TRIM14 Overexpression Induces Chemoresistance and Malignant Behaviors of Hepatocellular Carcinoma Cells by Activating the STAT3/HIF-1α Pathway
Abstract
1. Introduction
2. Results
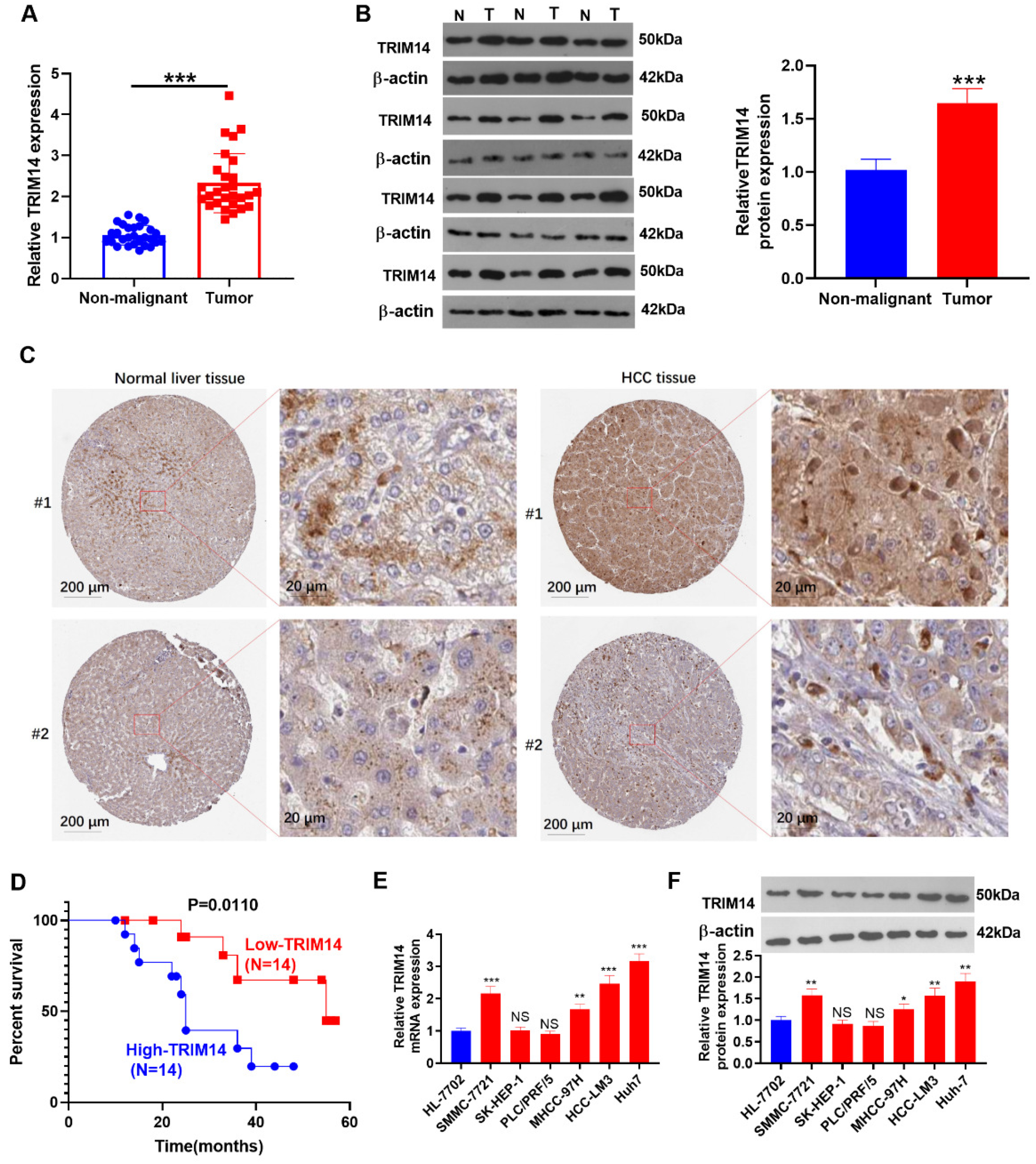
2.1. TRIM14 Upregulation Was Correlated with Poorer Prognosis of HCC Patients
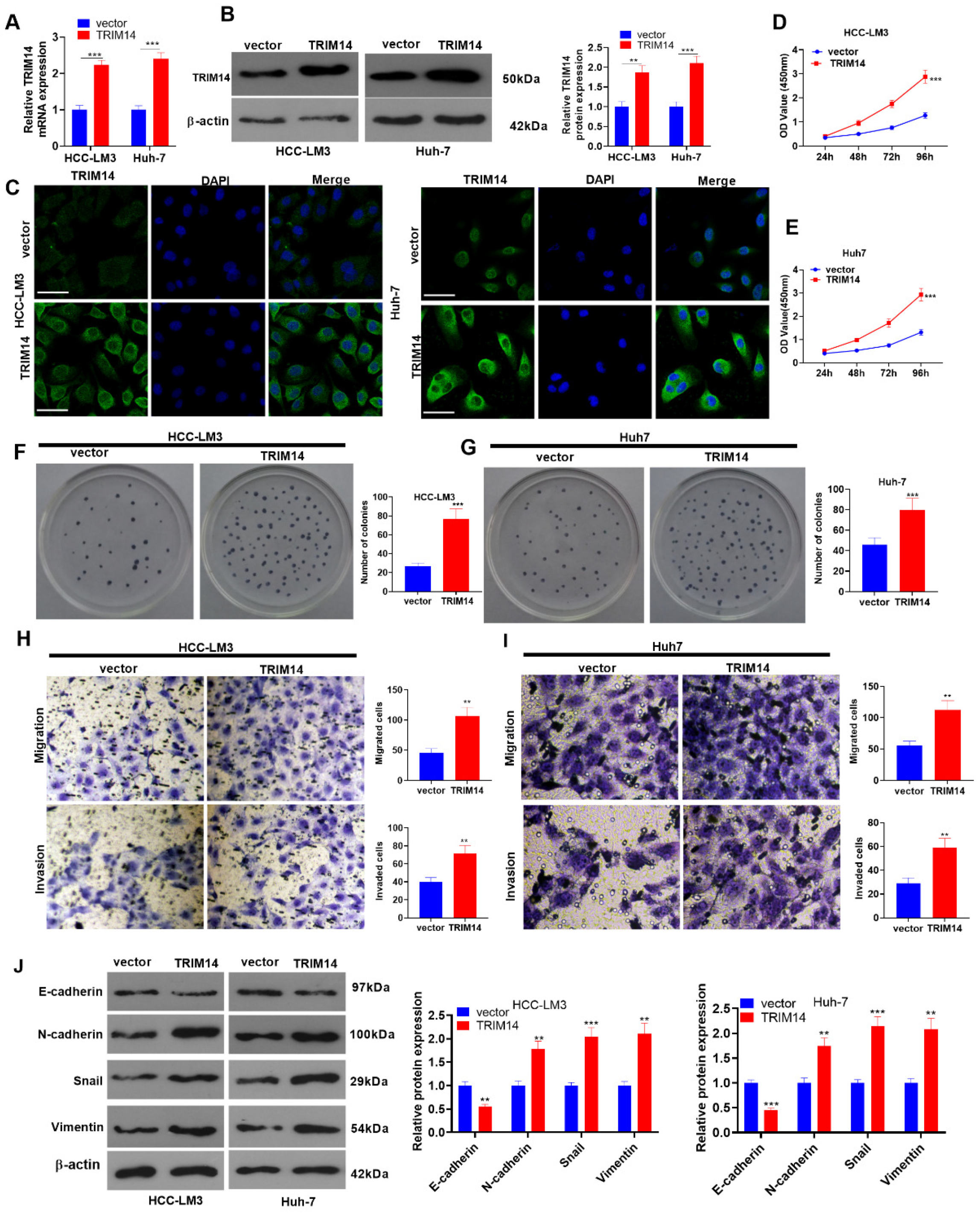
2.2. TRIM14 Overexpression Strengthened HCC Cell Proliferation and Metastasis
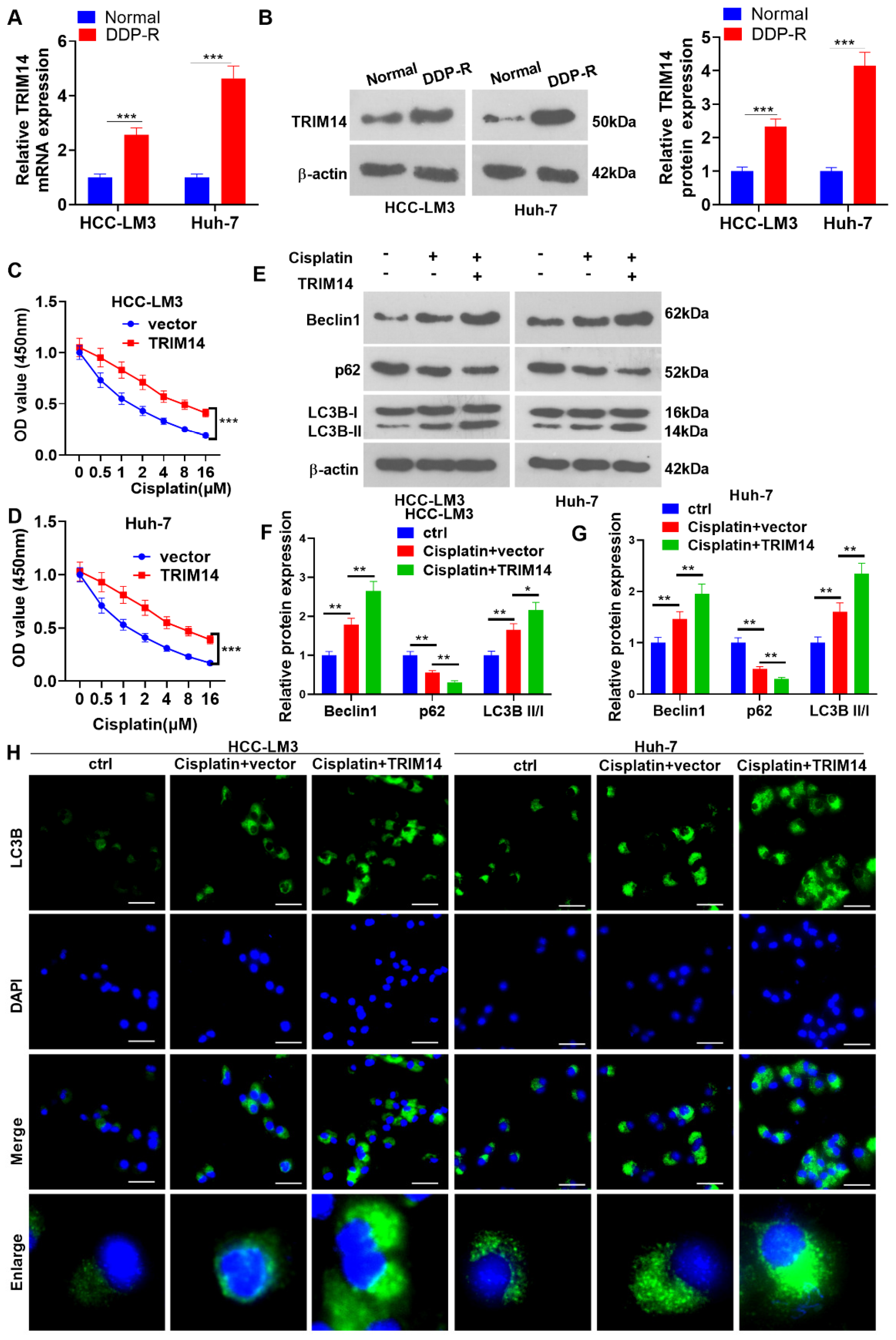
2.3. TRIM14 Overexpression Enhanced HCC Cell Cisplatin Resistance and Autophagy
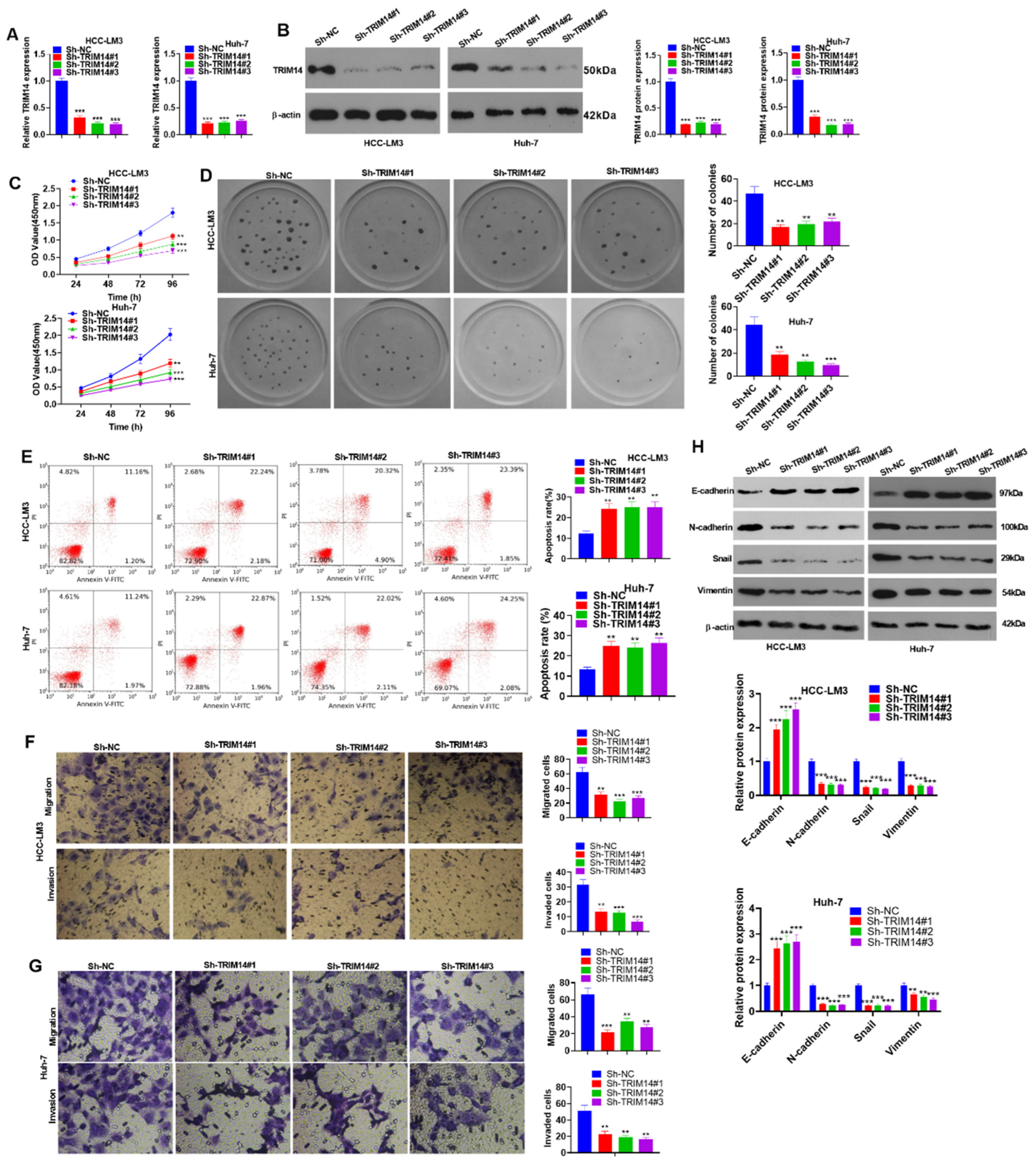
2.4. TRIM14 Downregulation Inhibited HCC Cell Proliferation and Augmented Apoptosis
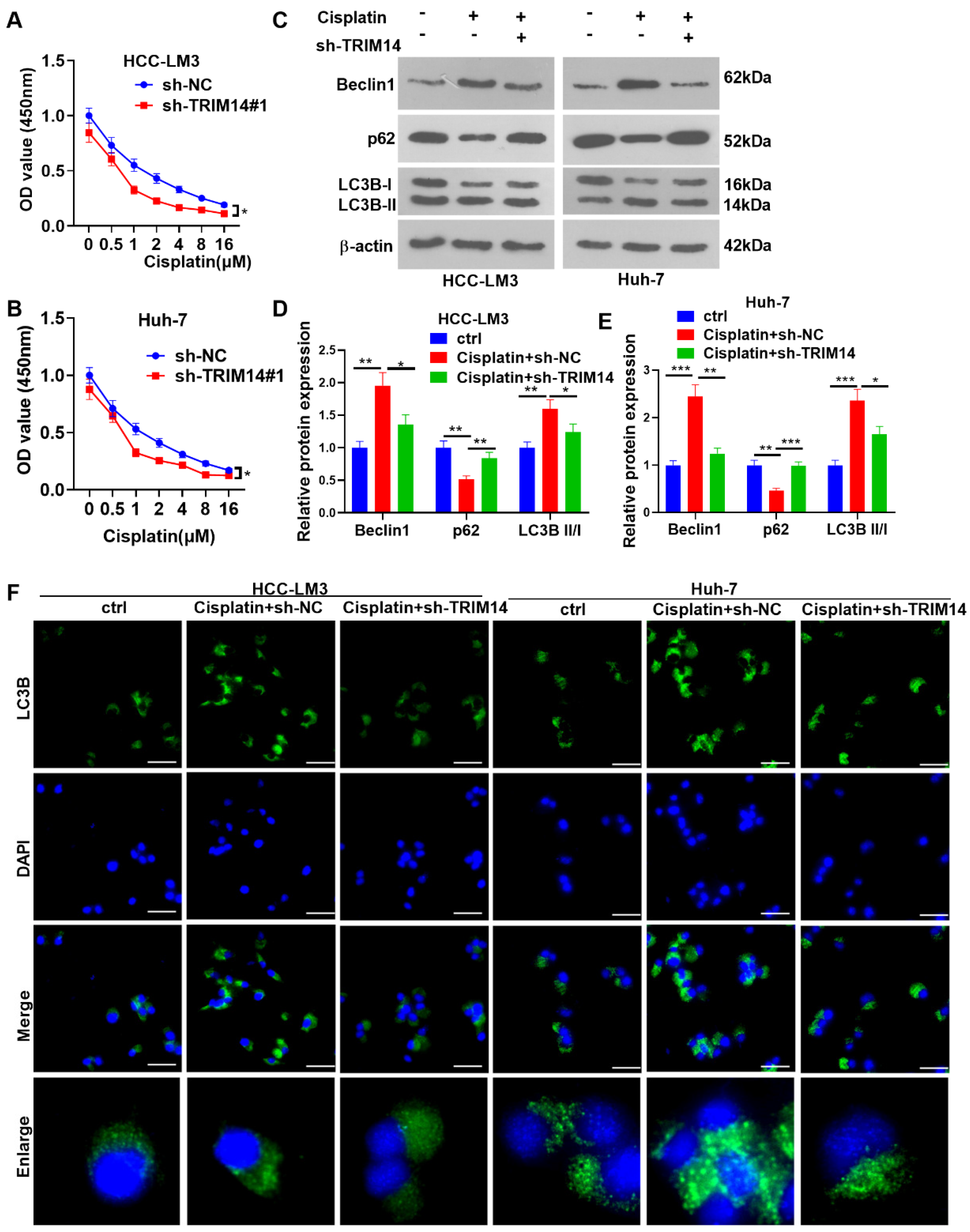
2.5. TRIM14 Inhibition Enhanced HCC Cell Sensitivity to Cisplatin and Reduced Autophagy
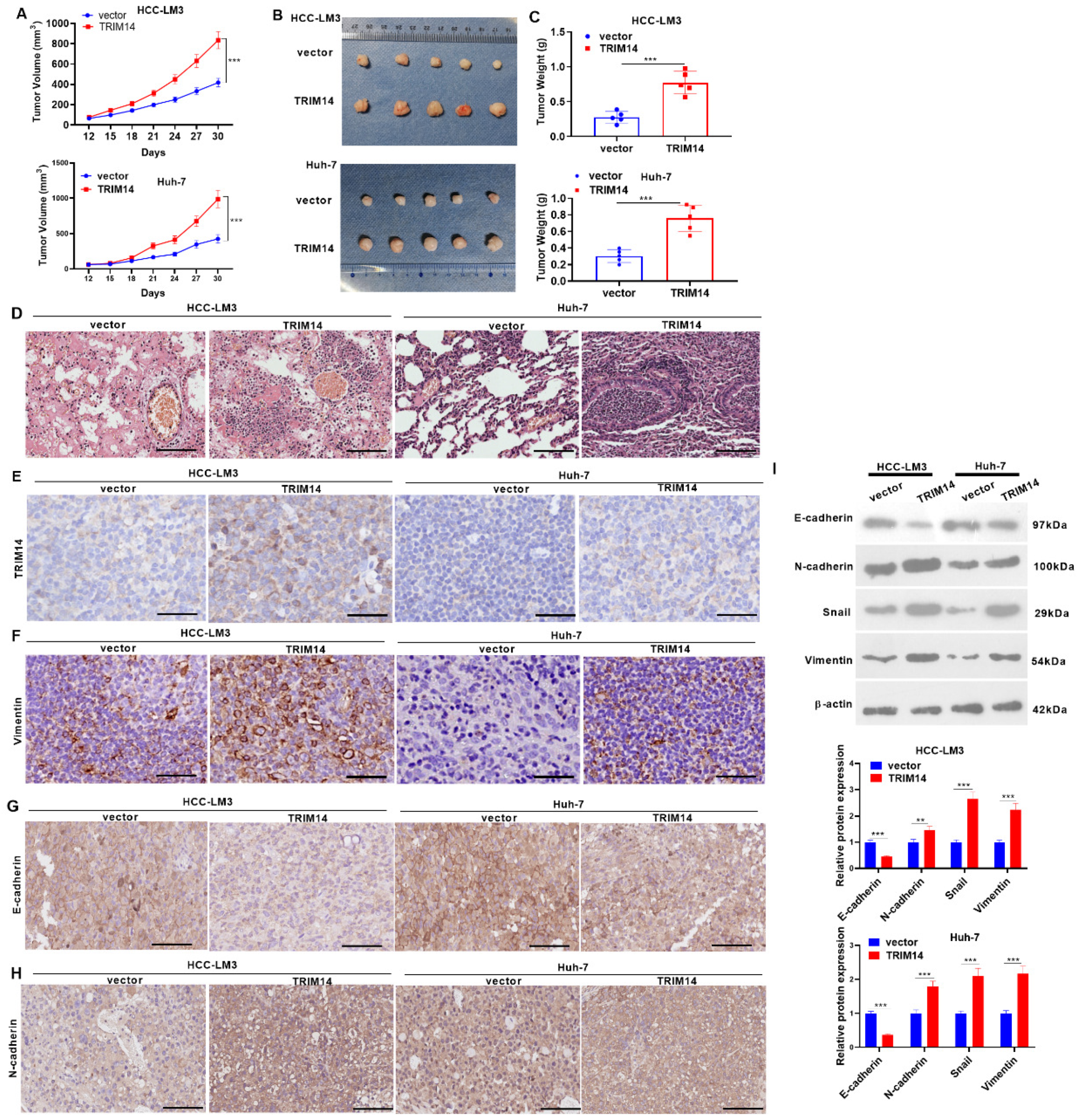
2.6. TRIM14 Overexpression Enhanced Tumor Growth In Vivo
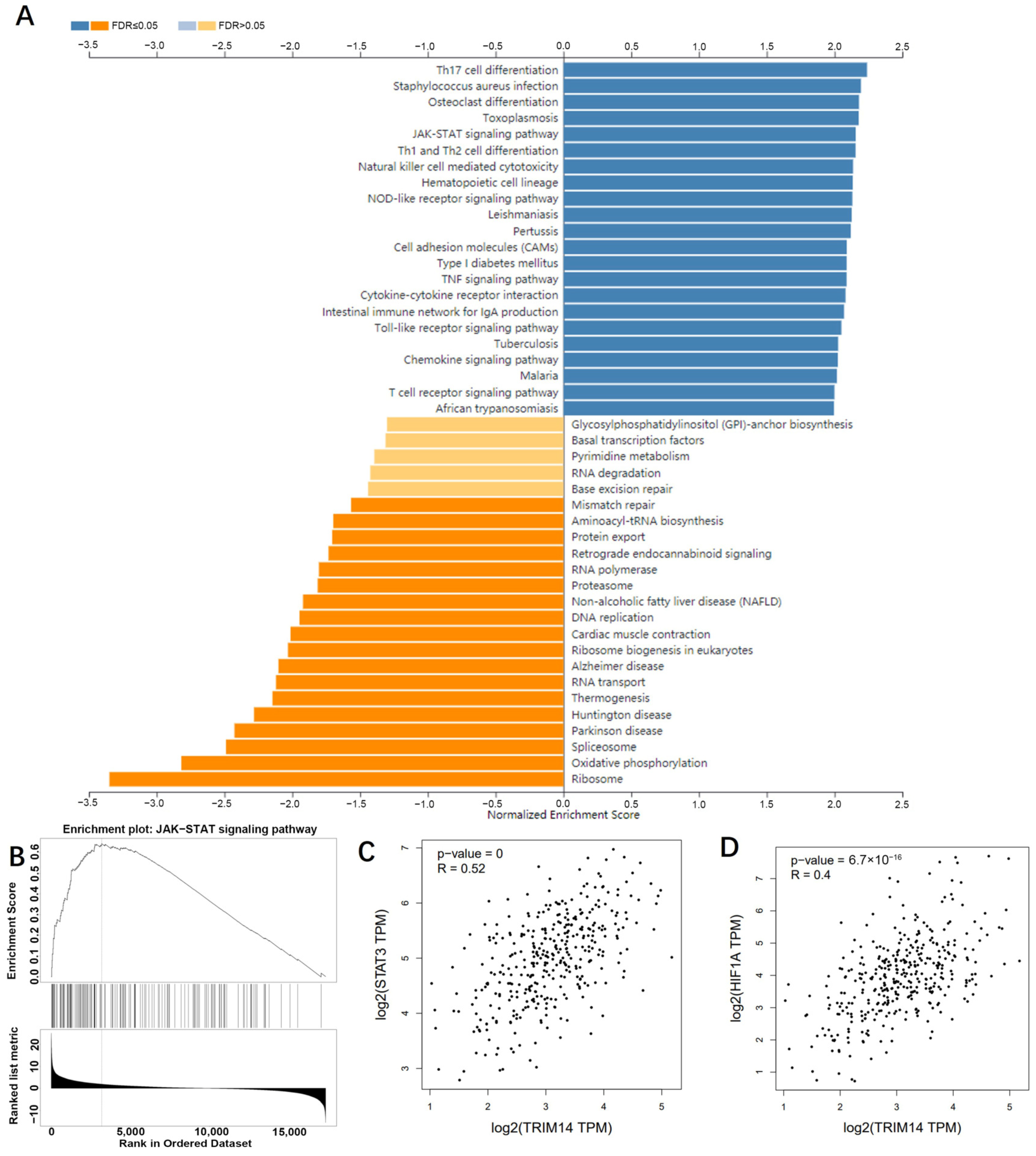
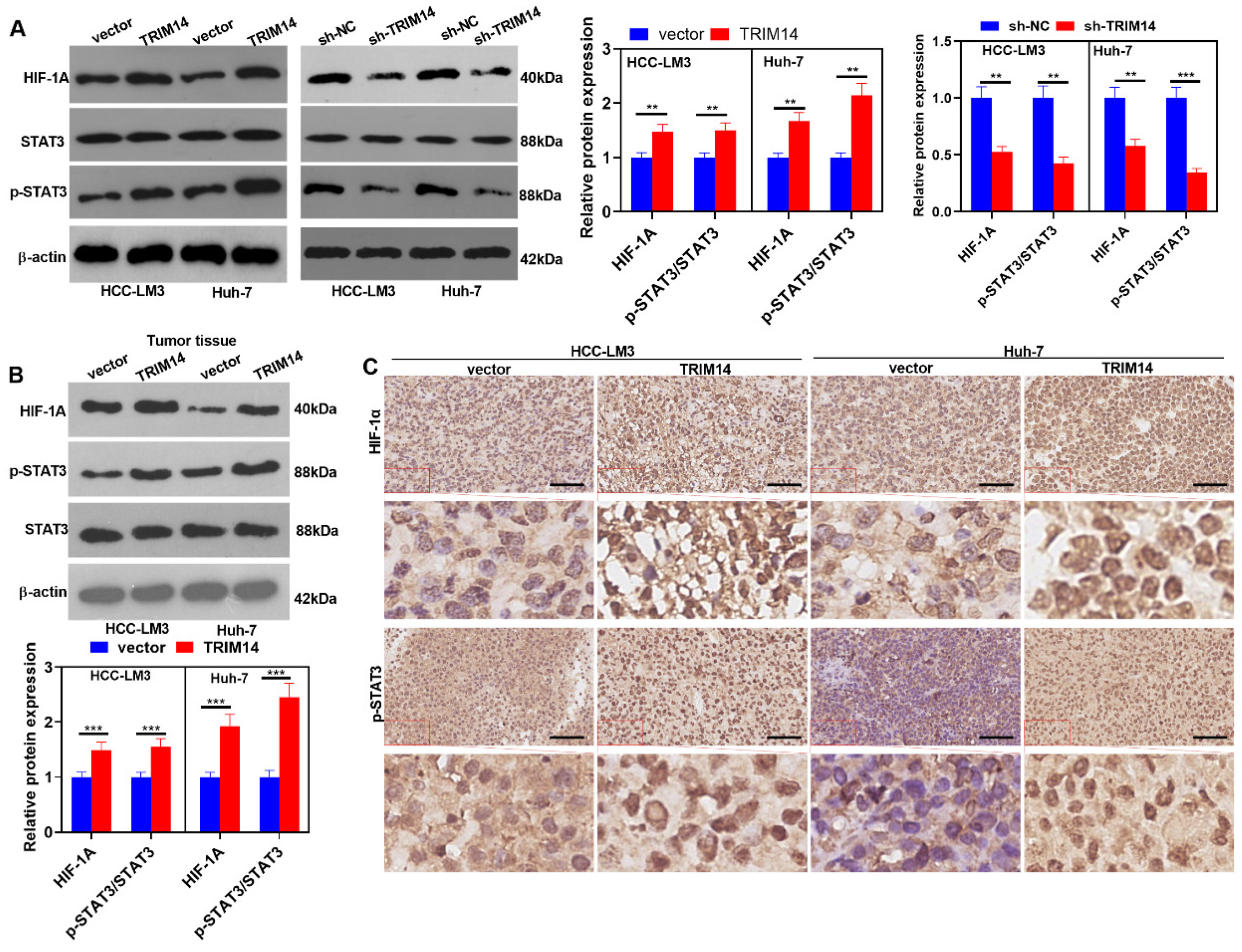
2.7. TRIM14 Overexpression Activated the STAT3/HIF-1α Pathway in HCC Tissues and Cells
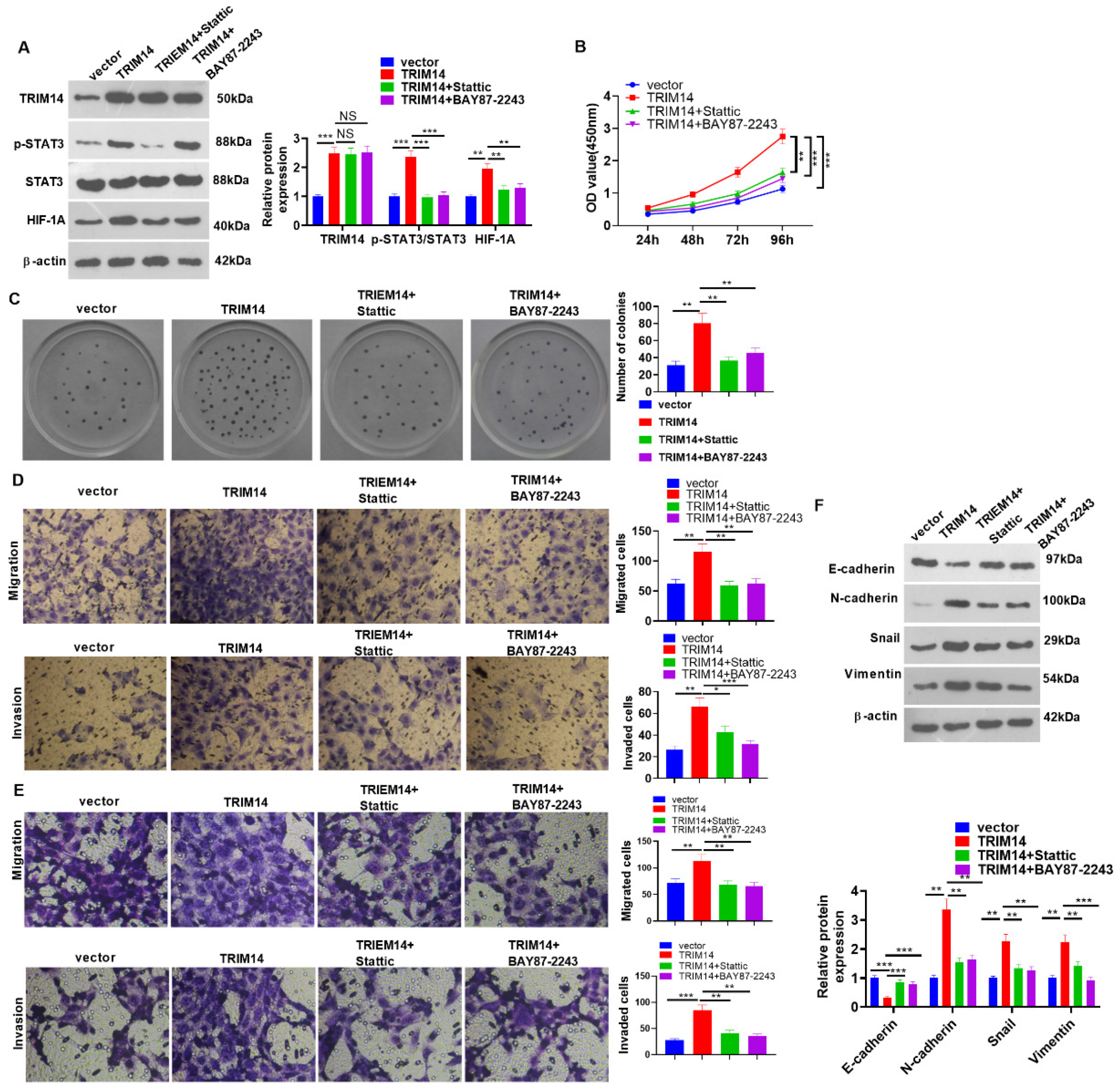
2.8. STAT3 or HIF-1α Inhibition Mitigated TRIM14-Mediated Malignancy
3. Discussion
4. Materials and Methods
4.1. Specimen Collection
4.2. Cell Culture
4.3. Cell Transfection
4.4. Quantitative Reverse Transcription PCR (qRT-PCR)
4.5. Western Blot
4.6. Cell Counting Kit-8 (CCK8) Assay
4.7. Clone Formation Assay
4.8. Transwell Assay
4.9. Flow Cytometry
4.10. Nude Mouse Tumor Formation Model
4.11. Immunohistochemistry
4.12. Immunofluorescence
4.13. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Anwanwan, D.; Singh, S.K.; Singh, S.; Saikam, V.; Singh, R. Challenges in liver cancer and possible treatment approaches. Biochim. Biophys. Acta Rev. Cancer 2020, 1873, 188314. [Google Scholar] [CrossRef] [PubMed]
- Torimura, T.; Iwamoto, H. Treatment and the prognosis of hepatocellular carcinoma in Asia. Liver Int. 2022, 42, 2042–2054. [Google Scholar] [CrossRef]
- Shu, X.-L.; Fan, C.-B.; Long, B.; Zhou, X.; Wang, Y. The anti-cancer effects of cisplatin on hepatic cancer are associated with modulation of miRNA-21 and miRNA-122 expression. Eur. Rev. Med. Pharmacol. Sci. 2016, 20, 4459–4465. [Google Scholar] [PubMed]
- Zheng, S.; Zhou, C.; Wang, Y.; Li, H.; Sun, Y.; Shen, Z. TRIM6 promotes colorectal cancer cells proliferation and response to thiostrepton by TIS21/FoxM1. J. Exp. Clin. Cancer Res. 2020, 39, 23. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.; Hu, J.; Zhang, T.; Jiang, C.; Wang, H.-Y. TRIM29 overexpression is associated with poor prognosis and promotes tumor progression by activating Wnt/β-catenin pathway in cervical cancer. Oncotarget 2016, 7, 28579–28591. [Google Scholar] [CrossRef] [PubMed]
- Hatakeyama, S. TRIM Family Proteins: Roles in Autophagy, Immunity, and Carcinogenesis. Trends Biochem. Sci. 2017, 42, 297–311. [Google Scholar] [CrossRef]
- Su, X.; Wang, J.; Chen, W.; Li, Z.; Fu, X.; Yang, A. Overexpression of TRIM14 promotes tongue squamous cell carcinoma aggressiveness by activating the NF-κB signaling pathway. Oncotarget 2016, 7, 9939–9950. [Google Scholar] [CrossRef]
- Xu, G.; Guo, Y.; Xu, D.; Wang, Y.; Shen, Y.; Wang, F.; Lv, Y.; Song, F.; Jiang, D.; Zhang, Y.; et al. TRIM14 regulates cell proliferation and invasion in osteosarcoma via promotion of the AKT signaling pathway. Sci. Rep. 2017, 7, srep42411. [Google Scholar] [CrossRef]
- Lee, H.; Jeong, A.J.; Ye, S.-K. Highlighted STAT3 as a potential drug target for cancer therapy. BMB Rep. 2019, 52, 415–423. [Google Scholar] [CrossRef]
- Ding, H.; Zhang, X.; Su, Y.; Jia, C.; Dai, C. GNAS promotes inflammation-related hepatocellular carcinoma progression by promoting STAT3 activation. Cell. Mol. Biol. Lett. 2020, 25, 8. [Google Scholar] [CrossRef]
- He, G.; Karin, M. NF-κB and STAT3—Key players in liver inflammation and cancer. Cell Res. 2011, 21, 159–168. [Google Scholar] [CrossRef]
- Movafagh, S.; Crook, S.; Vo, K. Regulation of Hypoxia-Inducible Factor-1a by Reactive Oxygen Species: New Developments in an Old Debate. J. Cell. Biochem. 2015, 116, 696–703. [Google Scholar] [CrossRef]
- Ding, L.; Chen, X.-P.; Wang, H.-P. Expression and clinical significance of HIF-1a protein in hepatocellular carcinoma tissues. Zhonghua Ganzangbing Zazhi Chin. J. Hepatol. 2004, 12, 656–659. [Google Scholar]
- Hamaguchi, Y.; Mori, A.; Fujimoto, Y.; Ito, T.; Iida, T.; Yagi, S.; Okajima, H.; Kaido, T.; Uemoto, S. Longer warm ischemia can accelerate tumor growth through the induction of HIF-1α and the IL-6-JAK-STAT3 signaling pathway in a rat hepatocellular carcinoma model. J. Hepato-Biliary-Pancreat. Sci. 2016, 23, 771–779. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Huang, L.; Quan, J.; Xiang, D. TRIM14 regulates melanoma malignancy via PTEN/PI3K/AKT and STAT3 pathways. Aging 2021, 13, 13225–13238. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Cheng, Y.; Zhang, S.; Fan, J.; Gao, Q. Changing epidemiology of hepatocellular carcinoma in Asia. Liver Int. 2022, 42, 2029–2041. [Google Scholar] [CrossRef]
- Wang, F.; Ruan, L.; Yang, J.; Zhao, Q.; Wei, W. TRIM14 promotes the migration and invasion of gastric cancer by regulating epithelial-to-mesenchymal transition via activation of AKT signaling regulated by miR-195-5p. Oncol. Rep. 2018, 40, 3273–3284. [Google Scholar] [CrossRef]
- Diao, W.; Zhu, C.; Guo, Q.; Cao, Y.; Song, Y.; Feng, H.; Li, J.; Xue, X.; Lu, P. Tripartite motif-containing 14 regulates cell proliferation and apoptosis in cervical cancer via the Akt signaling pathway. Mol. Med. Rep. 2020, 22, 5145–5154. [Google Scholar] [CrossRef]
- Hu, G.; Pen, W.; Wang, M. TRIM14 Promotes Breast Cancer Cell Proliferation by Inhibiting Apoptosis. Oncol. Res. 2019, 27, 439–447. [Google Scholar] [CrossRef]
- Jin, Z.; Li, H.; Hong, X.; Ying, G.; Lu, X.; Zhuang, L.; Wu, S. TRIM14 promotes colorectal cancer cell migration and invasion through the SPHK1/STAT3 pathway. Cancer Cell Int. 2018, 18, 202. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Wang, Y.; Zhang, C.; Yu, S.; Zhu, Q.; Hou, K.; Yan, B. RETRACTED ARTICLE: TRIM25 blockade by RNA interference inhibited migration and invasion of gastric cancer cells through TGF-β signaling. Sci. Rep. 2016, 6, 19070. [Google Scholar] [CrossRef] [PubMed]
- Qiu, F.; Xiong, J.-P.; Deng, J.; Xiang, X.-J. TRIM29 functions as an oncogene in gastric cancer and is regulated by miR-185. Int. J. Clin. Exp. Pathol. 2015, 8, 5053–5061. [Google Scholar] [PubMed]
- Zhang, Y.; Tao, R.; Wu, S.-S.; Xu, C.-C.; Wang, J.-L.; Chen, J.; Yu, Y.-S.; Tang, Z.-H.; Chen, X.-H.; Zang, G.-Q. TRIM52 up-regulation in hepatocellular carcinoma cells promotes proliferation, migration and invasion through the ubiquitination of PPM1A. J. Exp. Clin. Cancer Res. 2018, 37, 116. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Xu, C.; Zhang, X.; Huang, L.; Zheng, C.; Chen, H.; Wang, Y.; Ju, H.; Yao, Q. TRIM11 Upregulation Contributes to Proliferation, Invasion, and EMT of Hepatocellular Carcinoma Cells. Oncol. Res. Featur. Preclin. Clin. Cancer Ther. 2017, 25, 691–699. [Google Scholar] [CrossRef]
- Dong, B.; Zhang, W. High Levels of TRIM14 Are Associated with Poor Prognosis in Hepatocellular Carcinoma. Oncol. Res. Treat. 2018, 41, 129–134. [Google Scholar] [CrossRef]
- Zhu, H.; Chen, K.; Chen, Y.; Liu, J.; Zhang, X.; Zhou, Y.; Liu, Q.; Wang, B.; Chen, T.; Cao, X. RNA-binding protein ZCCHC4 promotes human cancer chemoresistance by disrupting DNA-damage-induced apoptosis. Signal Transduct. Target. Ther. 2022, 7, 240. [Google Scholar] [CrossRef]
- Li, D.; Yao, Y.; Rao, Y.; Huang, X.; Wei, L.; You, Z.; Zheng, G.; Hou, X.; Su, Y.; Varghese, Z.; et al. Cholesterol sensor SCAP contributes to sorafenib resistance by regulating autophagy in hepatocellular carcinoma. J. Exp. Clin. Cancer Res. 2022, 41, 116. [Google Scholar] [CrossRef]
- Zhou, M.; Zhang, G.; Hu, J.; Zhu, Y.; Lan, H.; Shen, X.; Lv, Y.; Huang, L. Rutin attenuates Sorafenib-induced Chemoresistance and Autophagy in Hepatocellular Carcinoma by regulating BANCR/miRNA-590-5P/OLR1 Axis. Int. J. Biol. Sci. 2021, 17, 3595–3607. [Google Scholar] [CrossRef]
- Li, X.; Wu, Q.; Ma, F.; Zhang, X.; Cai, L.; Yang, X. Mitochondrial fission factor promotes cisplatin resistancein hepatocellular carcinoma. Acta Biochim. Biophys. Sin. 2022, 54, 301–310. [Google Scholar] [CrossRef]
- Qu, X.; Gao, H.; Zhai, J.; Sun, J.; Tao, L.; Zhang, Y.; Song, Y.; Hu, T. Astragaloside IV enhances cisplatin chemosensitivity in hepatocellular carcinoma by suppressing MRP2. Eur. J. Pharm. Sci. 2020, 148, 105325. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.; Dong, Z.; Cai, X.; Shen, J.; Xu, Y.; Zhang, M.; Li, H.; Yu, W.; Chen, W. Hypoxia induces sorafenib resistance mediated by autophagy via activating FOXO3a in hepatocellular carcinoma. Cell Death Dis. 2020, 11, 1017. [Google Scholar] [CrossRef]
- Luo, L.; Sun, W.; Zhu, W.; Li, S.; Zhang, W.; Xu, X.; Fang, D.; Grahn, T.H.M.; Jiang, L.; Zheng, Y. BCAT1 decreases the sensitivity of cancer cells to cisplatin by regulating mTOR-mediated autophagy via branched-chain amino acid metabolism. Cell Death Dis. 2021, 12, 169. [Google Scholar] [CrossRef]
- Xiao, F.; Ouyang, B.; Zou, J.; Yang, Y.; Yi, L.; Yan, H. Trim14 promotes autophagy and chemotherapy resistance of gastric cancer cells by regulating AMPK/mTOR pathway. Drug Dev. Res. 2020, 81, 544–550. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Xing, H.; Guo, C.; Yang, Z.; Wang, Y.; Wang, Y. MiR-124 reversed the doxorubicin resistance of breast cancer stem cells through STAT3/HIF-1 signaling pathways. Cell Cycle 2019, 18, 2215–2227. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Liao, S.; Bennett, S.; Tang, H.; Song, D.; Wood, D.; Zhan, X.; Xu, J. STAT3 and its targeting inhibitors in osteosarcoma. Cell Prolif. 2021, 54, e12974. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, W.; Zhuang, L.; Zhu, H.; Mao, A.; Zhou, J.; Wang, L. TRIM14 Overexpression Induces Chemoresistance and Malignant Behaviors of Hepatocellular Carcinoma Cells by Activating the STAT3/HIF-1α Pathway. Int. J. Mol. Sci. 2023, 24, 12589. https://doi.org/10.3390/ijms241612589
Xu W, Zhuang L, Zhu H, Mao A, Zhou J, Wang L. TRIM14 Overexpression Induces Chemoresistance and Malignant Behaviors of Hepatocellular Carcinoma Cells by Activating the STAT3/HIF-1α Pathway. International Journal of Molecular Sciences. 2023; 24(16):12589. https://doi.org/10.3390/ijms241612589
Chicago/Turabian StyleXu, Weiqi, Lihong Zhuang, Hongxu Zhu, Anrong Mao, Jiamin Zhou, and Lu Wang. 2023. "TRIM14 Overexpression Induces Chemoresistance and Malignant Behaviors of Hepatocellular Carcinoma Cells by Activating the STAT3/HIF-1α Pathway" International Journal of Molecular Sciences 24, no. 16: 12589. https://doi.org/10.3390/ijms241612589
APA StyleXu, W., Zhuang, L., Zhu, H., Mao, A., Zhou, J., & Wang, L. (2023). TRIM14 Overexpression Induces Chemoresistance and Malignant Behaviors of Hepatocellular Carcinoma Cells by Activating the STAT3/HIF-1α Pathway. International Journal of Molecular Sciences, 24(16), 12589. https://doi.org/10.3390/ijms241612589




