tuberculosis. PLoS ONE 2016, 11, e0162220. [Google Scholar] [CrossRef]
- Coll, R.C.; Schroder, K.; Pelegrín, P. NLRP3 and pyroptosis blockers for treating inflammatory diseases. Trends Pharmacol. Sci. 2022, 43, 653–668. [Google Scholar] [CrossRef]
- Hu, Y.; Zhou, Q.; Liu, T.; Liu, Z. Coixol Suppresses NF-κB, MAPK Pathways and NLRP3 Inflammasome Activation in Lipopolysaccharide-Induced RAW 264.7 Cells. Molecules 2020, 25, 894. [Google Scholar] [CrossRef]
- Campillo-Gimenez, L.; Renaudin, F.; Jalabert, M.; Gras, P.; Gosset, M.; Rey, C.; Sarda, S.; Collet, C.; Cohen-Solal, M.; Combes, C.; et al. Inflammatory Potential of Four Different Phases of Calcium Pyrophosphate Relies on NF-κB Activation and MAPK Pathways. Front. Immunol. 2018, 9, 2248. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Wang, B.; Cao, S.; Wang, Y.; Wu, D. Silybin attenuates LPS-induced lung injury in mice by inhibiting NF-κB signaling and NLRP3 activation. Int. J. Mol. Med. 2017, 39, 1111–1118. [Google Scholar] [CrossRef] [PubMed]
- Cai, J.; Yi, M.; Tan, Y.; Li, X.; Li, G.; Zeng, Z.; Xiong, W.; Xiang, B. Natural product triptolide induces GSDME-mediated pyroptosis in head and neck cancer through suppressing mitochondrial hexokinase-II. J. Exp. Clin. Cancer Res. 2021, 40, 190. [Google Scholar] [CrossRef]
- Li, S.; Yan, D.; Huang, C.; Yang, F.; Cao, Y. TiO2 nanosheets promote the transformation of vascular smooth muscle cells into foam cells in vitro and in vivo through the up-regulation of nuclear factor kappa B subunit 2. J. Hazard Mater. 2022, 424, 127704. [Google Scholar] [CrossRef]
- Kang, D.W.; Lee, J.Y.; Oh, D.H.; Park, S.Y.; Woo, T.M.; Kim, M.K.; Park, M.H.; Jang, Y.H.; Min, D.S. Triptolide-induced suppression of phospholipase D expression inhibits proliferation of MDA-MB-231 breast cancer cells. Exp. Mol. Med. 2009, 41, 678–685. [Google Scholar] [CrossRef] [PubMed]
- Liebenberg, D.; Gordhan, B.G.; Kana, B.D. Drug resistant tuberculosis: Implications for transmission, diagnosis, and disease management. Front. Cell Infect. Microbiol. 2022, 12, 943545. [Google Scholar] [CrossRef] [PubMed]
- Joosten, S.A.; van Meijgaarden, K.E.; Arend, S.M.; Prins, C.; Oftung, F.; Korsvold, G.E.; Kik, S.V.; Arts, R.J.; van Crevel, R.; Netea, M.G.; et al. Mycobacterial growth inhibition is associated with trained innate immunity. J. Clin. Investig. 2018, 128, 1837–1851. [Google Scholar] [CrossRef]
- Ning, B.; Shen, J.; Liu, F.; Zhang, H.; Jiang, X. Baicalein Suppresses NLRP3 and AIM2 Inflammasome-Mediated Pyroptosis in Macrophages Infected by Mycobacterium tuberculosis via Induced Autophagy. Microbiol. Spectr. 2023, 11, e0471122. [Google Scholar] [CrossRef]
- Feng, Y.; Li, M.; Yangzhong, X.; Zhang, X.; Zu, A.; Hou, Y.; Li, L.; Sun, S. Pyroptosis in inflammation-related respiratory disease. J. Physiol. Biochem. 2022, 78, 721–737. [Google Scholar] [CrossRef]
- Chai, Q.; Yu, S.; Zhong, Y.; Lu, Z.; Qiu, C.; Yu, Y.; Zhang, X.; Zhang, Y.; Lei, Z.; Qiang, L.; et al. A bacterial phospholipid phosphatase inhibits host pyroptosis by hijacking ubiquitin. Science 2022, 378, eabq0132. [Google Scholar] [CrossRef]
- Hou, S.; Jiao, Y.; Yuan, Q.; Zhai, J.; Tian, T.; Sun, K.; Chen, Z.; Wu, Z.; Zhang, J. S100A4 protects mice from high-fat diet-induced obesity and inflammation. Lab. Investig. 2018, 98, 1025–1038. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Hou, S.; Gu, J.; Tian, T.; Yuan, Q.; Jia, J.; Qin, Z.; Chen, Z. S100A4 promotes colon inflammation and colitis-associated colon tumorigenesis. Oncoimmunology 2018, 7, e1461301. [Google Scholar] [CrossRef] [PubMed]
- Hou, S.; Tian, T.; Qi, D.; Sun, K.; Yuan, Q.; Wang, Z.; Qin, Z.; Wu, Z.; Chen, Z.; Zhang, J. S100A4 promotes lung tumor development through β-catenin pathway-mediated autophagy inhibition. Cell Death Dis. 2018, 9, 277. [Google Scholar] [CrossRef] [PubMed]
- Milani, M.; Mammarella, E.; Rossi, S.; Miele, C.; Lattante, S.; Sabatelli, M.; Cozzolino, M.; D’Ambrosi, N.; Apolloni, S. Targeting S100A4 with niclosamide attenuates inflammatory and profibrotic pathways in models of amyotrophic lateral sclerosis. J. Neuroinflamm. 2021, 18, 132. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Qu, D.; Liang, Y.; Huang, Q.; Li, M.; Hou, C. Elevated S100A4 in asthmatics and an allergen-induced mouse asthma model. J. Cell. Biochem. 2019, 120, 9667–9676. [Google Scholar] [CrossRef]
- Wang, C.; Ma, H.; Zhang, B.; Hua, T.; Wang, H.; Wang, L.; Han, L.; Li, Q.; Wu, W.; Sun, Y.; et al. Inhibition of IL1R1 or CASP4 attenuates spinal cord injury through ameliorating NLRP3 inflammasome-induced pyroptosis. Front. Immunol. 2022, 13, 963582. [Google Scholar] [CrossRef]
- Luo, B.; Li, B.; Wang, W.; Liu, X.; Xia, Y.; Zhang, C.; Zhang, M.; Zhang, Y.; An, F. NLRP3 gene silencing ameliorates diabetic cardiomyopathy in a type 2 diabetes rat model. PLoS ONE 2014, 9, e104771. [Google Scholar] [CrossRef]
- Wang, L.; Yan, H.; Chen, X.; Han, L.; Liu, G.; Yang, H.; Lu, D.; Liu, W.; Che, C. Thymol Ameliorates Aspergillus fumigatus Keratitis by Downregulating the TLR4/MyD88/NF-kB/IL-1β Signal Expression and Reducing Necroptosis and Pyroptosis. J. Microbiol. Biotechnol. 2023, 33, 43–50. [Google Scholar] [CrossRef]
- Pu, W.; Zhao, C.; Wazir, J.; Su, Z.; Niu, M.; Song, S.; Wei, L.; Li, L.; Zhang, X.; Shi, X.; et al. Comparative transcriptomic analysis of THP-1-derived macrophages infected with Mycobacterium tuberculosis H37Rv, H37Ra and BCG. J. Cell. Mol. Med. 2021, 25, 10504–10520. [Google Scholar] [CrossRef]


| Item | Content |
|---|---|
| Title | The transcriptional signature of active tuberculosis reflects symptom status in extrapulmonary and pulmonary tuberculosis |
| Organization name | National Institute for Medical Research (NIMR) |
| Status | Public on 4 November 2016 |
| Organism | Homo sapiens |
| Sample group | 61 healthy human controls, 47 humans with EPTB, 45 humans with PTB |
| Sample type | Peripheral blood mRNA transcriptome |
| Platforms | GPL10558 Illumina HumanHT-12 V4.0 expression bead chip |
| Gene Name | siRNA Sequences of S100A4 |
|---|---|
| Nontarget control (si-NC) | Sense: 5′-UUCUCCGAACGUGUCACGUTT-3′ Anti-sense:3′-ACGUGACACGUUCGGAGAATT-5′ |
| si-S100A4 | Sense: 5′-UCCAGAAGCUGAUGAGCAATT-3′ Anti-sense:3′-UUGCUCAUCAGCUUCUGGATT-5′ |
| Gene | Primer Sequence (5′-3′) | Tm (°C) | Product Size (bp) |
|---|---|---|---|
| β-actin | F: CCTGGCACCCAGCACAAT | 60 | 144 |
| R: GGGCCGGACTCGTCATAC | 59 | ||
| S100A4 | F: CGGGCAAAGAGGGTGACAAGTTC | 64 | 145 |
| R: TTGTCCCTGTTGCTGTCCAAGTTG | 64 | 145 | |
| GSDMD | F: TGGACCCTAACACCTGGCAGAC | 64 | 117 |
| R: GCACCTCAGTCACCACGTACAC | 63 | 117 | |
| NLRP3 | F: CTCGGTGACTTCGGAATCAGACTTC | 63 | 144 |
| R: CAGGGAATGGCTGGTGCTCAATAC | 64 | 144 | |
| ASC | F: TGGATGCTCTGTACGGGAAGGTC | 64 | 131 |
| R: CAAGTCCTTGCAGGTCCAGTTCC | 63 | 131 | |
| Caspase-1 | F: GGTGCTGAACAAGGAAGAGATGGAG | 63 | 104 |
| R: CCTGTGCCCCTTTCGGAATAACG | 64 | 104 | |
| IL-18 | F: TGACCAAGGAAATCGGCCTC | 60 | 116 |
| R: CCATACCTCTAGGCTGGCTATCT | 60 | 116 | |
| IL-1β | F: TACGAATCTCCGACCACCACTACAG | 64 | 137 |
| R: GGGAAAGAAGGTGCTCAGGTCATTC | 64 | 137 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, M.; Liu, Y.; Nie, X.; Ma, B.; Ma, Y.; Hou, Y.; Yang, Y.; Xu, J.; Wang, Y. S100A4 Promotes BCG-Induced Pyroptosis of Macrophages by Activating the NF-κB/NLRP3 Inflammasome Signaling Pathway. Int. J. Mol. Sci. 2023, 24, 12709. https://doi.org/10.3390/ijms241612709
Li M, Liu Y, Nie X, Ma B, Ma Y, Hou Y, Yang Y, Xu J, Wang Y. S100A4 Promotes BCG-Induced Pyroptosis of Macrophages by Activating the NF-κB/NLRP3 Inflammasome Signaling Pathway. International Journal of Molecular Sciences. 2023; 24(16):12709. https://doi.org/10.3390/ijms241612709
Chicago/Turabian StyleLi, Mengyuan, Yueyang Liu, Xueyi Nie, Boli Ma, Yabo Ma, Yuxin Hou, Yi Yang, Jinrui Xu, and Yujiong Wang. 2023. "S100A4 Promotes BCG-Induced Pyroptosis of Macrophages by Activating the NF-κB/NLRP3 Inflammasome Signaling Pathway" International Journal of Molecular Sciences 24, no. 16: 12709. https://doi.org/10.3390/ijms241612709
APA StyleLi, M., Liu, Y., Nie, X., Ma, B., Ma, Y., Hou, Y., Yang, Y., Xu, J., & Wang, Y. (2023). S100A4 Promotes BCG-Induced Pyroptosis of Macrophages by Activating the NF-κB/NLRP3 Inflammasome Signaling Pathway. International Journal of Molecular Sciences, 24(16), 12709. https://doi.org/10.3390/ijms241612709






