Novel Aspects of cAMP-Response Element Modulator (CREM) Role in Spermatogenesis and Male Fertility
Abstract
1. Introduction
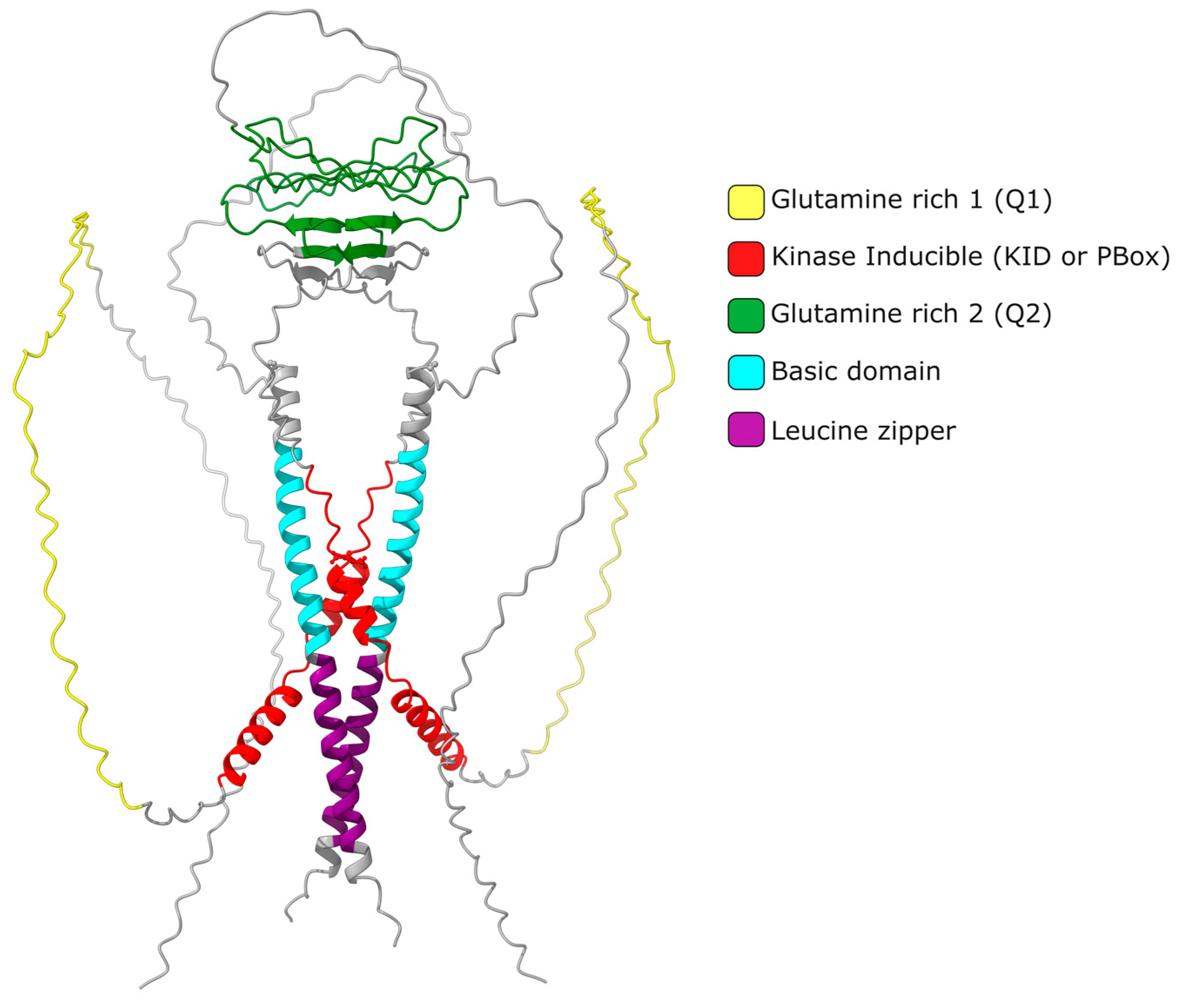
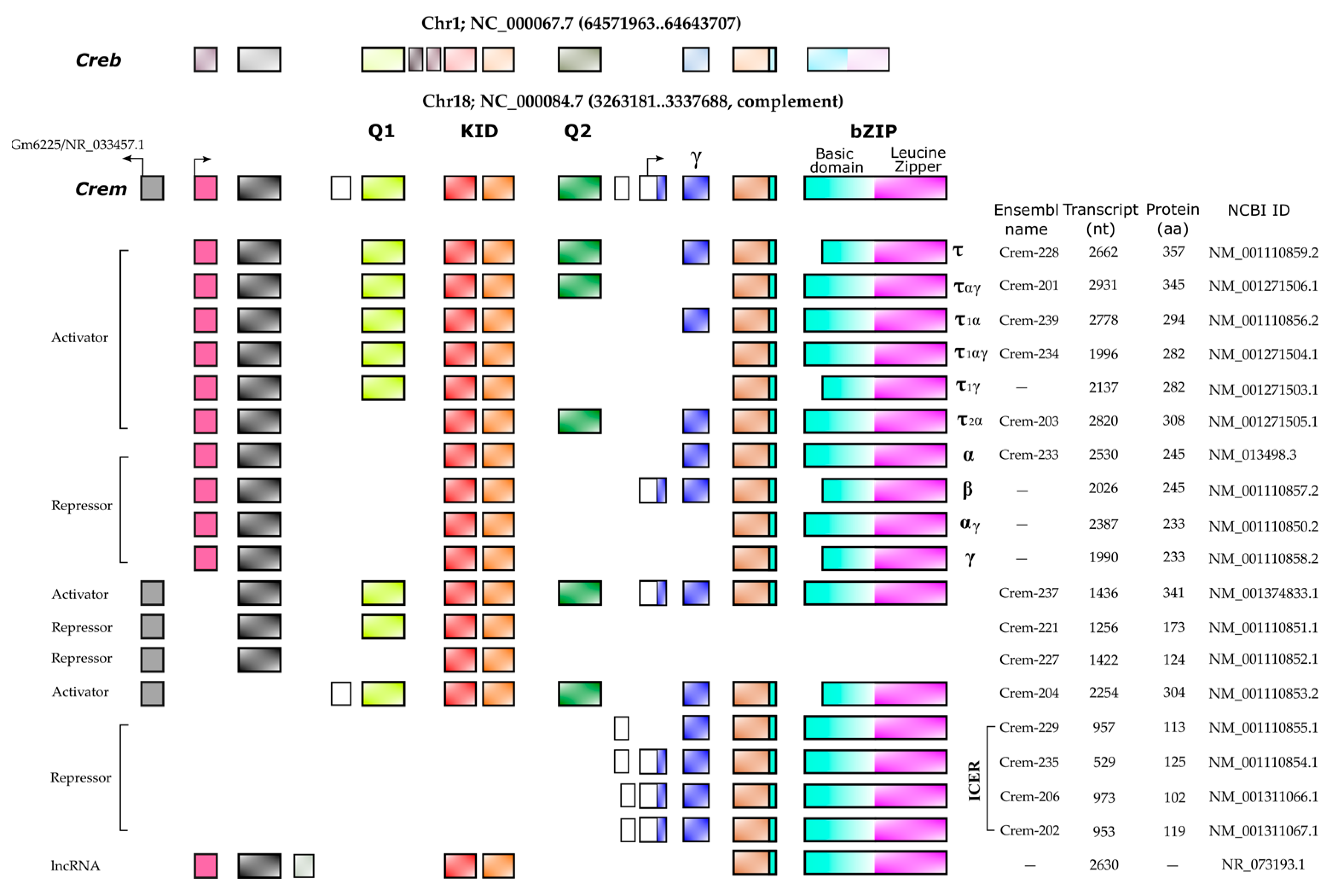
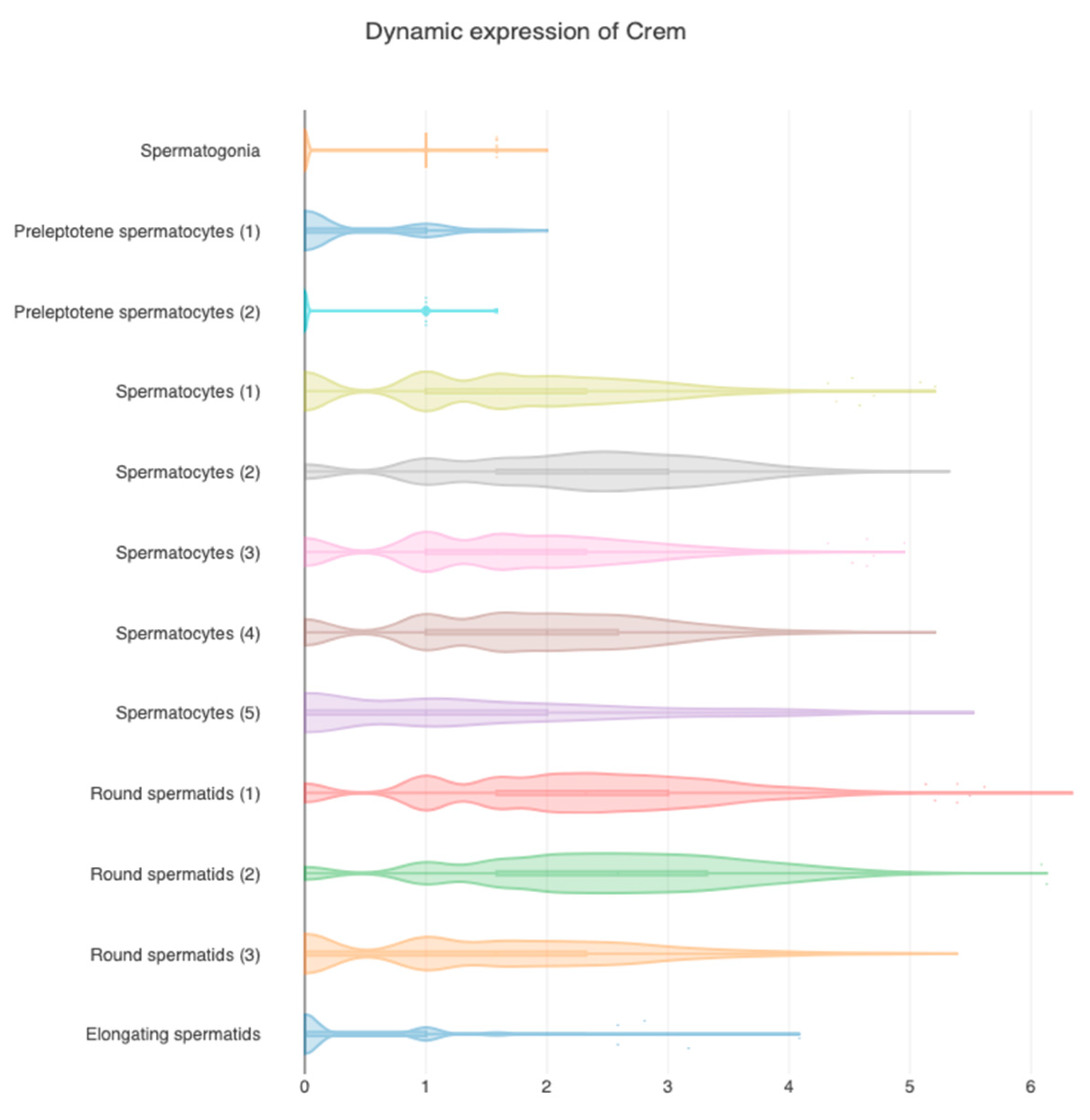
2. CREMτ Expression
3. CREM Regulation
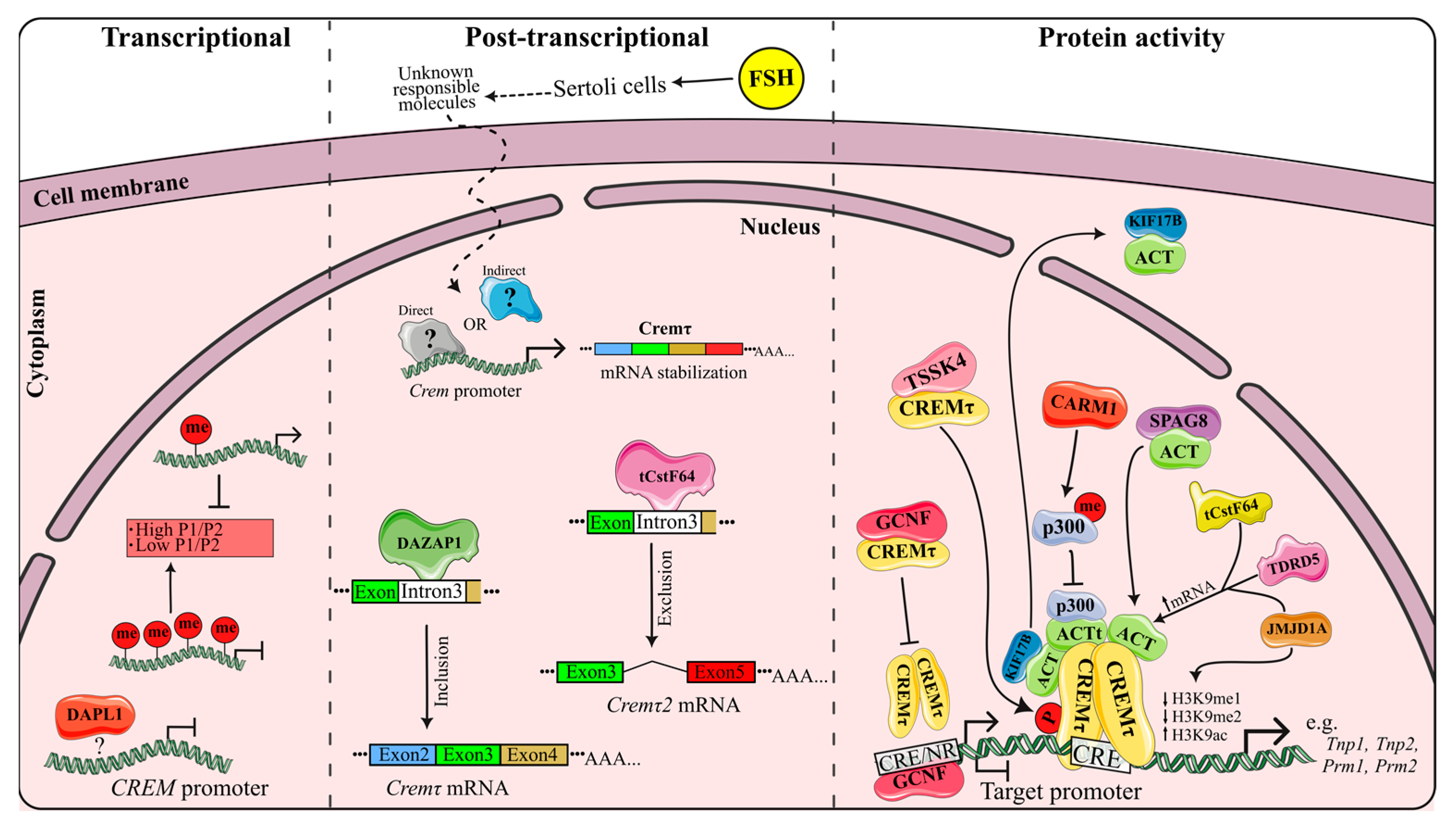
3.1. Transcriptional Regulation
3.2. Post-Transcriptional Regulation
3.3. Regulation of Protein Activity
4. Genes Regulated by CREM
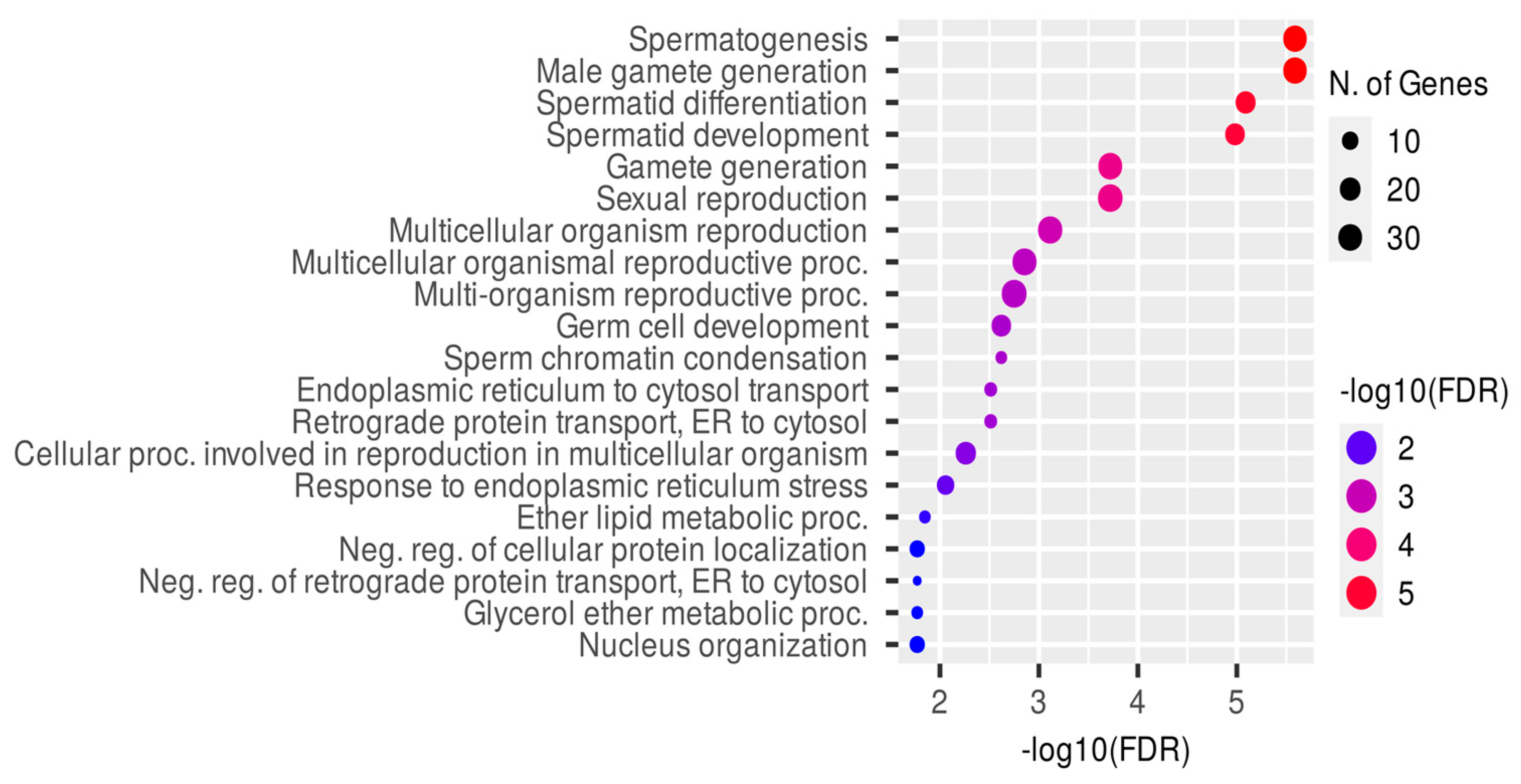
4.1. CREM Binding and Gene Expression Association
| Gene Symbol | Gene ID | Bound by CREM * | Log2FC * | Gene Symbol | Gene ID | Bound by CREM * | Log2FC * |
|---|---|---|---|---|---|---|---|
| Ace | 11421 | Yes | −2.42 | Prm3 | 19120 | No | −4.74 |
| Cdyl | 12593 | Yes | −2.72 | Rxra | 20181 | No | −1.24 |
| Cftr | 12638 | Yes | −1.33 | Spam1 | 20690 | No | −3.09 |
| Crem | 12916 | Yes | −1.71 | Tle3 | 21887 | No | −1.55 |
| Dbil5 | 13168 | Yes | −2.91 | Tssk1 | 22114 | No | −1.56 |
| Hspa1l | 15482 | Yes | −1.81 | Atp1a4 | 27222 | No | −4.83 |
| KIf17 | 16559 | Yes | −2.13 | Prdx4 | 53381 | No | −2.54 |
| Prm1 | 19118 | Yes | −5.53 | Fscn3 | 56223 | No | −4.86 |
| Tnp1 | 21958 | Yes | −4.86 | Tssk3 | 58864 | No | −4.65 |
| Tnp2 | 21959 | Yes | −5.46 | Efcab1 | 66793 | No | −3.57 |
| Tssk2 | 22115 | Yes | −4.15 | Txndc8 | 67402 | No | −4.72 |
| Abcg2 | 26357 | Yes | −1.32 | Rnf151 | 67504 | No | −1.13 |
| Pla2g10 | 26565 | Yes | −2.21 | Lyzl4 | 69032 | No | −2.26 |
| Oaz3 | 53814 | Yes | −5.04 | Lyzl6 | 69444 | No | −5.09 |
| Abhd2 | 54608 | Yes | −1.51 | Tekt5 | 70426 | No | −3.43 |
| Ube2j1 | 56228 | Yes | −1.7 | Tssk5 | 73542 | No | −2.26 |
| Tbc1d20 | 67231 | Yes | −1.46 | 1110017D15Rik | 73721 | No | −2.15 |
| Herc4 | 67345 | Yes | −2.68 | Paqr5 | 74090 | No | −3.12 |
| Spaca1 | 67652 | Yes | −4.29 | Tbc1d21 | 74286 | No | −4.62 |
| Galntl5 | 67909 | Yes | −6.03 | Spem1 | 74288 | No | −5.2 |
| Syce2 | 71846 | Yes | −1.68 | Hyal5 | 74468 | No | −4.84 |
| Calr3 | 73316 | Yes | −3 | Spata19 | 75469 | No | −5.1 |
| Izumo1 | 73456 | Yes | −1.75 | Spata9 | 75571 | No | −2.76 |
| Spata18 | 73472 | Yes | −2.6 | Spaca3 | 75622 | No | −1.45 |
| Iqcf1 | 74267 | Yes | −4.78 | Sun5 | 76407 | No | −5.08 |
| Osbp2 | 74309 | Yes | −2.15 | Catsper3 | 76856 | No | −5.44 |
| Wbp2nl | 74716 | Yes | −3.39 | 4930451I11Rik | 78118 | No | −1.66 |
| Ropn1 | 76378 | Yes | −5.73 | Creb3l4 | 78284 | Yes ** | −3.2 |
| Rhbdd1 | 76867 | Yes | −1.92 | Rdh10 | 98711 | No | −1.75 |
| Hook1 | 77963 | Yes | −1.43 | Tubg1 | 103733 | No | −1.14 |
| Herpud2 | 80517 | Yes | −1.85 | Fam170b | 105511 | No | −2.28 |
| Tssk6 | 83984 | Yes | −3.53 | Arrb1 | 109689 | No | −1.05 |
| Spag4 | 245865 | Yes | −1.85 | Plcz1 | 114875 | No | −4.77 |
| Chd5 | 269610 | Yes | −2.63 | Gm4787 | 214321 | No | −4.36 |
| Ccin | 442829 | Yes | −4.32 | Tppp2 | 219038 | No | −3.66 |
| Adam4 | 11498 | No | −3 | Catsper1 | 225865 | No | −4.38 |
| Akap4 | 11643 | No | −5.49 | Adam6b | 238405 | No | −4.07 |
| Capza3 | 12344 | No | −5.62 | Adam6a | 238406 | No | −3.74 |
| Cast | 12380 | No | −1.75 | Lrrc52 | 240899 | No | −2.97 |
| Cd46 | 17221 | No | −2.31 | Adcy10 | 271639 | No | −1.6 |
| Smcp | 17235 | No | −3.16 | Acsbg2 | 328845 | No | −4.72 |
| Odf1 | 18285 | No | −2.64 | Catsper4 | 329954 | No | −2.07 |
| Pappa | 18491 | No | −1.99 | Ccdc33 | 382077 | No | −2.66 |
| Pgk2 | 18663 | No | −1.64 | Plb1 | 665270 | No | −1.19 |
| Prm2 | 19119 | No | −4.22 | Prrs37 | None | No | −5.55 |
4.2. Confirmed CREM Target Genes
5. Male Fertility and CREM
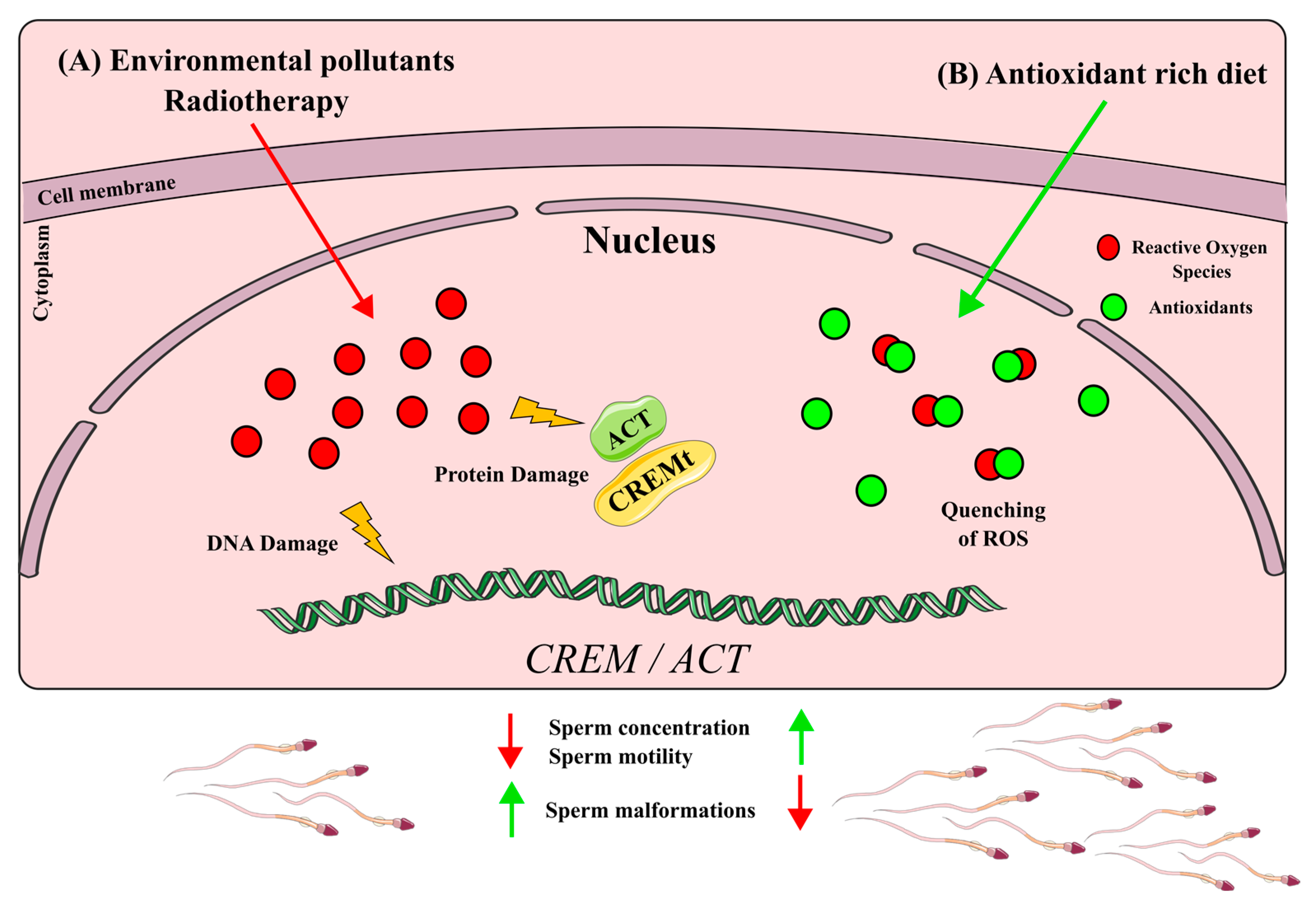
6. Impact of Xenobiotics in CREMτ Regulation
6.1. CREM Disruption by ROS Production Xenobiotics
6.2. CREM Up-Regulation by Antioxidants
7. Other CREM Implications in Health and Disease
8. Conclusions
9. Methods and Searching Strategy
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Kendall, S.K.; Samuelson, L.C.; Saunders, T.L.; Wood, R.I.; Camper, S.A. Targeted disruption of the pituitary glycoprotein hormone alpha-subunit produces hypogonadal and hypothyroid mice. Genes Dev. 1995, 9, 2007–2019. [Google Scholar] [CrossRef]
- De Kretser, D.M.; Loveland, K.L.; Meinhardt, A.; Simorangkir, D.; Wreford, N. Spermatogenesis. Hum. Reprod. 1998, 13 (Suppl. 1), 1–8. [Google Scholar] [CrossRef]
- Griswold, M.D. Cellular and molecular basis for the action of retinoic acid in spermatogenesis. J. Mol. Endocrinol. 2022, 69, T51–T57. [Google Scholar] [CrossRef]
- Schultz, N.; Hamra, F.K.; Garbers, D.L. A multitude of genes expressed solely in meiotic or postmeiotic spermatogenic cells offers a myriad of contraceptive targets. Proc. Natl. Acad. Sci. USA 2003, 100, 12201–12206. [Google Scholar] [CrossRef] [PubMed]
- Shima, J.E.; McLean, D.J.; McCarrey, J.R.; Griswold, M.D. The murine testicular transcriptome: Characterizing gene expression in the testis during the progression of spermatogenesis. Biol. Reprod. 2004, 71, 319–330. [Google Scholar] [CrossRef] [PubMed]
- Foulkes, N.S.; Mellström, B.; Benusiglio, E.; Sassone-Corsi, P. Developmental switch of CREM function during spermatogenesis: From antagonist to activator. Nature 1992, 355, 80–84. [Google Scholar] [CrossRef] [PubMed]
- Don, J.; Stelzer, G. The expanding family of CREB/CREM transcription factors that are involved with spermatogenesis. Mol. Cell. Endocrinol. 2002, 187, 115–124. [Google Scholar] [CrossRef]
- Foulkes, N.S.; Schlotter, F.; Pévet, P.; Sassone-Corsi, P. Pituitary hormone FSH directs the CREM functional switch during spermatogenesis. Nature 1993, 362, 264–267. [Google Scholar] [CrossRef] [PubMed]
- Foulkes, N.S.; Borrelli, E.; Sassone-Corsi, P. CREM gene: Use of alternative DNA-binding domains generates multiple antagonists of cAMP-induced transcription. Cell 1991, 64, 739–749. [Google Scholar] [CrossRef]
- Behr, R.; Weinbauer, G.F. cAMP response element modulator (CREM): An essential factor for spermatogenesis in primates? Int. J. Androl. 2001, 24, 126–135. [Google Scholar] [CrossRef] [PubMed]
- De Cesare, D.; Fimia, G.M.; Sassone-Corsi, P. Signaling routes to CREM and CREB: Plasticity in transcriptional activation. Trends. Biochem. Sci. 1999, 24, 281–285. [Google Scholar] [CrossRef] [PubMed]
- Fimia, G.M.; De Cesare, D.; Sassone-Corsi, P. CBP-independent activation of CREM and CREB by the LIM-only protein ACT. Nature 1999, 398, 165–169. [Google Scholar] [CrossRef]
- Fimia, G.M.; Morlon, A.; Macho, B.; De Cesare, D.; Sassone-Corsi, P. Transcriptional cascades during spermatogenesis: Pivotal role of CREM and ACT. Mol. Cell. Endocrinol. 2001, 179, 17–23. [Google Scholar] [CrossRef] [PubMed]
- Macho, B.; Brancorsini, S.; Fimia, G.M.; Setou, M.; Hirokawa, N.; Sassone-Corsi, P. CREM-dependent transcription in male germ cells controlled by a kinesin. Science 2002, 298, 2388–2390. [Google Scholar] [CrossRef] [PubMed]
- Kotaja, N.; De Cesare, D.; Macho, B.; Monaco, L.; Brancorsini, S.; Goossens, E.; Tournaye, H.; Gansmuller, A.; Sassone-Corsi, P. Abnormal sperm in mice with targeted deletion of the act (activator of cAMP-responsive element modulator in testis) gene. Proc. Natl. Acad. Sci. USA 2004, 101, 10620–10625. [Google Scholar] [CrossRef] [PubMed]
- Hogeveen, K.N.; Sassone-Corsi, P. Regulation of gene expression in post-meiotic male germ cells: CREM-signalling pathways and male fertility. Hum. Fertil. 2006, 9, 73–79. [Google Scholar] [CrossRef]
- Monaco, L.; Foulkes, N.S.; Sassone-Corsi, P. Pituitary follicle-stimulating hormone (FSH) induces CREM gene expression in Sertoli cells: Involvement in long-term desensitization of the FSH receptor. Proc. Natl. Acad. Sci. USA 1995, 92, 10673–10677. [Google Scholar] [CrossRef]
- Delmas, V.; Sassone-Corsi, P. The key role of CREM in the cAMP signaling pathway in the testis. Mol. Cell. Endocrinol. 1994, 100, 121–124. [Google Scholar] [CrossRef]
- Ruppert, S.; Cole, T.J.; Boshart, M.; Schmid, E.; Schütz, G. Multiple mRNA isoforms of the transcription activator protein CREB: Generation by alternative splicing and specific expression in primary spermatocytes. EMBO J. 1992, 11, 1503–1512. [Google Scholar] [CrossRef]
- Walker, W.H.; Sanborn, B.M.; Habener, J.F. An isoform of transcription factor CREM expressed during spermatogenesis lacks the phosphorylation domain and represses cAMP-induced transcription. Proc. Natl. Acad. Sci. USA 1994, 91, 12423–12427. [Google Scholar] [CrossRef]
- Daniel, P.B.; Rohrbach, L.; Habener, J.F. Novel cyclic adenosine 3′,5′-monophosphate (cAMP) response element modulator theta isoforms expressed by two newly identified cAMP-responsive promoters active in the testis. Endocrinology 2000, 141, 3923–3930. [Google Scholar] [CrossRef] [PubMed]
- Gellersen, B.; Kempf, R.; Sandhowe, R.; Weinbauer, G.F.; Behr, R. Novel leader exons of the cyclic adenosine 3′,5′-monophosphate response element modulator (CREM) gene, transcribed from promoters P3 and P4, are highly testis-specific in primates. Mol. Hum. Reprod. 2002, 8, 965–976. [Google Scholar] [CrossRef]
- Fiszer, D.; Białas, M.; Rozwadowska, N.; Kosicki, W.; Jedrzejczak, P.; Kurpisz, M. Crem activator isoforms in normal and impaired human spermatogenesis analyzed by real time RT-PCR. Arch. Androl. 2007, 53, 257–265. [Google Scholar] [CrossRef] [PubMed]
- Guan, Y.; Liang, G.; Martin, G.B.; Guan, L.L. Functional changes in mRNA expression and alternative pre-mRNA splicing associated with the effects of nutrition on apoptosis and spermatogenesis in the adult testis. BMC Genom. 2017, 18, 64. [Google Scholar] [CrossRef] [PubMed]
- Grozdanov, P.N.; Amatullah, A.; Graber, J.H.; MacDonald, C.C. TauCstF-64 Mediates Correct mRNA Polyadenylation and Splicing of Activator and Repressor Isoforms of the Cyclic AMP-Responsive Element Modulator (CREM) in Mouse Testis. Biol. Reprod. 2016, 94, 34. [Google Scholar] [CrossRef]
- Gene [Internet]. Bethesda (MD): National Library of Medicine (US), National Center for Biotechnology Information; 2004. Available online: https://www.ncbi.nlm.nih.gov/gene/1390 (accessed on 27 June 2023).
- Gene [Internet]. Bethesda (MD): National Library of Medicine (US), National Center for Biotechnology Information; 2004. Available online: https://www.ncbi.nlm.nih.gov/gene/12916 (accessed on 27 June 2023).
- Green, C.D.; Ma, Q.; Manske, G.L.; Shami, A.N.; Zheng, X.; Marini, S.; Moritz, L.; Sultan, C.; Gurczynski, S.J.; Moore, B.B.; et al. A Comprehensive Roadmap of Murine Spermatogenesis Defined by Single-Cell RNA-Seq. Dev. Cell. 2018, 46, 651–667. [Google Scholar] [CrossRef] [PubMed]
- Peri, A.; Krausz, C.; Cioppi, F.; Granchi, S.; Forti, G.; Francavilla, S.; Serio, M. Cyclic adenosine 3′,5′-monophosphate-responsive element modulator gene expression in germ cells of normo- and oligoazoospermic men. J. Clin. Endocrinol. Metab. 1998, 83, 3722–3726. [Google Scholar]
- Noda, T.; Shidara, O.; Harayama, H.; Noda, T.; Shidara, O.; Harayama, H. Detection of the activator cAMP responsive element modulator (CREM) isoform ortholog proteins in porcine spermatids and sperm. Theriogenology 2012, 77, 1360–1368. [Google Scholar] [CrossRef]
- Jazireian, P.; Favaedi, R.; Sadighi Gilani, M.A.; Shahhoseini, M. Dynamic Expression and Chromatin Incorporation of ACT and CREM Transcription Factors in Testis Tissues of Infertile Men. Cell J. 2021, 23, 736–741. [Google Scholar]
- Kaprio, H.; Heuser, V.D.; Orte, K.; Tukiainen, M.; Leivo, I.; Gardberg, M. Expression of Transcription Factor CREM in Human Tissues. J. Histochem. Cytochem. 2021, 69, 495–509. [Google Scholar] [CrossRef]
- Kumar, T.R.; Low, M.J.; Matzuk, M.M. Genetic rescue of follicle-stimulating hormone beta-deficient mice. Endocrinology. 1998, 139, 3289–3295. [Google Scholar] [CrossRef] [PubMed]
- Dierich, A.; Sairam, M.R.; Monaco, L.; Fimia, G.M.; Gansmuller, A.; LeMeur, M.; Sassone-Corsi, P. Impairing follicle-stimulating hormone (FSH) signaling in vivo: Targeted disruption of the FSH receptor leads to aberrant gametogenesis and hormonal imbalance. Proc. Natl. Acad. Sci. USA 1998, 95, 13612–13617. [Google Scholar] [CrossRef] [PubMed]
- Khanehzad, M.; Abbaszadeh, R.; Holakuyee, M.; Modarressi, M.H.; Nourashrafeddin, S.M. FSH regulates RA signaling to commit spermatogonia into differentiation pathway and meiosis. Reprod. Biol. Endocrinol. 2021, 19, 4. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.M.; Li, Z.F.; Yang, W.X.; Tan, F.Q. Follicle-stimulating hormone signaling in Sertoli cells: A licence to the early stages of spermatogenesis. Reprod. Biol. Endocrinol. 2022, 20, 97. [Google Scholar] [CrossRef]
- Chen, H.B.; Pineda, J.C.; Arizono, S.; Takeda, T.; Li, R.S.; Hattori, Y.; Sano, H.; Miyauchi, Y.; Hirota, Y.; Tanaka, Y.; et al. DAPL1 is a novel regulator of testosterone production in Leydig cells of mouse testis. Sci. Rep. 2021, 11, 18532. [Google Scholar] [CrossRef]
- Rotondo, J.C.; Lanzillotti, C.; Mazziotta, C.; Tognon, M.; Martini, F. Epigenetics of Male Infertility: The Role of DNA Methylation. Front. Cell Dev. Biol. 2021, 9, 689624. [Google Scholar] [CrossRef]
- Nanassy, L.; Carrell, D.T. Analysis of the methylation pattern of six gene promoters in sperm of men with abnormal protamination. Asian J. Androl. 2011, 13, 342–346. [Google Scholar] [CrossRef]
- Nanassy, L.; Carrell, D.T. Abnormal methylation of the promoter of CREM is broadly associated with male factor infertility and poor sperm quality but is improved in sperm selected by density gradient centrifugation. Fertil. Steril. 2011, 95, 2310–2314. [Google Scholar] [CrossRef]
- Sang, Y.; Liu, J.; Li, X.; Zhou, G.; Zhang, Y.; Gao, L.; Zhao, Y.; Zhou, X. The effect of SiNPs on DNA methylation of genome in mouse spermatocytes. Environ. Sci. Pollut. Res. Int. 2021, 28, 43684–43697. [Google Scholar] [CrossRef]
- Chen, H.Y.; Yu, Y.H.; Yen, P.H. DAZAP1 regulates the splicing of Crem, Crisp2 and Pot1a transcripts. Nucleic Acids Res. 2013, 41, 9858–9869. [Google Scholar] [CrossRef]
- Liu, Z.; Zhou, S.; Liao, L.; Chen, X.; Meistrich, M.; Xu, J. Jmjd1a demethylase-regulated histone modification is essential for cAMP-response element modulator-regulated gene expression and spermatogenesis. J. Biol. Chem. 2010, 285, 2758–2770. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Lin, G.; Wei, Y.; Hexige, S.; Niu, Y.; Liu, L.; Yang, C.; Yu, L. TSSK5, a novel member of the testis-specific serine/threonine kinase family, phosphorylates CREB at Ser-133, and stimulates the CRE/CREB responsive pathway. Biochem. Biophys. Res. Commun. 2005, 333, 742–749. [Google Scholar] [CrossRef] [PubMed]
- Fu, G.; Wei, Y.; Wang, X.; Yu, L. Phosphorylated testis-specific serine/threonine kinase 4 may phosphorylate Crem at Ser-117. Biosci. Biotechnol. Biochem. 2016, 80, 1088–1094. [Google Scholar] [CrossRef] [PubMed]
- Bao, J.; Rousseaux, S.; Shen, J.; Lin, K.; Lu, Y.; Bedford, M.T. The arginine methyltransferase CARM1 represses p300•ACT•CREMτ activity and is required for spermiogenesis. Nucleic Acids Res. 2018, 46, 4327–4343. [Google Scholar] [CrossRef]
- Wu, H.; Chen, Y.; Miao, S.; Zhang, C.; Zong, S.; Koide, S.S.; Wang, L. Sperm associated antigen 8 (SPAG8), a novel regulator of activator of CREM in testis during spermatogenesis. FEBS Lett. 2010, 584, 2807–2815. [Google Scholar] [CrossRef]
- Kotaja, N.; Macho, B.; Sassone-Corsi, P. Microtubule-independent and protein kinase A-mediated function of kinesin KIF17b controls the intracellular transport of activator of CREM in testis (ACT). J. Biol. Chem. 2005, 280, 31739–31745. [Google Scholar] [CrossRef]
- Yabuta, Y.; Ohta, H.; Abe, T.; Kurimoto, K.; Chuma, S.; Saitou, M. TDRD5 is required for retrotransposon silencing, chromatoid body assembly, and spermiogenesis in mice. J. Cell Biol. 2011, 192, 781–795. [Google Scholar] [CrossRef]
- Rajkovic, M.; Iwen, K.A.; Hofmann, P.J.; Harneit, A.; Weitzel, J.M. Functional cooperation between CREM and GCNF directs gene expression in haploid male germ cells. Nucleic Acids Res. 2010, 38, 2268–2278. [Google Scholar] [CrossRef]
- Lardenois, A.; Chalmel, F.; Demougin, P.; Kotaja, N.; Sassone-Corsi, P.; Primig, M. Fhl5/Act, a CREM-binding transcriptional activator required for normal sperm maturation and morphology, is not essential for testicular gene expression. Reprod. Biol. Endocrinol. 2009, 7, 133. [Google Scholar] [CrossRef]
- Blendy, J.A.; Kaestner, K.H.; Weinbauer, G.F.; Nieschlag, E.; Schütz, G. Severe impairment of spermatogenesis in mice lacking the CREM gene. Nature 1996, 380, 102–105. [Google Scholar] [CrossRef]
- Nantel, F.; Monaco, L.; Foulkes, N.S.; Masquilier, D.; LeMeur, M.; Henriksén, K.; Dierich, A.; Parvinen, M.; Sassone-Corsi, P. Spermiogenesis deficiency and germ-cell apoptosis in CREM-mutant mice. Nature 1996, 380, 159–162. [Google Scholar] [CrossRef] [PubMed]
- Martianov, I.; Choukrallah, M.A.; Krebs, A.; Ye, T.; Legras, S.; Rijkers, E.; Van Ijcken, W.; Jost, B.; Sassone-Corsi, P.; Davidson, I. Cell-specific occupancy of an extended repertoire of CREM and CREB binding loci in male germ cells. BMC Genom. 2010, 11, 530. [Google Scholar] [CrossRef]
- Kosir, R.; Juvan, P.; Perse, M.; Budefeld, T.; Majdic, G.; Fink, M.; Sassone-Corsi, P.; Rozman, D. Novel insights into the downstream pathways and targets controlled by transcription factors CREM in the testis. PLoS ONE 2012, 7, e31798. [Google Scholar] [CrossRef]
- Yoshioka, H.; Geyer, C.B.; Hornecker, J.L.; Patel, K.T.; McCarrey, J.R. In vivo analysis of developmentally and evolutionarily dynamic protein-DNA interactions regulating transcription of the Pgk2 gene during mammalian spermatogenesis. Mol. Cell. Biol. 2007, 27, 7871–7885. [Google Scholar] [CrossRef]
- El-Alfy, M.; Azzi, L.; Lessard, J.; Lavergne, E.; Pelletier, M.; Labrie, C. Stage-specific expression of the Atce1/Tisp40alpha isoform of CREB3L4 in mouse spermatids. J. Androl. 2006, 27, 686–694. [Google Scholar] [CrossRef] [PubMed]
- Nagamori, I.; Yomogida, K.; Adams, P.D.; Sassone-Corsi, P.; Nojima, H. Transcription factors, cAMP-responsive element modulator (CREM) and Tisp40, act in concert in postmeiotic transcriptional regulation. J. Biol. Chem. 2006, 281, 15073–15081. [Google Scholar] [CrossRef]
- Oviedo, N.; Ortiz-Borrayo, L.; Hernández-Sánchez, J.; Jiménez-Badillo, S.E.; Tesoro-Cruz, E.; Moreno-Navor, E.; Aguirre-Alvarado, C.; Bekker-Méndez, V.C. Human CATSPER1 Promoter Is Regulated by CREB1 and CREMτ Transcriptional Factors In Vitro. Arch. Med. Res. 2018, 49, 135–146. [Google Scholar] [CrossRef] [PubMed]
- Contreras-Marciales, A.D.P.; López-Guzmán, S.F.; Benítez-Hess, M.L.; Oviedo, N.; Hernández-Sánchez, J. Characterization of the promoter region of the murine Catsper2 gene. FEBS Open Bio 2022, 12, 2236–2249. [Google Scholar] [CrossRef] [PubMed]
- Tramer, F.; Vetere, A.; Martinelli, M.; Paroni, F.; Marsich, E.; Boitani, C.; Sandri, G.; Panfili, E. cAMP-response element modulator-tau activates a distinct promoter element for the expression of the phospholipid hydroperoxide/sperm nucleus glutathione peroxidase gene. Biochem. J. 2004, 383 Pt 1, 179–185. [Google Scholar] [CrossRef]
- Ghanayem, B.I.; Bai, R.; Kissling, G.E.; Travlos, G.; Hoffler, U. Diet-induced obesity in male mice is associated with reduced fertility and potentiation of acrylamide-induced reproductive toxicity. Biol. Reprod. 2010, 82, 96–104. [Google Scholar] [CrossRef]
- Christensen, G.L.; Wooding, S.P.; Ivanov, I.P.; Atkins, J.F.; Carrell, D.T. Sequencing and haplotype analysis of the activator of CREM in the testis (ACT) gene in populations of fertile and infertile males. Mol. Hum. Reprod. 2006, 12, 257–262. [Google Scholar] [CrossRef] [PubMed]
- He, X.J.; Song, B.; Du, W.D.; Cao, Y.X.; Zhang, Y.; Ruan, J.; Tian, H.; Zhou, F.S.; Zuo, X.B.; Wu, H.; et al. CREM variants rs4934540 and rs2295415 conferred susceptibility to nonobstructive azoospermia risk in the Chinese population. Biol. Reprod. 2014, 91, 52. [Google Scholar] [CrossRef] [PubMed]
- Abofoul-Azab, M.; Lunenfeld, E.; Kleiman, S.; Barda, S.; Hauser, R.; Huleihel, M. Determining the expression levels of CSF-1 and OCT4, CREM-1, and protamine in testicular biopsies of adult Klinefelter patients: Their possible correlation with spermatogenesis. Andrologia 2022, 54, e14558. [Google Scholar] [CrossRef] [PubMed]
- Song, B.; Wang, C.; Chen, Y.; Li, G.; Gao, Y.; Zhu, F.; Wu, H.; Lv, M.; Zhou, P.; Wei, Z.; et al. Sperm DNA integrity status is associated with DNA methylation signatures of imprinted genes and non-imprinted genes. J. Assist. Reprod. Genet. 2021, 38, 2041–2048. [Google Scholar] [CrossRef] [PubMed]
- Dvorakova-Hortova, K.; Sidlova, A.; Ded, L.; Hladovcova, D.; Vieweg, M.; Weidner, W.; Steger, K.; Stopka, P.; Paradowska-Dogan, A. Toxoplasma gondii decreases the reproductive fitness in mice. PLoS ONE 2014, 9, e96770. [Google Scholar] [CrossRef] [PubMed]
- Boekelheide, K. Mechanisms of toxic damage to spermatogenesis. J. Natl. Cancer. Inst. Monogr. 2005, 34, 6–8. [Google Scholar] [CrossRef] [PubMed]
- Levine, H.; Jørgensen, N.; Martino-Andrade, A.; Mendiola, J.; Weksler-Derri, D.; Mindlis, I.; Pinotti, R.; Swan, S.H. Temporal trends in sperm count: A systematic review and meta-regression analysis. Hum. Reprod. Update 2017, 23, 646–659. [Google Scholar] [CrossRef]
- Centola, G.M.; Blanchard, A.; Demick, J.; Li, S.; Eisenberg, M.L. Decline in sperm count and motility in young adult men from 2003 to 2013: Observations from a U.S. Sperm Bank. Andrology 2017, 4, 270–276. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, G.; Zhong, Y.; Huang, M.; Wu, J.; Zheng, J.; Rong, W.; Zeng, L.; Yin, X.; Lu, F.; et al. 1,2-Dichloroethane Induces Reproductive Toxicity Mediated by the CREM/CREB Signaling Pathway in Male NIH Swiss Mice. Toxicol. Sci. 2017, 160, 299–314. [Google Scholar] [CrossRef]
- Wang, C.; Chen, Y.; Manthar, R.K.; Wang, J. Abnormal spermatogenesis following sodium fluoride exposure is associated with the downregulation of CREM and ACT in the mouse testis. Toxicol. Ind. Health 2018, 34, 219–227. [Google Scholar] [CrossRef]
- Liu, J.; Zhang, P.; Zhao, Y.; Zhang, H. Low dose carbendazim disrupts mouse spermatogenesis might Be through estrogen receptor related histone and DNA methylation. Ecotoxicol. Environ. Saf. 2019, 176, 242–249. [Google Scholar] [CrossRef]
- Kim, S.W.; Yoo, S.H.; Lee, H.J.; Kim, K.; Kim, D.R.; Park, S.K.; Chang, M.S. Cistanches herba induces testis cytotoxicity in male mice. Bull. Environ. Contam. Toxicol. 2012, 88, 112–117. [Google Scholar] [CrossRef]
- Nagahori, K.; Qu, N.; Kuramasu, M.; Ogawa, Y.; Kiyoshima, D.; Suyama, K.; Hayashi, S.; Sakabe, K.; Yoshimoto, T.; Itoh, M. Changes in Expression of Specific mRNA Transcripts after Single- or Re-Irradiation in Mouse Testes. Genes 2022, 13, 151. [Google Scholar] [CrossRef] [PubMed]
- Guerriero, G.; Trocchia, S.; Abdel-Gawad, F.K.; Ciarcia, G. Roles of reactive oxygen species in the spermatogenesis regulation. Front. Endocrinol. 2014, 5, 56. [Google Scholar] [CrossRef]
- Lee, S.J.; Sŏn-ju, Y. Korean Folk Medicine; Seoul National University Press: Seoul, Republic of Korea, 1966; p. 75. [Google Scholar]
- Kim, E.J.; Lee, Y.J.; Shin, H.K.; Park, J.H. Induction of apoptosis by the aqueous extract of Rubus coreanum in HT-29 human colon cancer cells. Nutrition 2005, 21, 1141–1148. [Google Scholar] [CrossRef]
- Bensky, D.; Barolet, R. Chinese Herbal Medicine Formulas and Strategies, Vol. 7–8; Eastland Press: Seattle, WA, USA, 1990; pp. 263–264. [Google Scholar]
- Pang, G.C.; Kim, M.S.; Lee, M.W. Hydrolyzable tannins from the fruits of Rubus coreanum. Korean J. Pharmacogn. 1996, 27, 366–370. [Google Scholar]
- Kim, S.W.; Shin, M.H.; Jung, J.H.; Kim, N.D.; Im, K.S. A triterpene glucosyl ester from the roots of Rubus crataegifolius. Arch. Pharmacal Res. 2001, 24, 412–415. [Google Scholar]
- Patel, A.V.; Rojas-Vera, J.; Dacke1, C.G. Therapeutic Constituents and Actions of Rubus Species. Curr. Med. Chem. 2004, 11, 1501–1512. [Google Scholar] [CrossRef] [PubMed]
- Oh, M.S.; Yang, W.M.; Chang, M.S.; Park, W.; Kim, D.R.; Lee, H.K.; Kim, W.N.; Park, S.K. Effects of Rubus coreanus on sperm parameters and cAMP-responsive element modulator (CREM) expression in rat testes. J. Ethnopharmacol. 2007, 114, 463–467. [Google Scholar] [CrossRef]
- Oh, M.S.; Chang, M.S.; Park, W.; Kim, D.; Bae, H.; Huh, Y.; Park, S.K. Yukmijihwang-tang protects against cyclophosphamide-induced reproductive toxicity. Reprod. Toxicol. 2007, 24, 365–370. [Google Scholar] [CrossRef]
- Arafa, M.; Agarwal, A.; Majzoub, A.; Khalafalla, K.; Alsaid, S.; Elbardisi, H. Efficacy of antioxidant supplementation on conventional and advanced sperm function tests in patients with idiopathic male infertility. Fertil. Steril. 2019, 112, e362. [Google Scholar] [CrossRef]
- Al-Maghrebi, M.; Renno, W.M.; Al-Somali, H.F.; Botras, M.S.; Qadhi, I.N. Lutein modulates transcription dysregulation of adhesion molecules and spermatogenesis transcription factors induced by testicular ischemia reperfusion injury: It could be SAFE. Naunyn-Schmiedeberg’s Arch. Pharmacol. 2016, 389, 539–551. [Google Scholar] [CrossRef] [PubMed]
- Ye, N.; Lv, Z.; Huang, Z.; Cheng, Y.; Wei, Q.; Shi, F. Dietary folic acid supplementation improves semen quality and spermatogenesis through altering autophagy and histone methylation in the testis of aged broiler breeder roosters. Theriogenology 2022, 181, 8–15. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, A.; Panner Selvam, M.K.; Samanta, L.; Vij, S.C.; Parekh, N.; Sabanegh, E.; Tadros, N.N.; Arafa, M.; Sharma, R. Effect of Antioxidant Supplementation on the Sperm Proteome of Idiopathic Infertile Men. Antioxidants 2019, 8, 488. [Google Scholar] [CrossRef]
- Mioduszewska, B.; Jaworski, J.; Szklarczyk, A.W.; Klejman, A.; Kaczmarek, L. Inducible cAMP early repressor (ICER)-evoked delayed neuronal death in the organotypic hippocampal culture. J. Neurosci. Res. 2008, 86, 61–70. [Google Scholar] [CrossRef] [PubMed]
- Kojima, N.; Borlikova, G.; Sakamoto, T.; Yamada, K.; Ikeda, T.; Itohara, S.; Niki, H.; Endo, S. Inducible cAMP early repressor acts as a negative regulator for kindling epileptogenesis and long-term fear memory. J. Neurosci. 2008, 28, 6459–6472. [Google Scholar] [CrossRef]
- Porter, B.E.; Lund, I.V.; Varodayan, F.P.; Wallace, R.W.; Blendy, J.A. The role of transcription factors cyclic-AMP responsive element modulator (CREM) and inducible cyclic-AMP early repressor (ICER) in epileptogenesis. Neuroscience 2008, 152, 829–836. [Google Scholar] [CrossRef]
- Hu, Y.; Lund, I.V.; Gravielle, M.C.; Farb, D.H.; Brooks-Kayal, A.R.; Russek, S.J. Surface expression of GABAA receptors is transcriptionally controlled by the interplay of cAMP-response element-binding protein and its binding partner inducible cAMP early repressor. J. Biol. Chem. 2008, 283, 9328–9340. [Google Scholar] [CrossRef]
- Lund, I.V.; Hu, Y.; Raol, Y.H.; Benham, R.S.; Faris, R.; Russek, S.J.; Brooks-Kayal, A.R. BDNF selectively regulates GABAA receptor transcription by activation of the JAK/STAT pathway. Sci. Signal. 2008, 1, ra9. [Google Scholar] [CrossRef]
- Favre, D.; Le Gouill, E.; Fahmi, D.; Verdumo, C.; Chinetti-Gbaguidi, G.; Staels, B.; Caiazzo, R.; Pattou, F.; Lê, K.A.; Tappy, L.; et al. Impaired expression of the inducible cAMP early repressor accounts for sustained adipose CREB activity in obesity. Diabetes 2011, 60, 3169–3174. [Google Scholar] [CrossRef]
- Brajkovic, S.; Marenzoni, R.; Favre, D.; Guérardel, A.; Salvi, R.; Beeler, N.; Froguel, P.; Vollenweider, P.; Waeber, G.; Abderrahmani, A. Evidence for tuning adipocytes ICER levels for obesity care. Adipocyte 2012, 1, 157–160. [Google Scholar] [CrossRef]
- Abderrahmani, A.; Cheviet, S.; Ferdaoussi, M.; Coppola, T.; Waeber, G.; Regazzi, R. ICER induced by hyperglycemia represses the expression of genes essential for insulin exocytosis. EMBO J. 2006, 25, 977–986. [Google Scholar] [CrossRef]
- Favre, D.; Niederhauser, G.; Fahmi, D.; Plaisance, V.; Brajkovic, S.; Beeler, N.; Allagnat, F.; Haefliger, J.A.; Regazzi, R.; Waeber, G.; et al. Role for inducible cAMP early repressor in promoting pancreatic beta cell dysfunction evoked by oxidative stress in human and rat islets. Diabetologia 2011, 54, 2337–2346. [Google Scholar] [CrossRef][Green Version]
- Zmrzljak, U.P.; Korenčič, A.; Košir, R.; Goličnik, M.; Sassone-Corsi, P.; Rozman, D. Inducible cAMP early repressor regulates the Period 1 gene of the hepatic and adrenal clocks. J. Biol. Chem. 2013, 288, 10318–10327. [Google Scholar] [CrossRef]
- Ohtsubo, H.; Ichiki, T.; Miyazaki, R.; Inanaga, K.; Imayama, I.; Hashiguchi, Y.; Sadoshima, J.; Sunagawa, K. Inducible cAMP early repressor inhibits growth of vascular smooth muscle cell. Arter. Thromb. Vasc. Biol. 2007, 27, 1549–1555. [Google Scholar] [CrossRef][Green Version]
- Ahlmann, M.; Varga, G.; Sturm, K.; Lippe, R.; Benedyk, K.; Viemann, D.; Scholzen, T.; Ehrchen, J.; Müller, F.U.; Seidl, M.; et al. The cyclic AMP response element modulator {alpha} suppresses CD86 expression and APC function. J. Immunol. 2009, 182, 4167–4174. [Google Scholar] [CrossRef] [PubMed]
- Hedrich, C.M.; Crispin, J.C.; Rauen, T.; Ioannidis, C.; Apostolidis, S.A.; Lo, M.S.; Kyttaris, V.C.; Tsokos, G.C. cAMP response element modulator α controls IL2 and IL17A expression during CD4 lineage commitment and subset distribution in lupus. Proc. Natl. Acad. Sci. USA 2012, 109, 16606–16611. [Google Scholar] [CrossRef]
- Juang, Y.T.; Rauen, T.; Wang, Y.; Ichinose, K.; Benedyk, K.; Tenbrock, K.; Tsokos, G.C. Transcriptional activation of the cAMP-responsive modulator promoter in human T cells is regulated by protein phosphatase 2A-mediated dephosphorylation of SP-1 and reflects disease activity in patients with systemic lupus erythematosus. J. Biol. Chem. 2011, 286, 1795–1801. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Ding, S.; Zhang, H.; Long, H.; Wu, H.; Zhao, M.; Chan, V.; Lau, C.S.; Lu, Q. Increased Set1 binding at the promoter induces aberrant epigenetic alterations and up-regulates cyclic adenosine 5′-monophosphate response element modulator alpha in systemic lupus erythematosus. Clin. Epigenet. 2016, 8, 126. [Google Scholar] [CrossRef] [PubMed]
- Hedrich, C.M.; Rauenl, T.; Kis-Tothl, K.; Kyttaris, V.C.; Tsokos, G.C. cAMP-responsive element modulator α (CREMα) suppresses IL-17F protein expression in T lymphocytes from patients with systemic lupus erythematosus (SLE). J. Biol. Chem. 2012, 287, 4715–4725. [Google Scholar] [CrossRef]
- Lippe, R.; Ohl, K.; Varga, G.; Rauen, T.; Crispin, J.C.; Juang, Y.T.; Kuerten, S.; Tacke, F.; Wolf, M.; Roebrock, K.; et al. CREMα overexpression decreases IL-2 production, induces a T(H)17 phenotype and accelerates autoimmunity. J. Mol. Cell. Biol. 2012, 4, 121–123. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mirdita, M.; Schütze, K.; Moriwaki, Y.; Heo, L.; Ovchinnikov, S.; Steinegger, M. ColabFold: Making protein folding accessible to all. Nat. Methods 2022, 19, 679–682. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Meng, E.C.; Couch, G.S.; Croll, T.I.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 2021, 30, 70–82. [Google Scholar] [CrossRef] [PubMed]
- Darde, T.A.; Sallou, O.; Becker, E.; Evrard, B.; Monjeaud, C.; Le Bras, Y.; Jégou, B.; Collin, O.; Rolland, A.D.; Chalmel, F. The ReproGenomics Viewer: An integrative cross-species toolbox for the reproductive science community. Nucleic Acids Res. 2015, 43, W109–W116. [Google Scholar] [CrossRef] [PubMed]
- Darde, T.A.; Lecluze, E.; Lardenois, A.; Stévant, I.; Alary, N.; Tüttelmann, F.; Collin, O.; Nef, S.; Jégou, B.; Rolland, A.D.; et al. The ReproGenomics Viewer: A multi-omics and cross-species resource compatible with single-cell studies for the reproductive science community. Bioinformatics 2019, 35, 3133–3139. [Google Scholar] [CrossRef] [PubMed]
- Ge, S.X.; Jung, D.; Yao, R. ShinyGO: A graphical gene-set enrichment tool for animals and plants. Bioinformatics 2020, 36, 2628–2629. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sánchez-Jasso, D.E.; López-Guzmán, S.F.; Bermúdez-Cruz, R.M.; Oviedo, N. Novel Aspects of cAMP-Response Element Modulator (CREM) Role in Spermatogenesis and Male Fertility. Int. J. Mol. Sci. 2023, 24, 12558. https://doi.org/10.3390/ijms241612558
Sánchez-Jasso DE, López-Guzmán SF, Bermúdez-Cruz RM, Oviedo N. Novel Aspects of cAMP-Response Element Modulator (CREM) Role in Spermatogenesis and Male Fertility. International Journal of Molecular Sciences. 2023; 24(16):12558. https://doi.org/10.3390/ijms241612558
Chicago/Turabian StyleSánchez-Jasso, Diego Eduardo, Sergio Federico López-Guzmán, Rosa Maria Bermúdez-Cruz, and Norma Oviedo. 2023. "Novel Aspects of cAMP-Response Element Modulator (CREM) Role in Spermatogenesis and Male Fertility" International Journal of Molecular Sciences 24, no. 16: 12558. https://doi.org/10.3390/ijms241612558
APA StyleSánchez-Jasso, D. E., López-Guzmán, S. F., Bermúdez-Cruz, R. M., & Oviedo, N. (2023). Novel Aspects of cAMP-Response Element Modulator (CREM) Role in Spermatogenesis and Male Fertility. International Journal of Molecular Sciences, 24(16), 12558. https://doi.org/10.3390/ijms241612558






