1. Introduction
The purinergic P2X7 receptor (P2X7R) is a trimeric ligand-gated cation channel that is activated by extracellular adenosine triphosphate (ATP). Receptor activation is accompanied by a rapid Na
+ and Ca
2+ influx into and K
+ efflux out of the cell cytoplasm, as well as the opening of large pores (macropores) to facilitate the passage of organic ions and hydrophilic solutes of MW up to 900 Da. This property of P2X7R distinguishes them from other subtypes of the P2X receptor family and allows the use of a number of fluorescent molecules, including ethidium bromide, YO-PRO-1, or Lucifer yellow, to assess the functioning of the macropore [
1,
2]. The opening of the P2X7R macropore in response to the ATP action triggers some cellular processes, such as the release of cathepsins, the production of reactive oxygen species (ROS), the plasma membrane blebbing, and apoptosis, while prolonged activation of the receptor leads to cell death [
3].
This type of receptor is expressed by different types of cells, including dendritic and mast cells, T and B lymphocytes and their precursors, macrophages, and microglia. Because the P2X7R is highly expressed in immune cells, its signaling is important for modulating both the innate and adaptive immune response and, in particular, for the regulation of inflammation [
3,
4]. Dysfunction or overexpression of P2X7R causes numerous pathological conditions associated with pain and inflammation. It is known that P2X7R activates the assembly of the NLRP3 inflammasome, control the release of pro-inflammatory cytokines and the activation of immune cells, thereby modifying immune and inflammatory responses, which leads to a wide range of diseases [
4].
Currently, the search and development of P2X7R blockers with high selectivity, efficacy, and bioavailability is an urgent pharmacological task. A series of P2X7R blockers with high inhibitory activity has already been synthesized. Several P2X7R antagonist compounds are in various stages of preclinical and clinical trials. However, despite the intensive search for highly selective P2X7R blockers, as well as modulators of other receptors of this family, so far, none of them has been brought to the pharmaceutical market [
1,
5].
1,4-Naphthoquinones (1,4-NQs) are a class of natural and synthetic compounds with a broad variety of biological activities, including analgesic and anti-inflammatory properties [
6]. It has been proven that some synthetic derivatives of the 1,4-naphthoquinoid nucleus are able to effectively bind P2X7 receptor and inhibit its functions in in vitro, in vivo, and in silico models [
7,
8,
9,
10,
11]. As a result, this class of compounds has been considered the scaffold for the development of effective drugs blocking the functions of P2X7R.
This work continues a series of studies of the biological activity of a number of 1,4-naphthoquinones synthesized by us and their ability to influence the functioning of purinergic P2X7 receptors in various cell types. We have already evaluated the effect of these compounds on Neuro-2a neuronal cells [
12] and the effect of some of them on the functioning of P2X7R in macrophage cells [
13]. In this study, we focused on the extended investigation of the synthetic tetracyclic thioglucoside conjugate,
U-556, impact on RAW 264.7 macrophage P2X7R functioning, data for which have not been presented before. Herein, we report that this compound is able to inhibit ATP-induced Ca
2+ influx and YO-PRO-1 uptake into mouse macrophage cells, reduce ROS production, and protect macrophages from the cytotoxic ATP impact. Above all, the novelty of this work is new data on the effect of
U-556 on the functioning of the receptor macropore upon modulation by a specific P2X7R agonist and antagonist. The work expands studies of the cytoprotective effect of naphthoquinone in the toxic effect of high concentrations of ATP. Direct binding of
U-556 to P2X7R was confirmed by surface plasmon resonance (SPR). Finally, the anti-inflammatory activity of
U-556 in in vivo experiments was carried out in a model of carrageenan-induced paw edema in mice.
3. Discussion
Macrophages (or the “big eaters”) are specialized cells in the body of animals and humans capable of actively capturing and digesting bacteria, the remains of dead cells, and other particles that are foreign or toxic to the body. Macrophages are present in virtually every organ/tissue, where they act as the first line of immune defense against pathogens and play an important role in maintaining tissue homeostasis, and are directly involved in the processes of inflammation [
15]. One of the key targets of the signaling pathway leading to inflammation is the P2X7 receptor of macrophages. Activation of P2X7R by high concentrations of ATP under pathological conditions provides a non-selective flow of a number of mono- and divalent cations and some low-molecular chemicals through the formed receptor macropore. This process is coupled in macrophages with LPS-recognizing toll-like receptor 4 (TLR4), which co-activation leads to the formation of inflammasome 3 (NLRP3), followed by activation of (pro-IL-1β)-degrading caspase-1, and release of the inflammatory inducer, the pro-inflammatory cytokine IL-1β [
16].
It has now become clear that a number of inflammatory diseases, such as rheumatoid arthritis, Crohn’s disease, liver fibrosis, sepsis, kidney and lungs inflammation, tumor-associated inflammation, and some other, is largely due to the activation of P2X7R in macrophages. The same pattern is found in the study of the causes of some neurodegenerative disorders related to neuroinflammation, such as Alzheimer’s disease, Parkinson’s disease, Huntington’s disease, amyotrophic lateral sclerosis, multiple sclerosis, postischemic conditions, and epilepsy. The important role of P2X7R in the processes of inflammation has led to an intensive search for highly effective selective blockers of this target. However, despite the fact that a number of pharmaceutical companies have already created a series of promising ligands with pronounced P2X7R inhibitory properties, none of these compounds has successfully passed clinical trials yet [
4,
17].
The anti-inflammatory properties of 1,4-NQs are well known. Some of the natural and synthetic compounds were found to inhibit histamine, IL-1β, IL-6, and TNF-α release, cyclooxygenase-2 (COX-2) activity, and reduce edema in the carrageenan model of inflammation [
6]. Some synthetic 1,4-NQs were observed to effectively block the P2X7R functions in mouse neuronal and macrophage cells [
7,
8,
9,
10,
11,
12,
13]. In this study, we proved that synthetic tetracyclic naphthoquinone-thioglucoside conjugate
U-556, is able to effectively block the functioning of P2X7R in mouse macrophages. This effect manifested itself primarily in the inhibition of ATP-induced Ca
2+ influx into macrophage cells primed with bacterial LPS. The response of macrophages to LPS is associated with the triggering of a cascade of pro-inflammatory reactions, mainly through the TLR4 receptor which stimulates additional ATP release and enhances the P2X7R response [
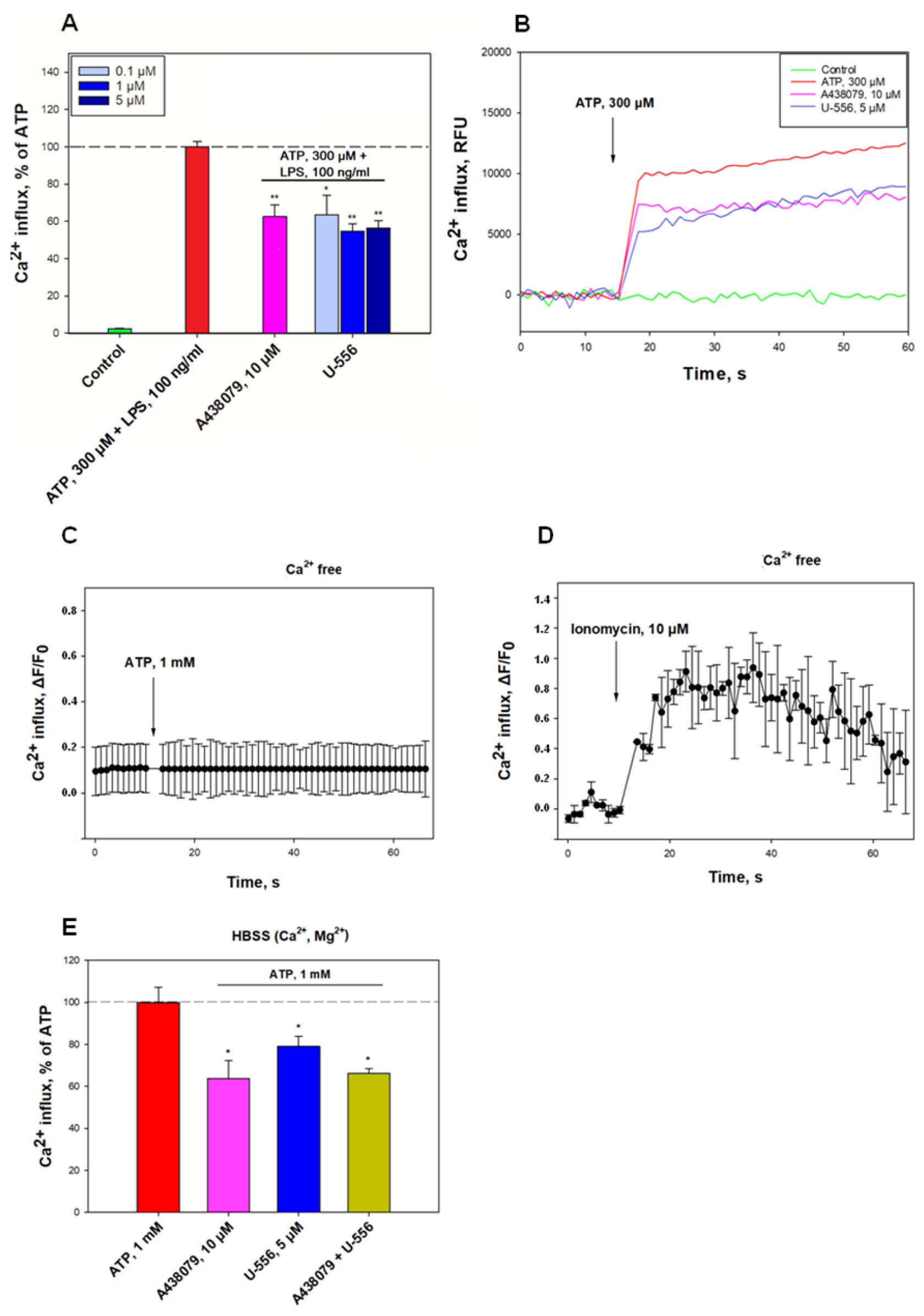
18]. Moreover, such stimulation, in turn, increases the percentage of death cells exposed to ATP. The level of ion transport inhibition for
U-556 was comparable to the selective P2X7R blocker A438079.
To exclude the participation of P2Y receptors also present in macrophages, the ATP-induced changes in intracellular calcium concentration Ca2+ transport was measured in a calcium-free medium. Under these conditions, ATP did not cause an increase in [Ca2+]i. Since metabotropic P2Y receptors are not able to provide calcium current from the extracellular medium through cell membranes, unlike P2X, the participation of P2Y receptors was excluded. Ionomycin is a selective calcium ionophore that transports Ca2+ through biomembranes along a concentration gradient. In a calcium-free medium, ionomycin increases [Ca2+]i in the cytoplasm due to Ca2+ transport from intracellular depots. In our study, the use of the Ca-selective ionophore ionomycin in Ca2+-free media resulted in a marked elevation of [Ca2+]i. This suggests that intracellular calcium stores remain full after ATP application and are not involved in the Ca-response of cells to the ATP.
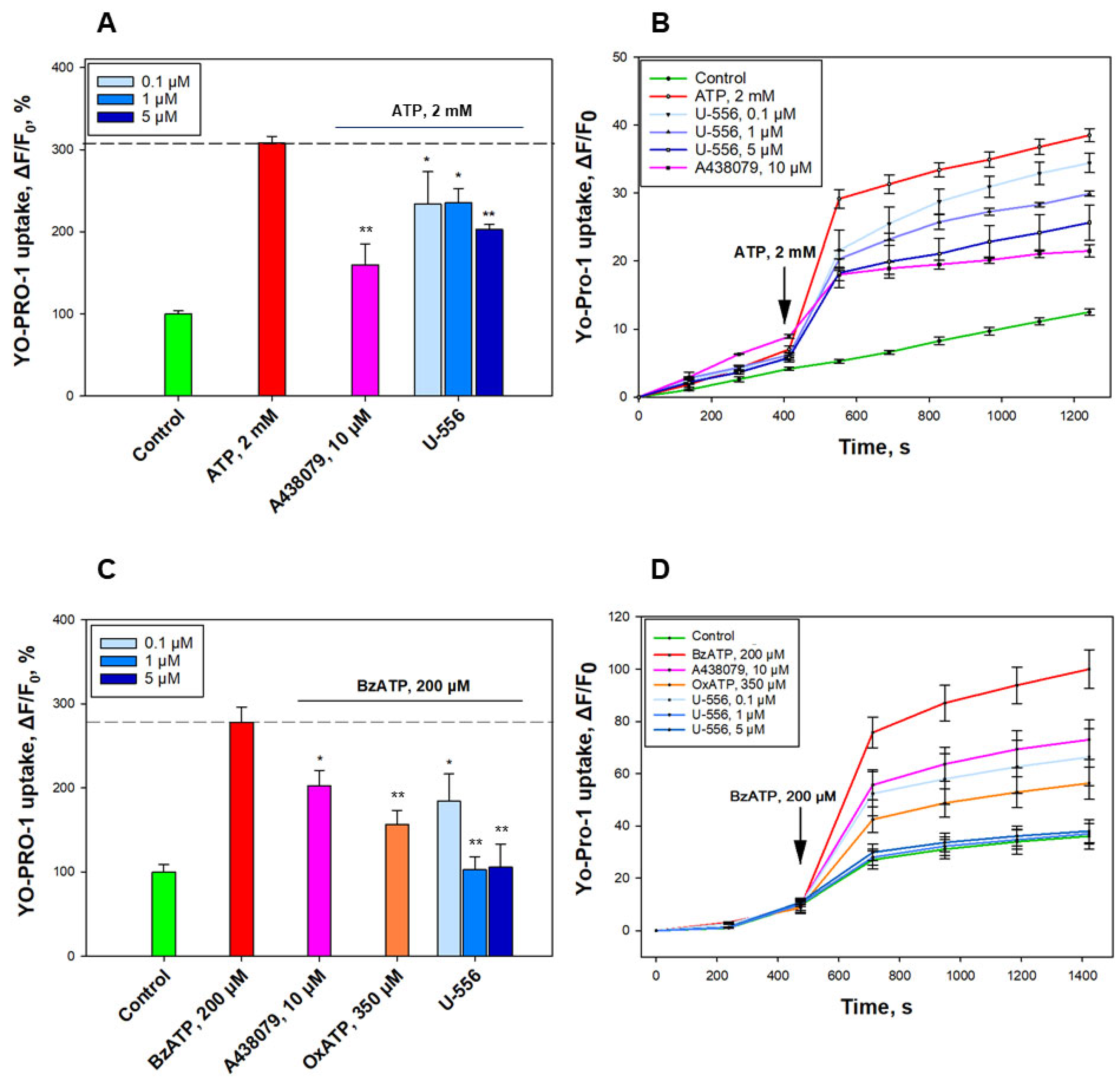
A significant increase in the YO-PRO-1 uptake into macrophages in the presence of ATP confirms the opening of the P2X7R macropore under these conditions. The data obtained on reducing the dye uptake after pre-incubation of cells with U-556 corroborate a direct inhibitory effect of the studied conjugate U-556 on the ATP-induced P2X7R macropore formation compatible with A438079 blocker action. Experiments with a selective P2X7R agonist convincingly demonstrate a pronounced entry of the dye through the macropores of this receptor formed under BzATP action. Noteworthy, under these conditions, U-556 actually completely suppresses the dye uptake by cells, indicating a complete blocking of the P2X7R function. Surprisingly, the selective blocker A438079 and P2X receptor antagonist OxATP did not show a complete blocking effect on the functioning of the receptor macropore. This is probably due to the peculiarities of the structure or functioning of P2X7R in RAW 264.7 macrophage cells.
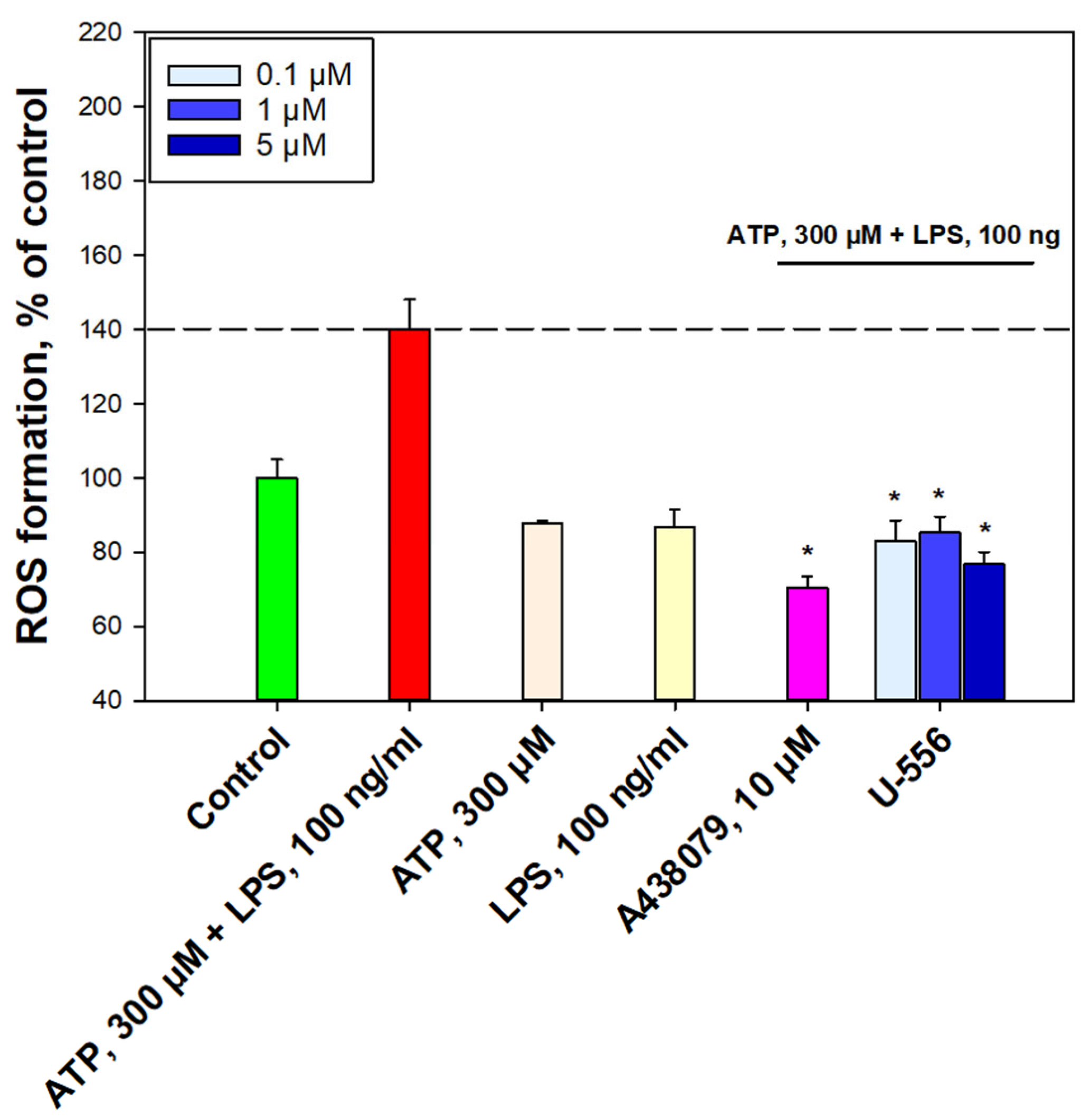
The ability of the investigated conjugate U-556 to significantly suppress the elevation of ROS production in cells and to protect and maintain the macrophage metabolic activity and viability in the presence of a cytotoxic ATP concentration proves that this compound can block the formation of P2X7R macropores the opening of which leads to cell death due to oxidative stress. However, no significant anti-radical activity of the studied compound was observed in the DPPH test and in the test of peroxidation in the mouse brain homogenate. This suggests that the studied conjugate U-556 has no pronounced antioxidant power and does not additionally contribute to diminishing intracellular ROS levels under ATP influence. A diminution in the ATP cytotoxic effect expressed as the elimination of the biomembrane integrity, permeability disturbance, and LDH release from cells, as well as in the abolition of the intracellular esterase inhibition, an increase in cell metabolic activity is an additional verification of compound U-556 capability to retain the viability of macrophages by blocking of P2X7R functioning.
In this study, we did not find a pronounced dose-effect relationship for compound
U-556. The same lack of clear dose dependence has been previously described for other 1,4-NQs, which is likely due to the non-linear mode of P2X7R inhibition [
12,
13].
Compound U-556 selective binding to P2X7 subunits and the formation of a stable complex was demonstrated by SPR. Indeed, this is direct evidence of the ability of the studied 1,4-NQ derivative to specifically interact with the extracellular part of the P2X7 subtype purine receptors, which can affect their functioning. The parameters of naphthoquinone/P2X7R interaction estimated using the SPR technique may differ from those observed in experiments with live cultured macrophages since the conditions for these two types of studies are significantly different.
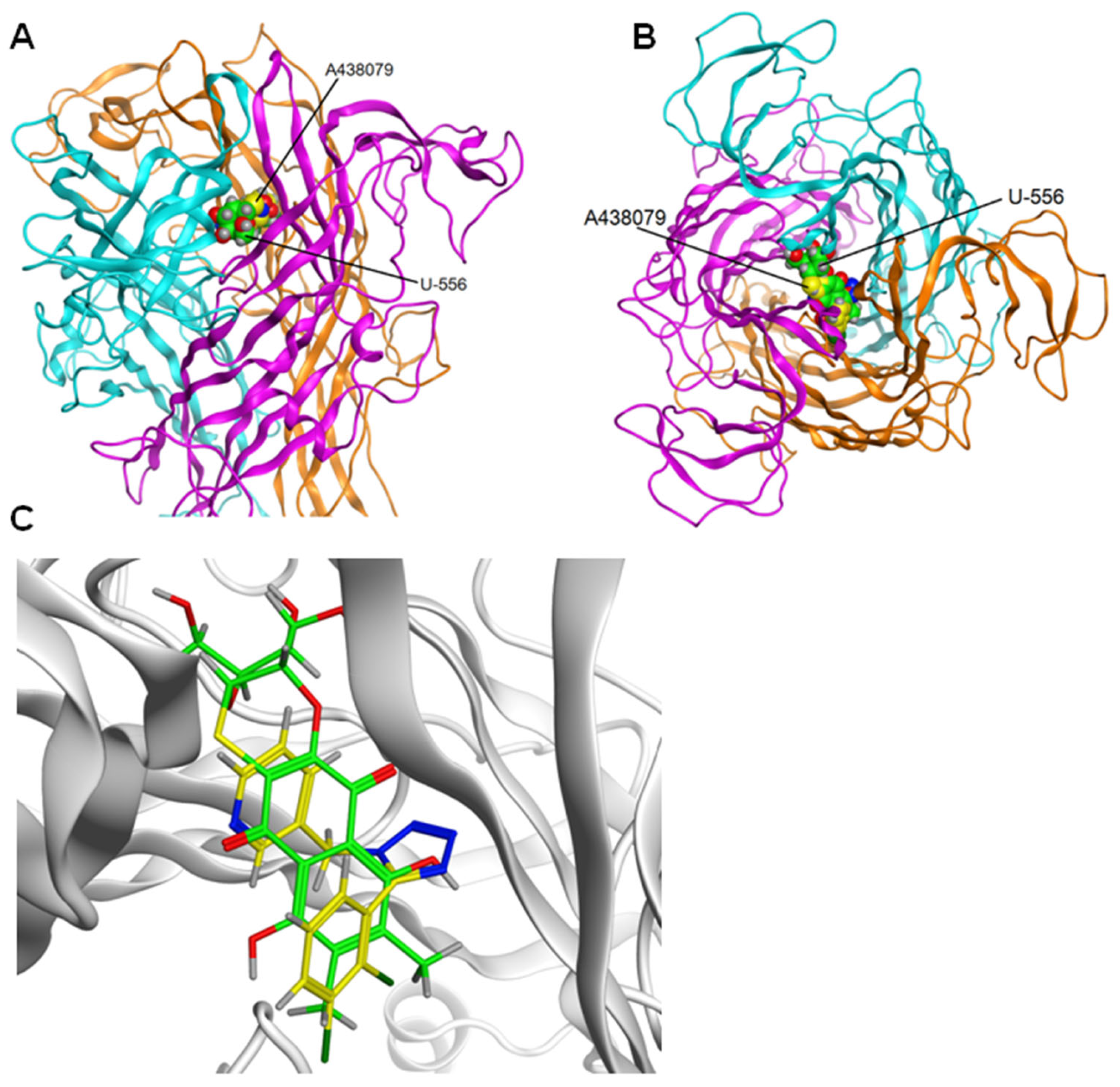
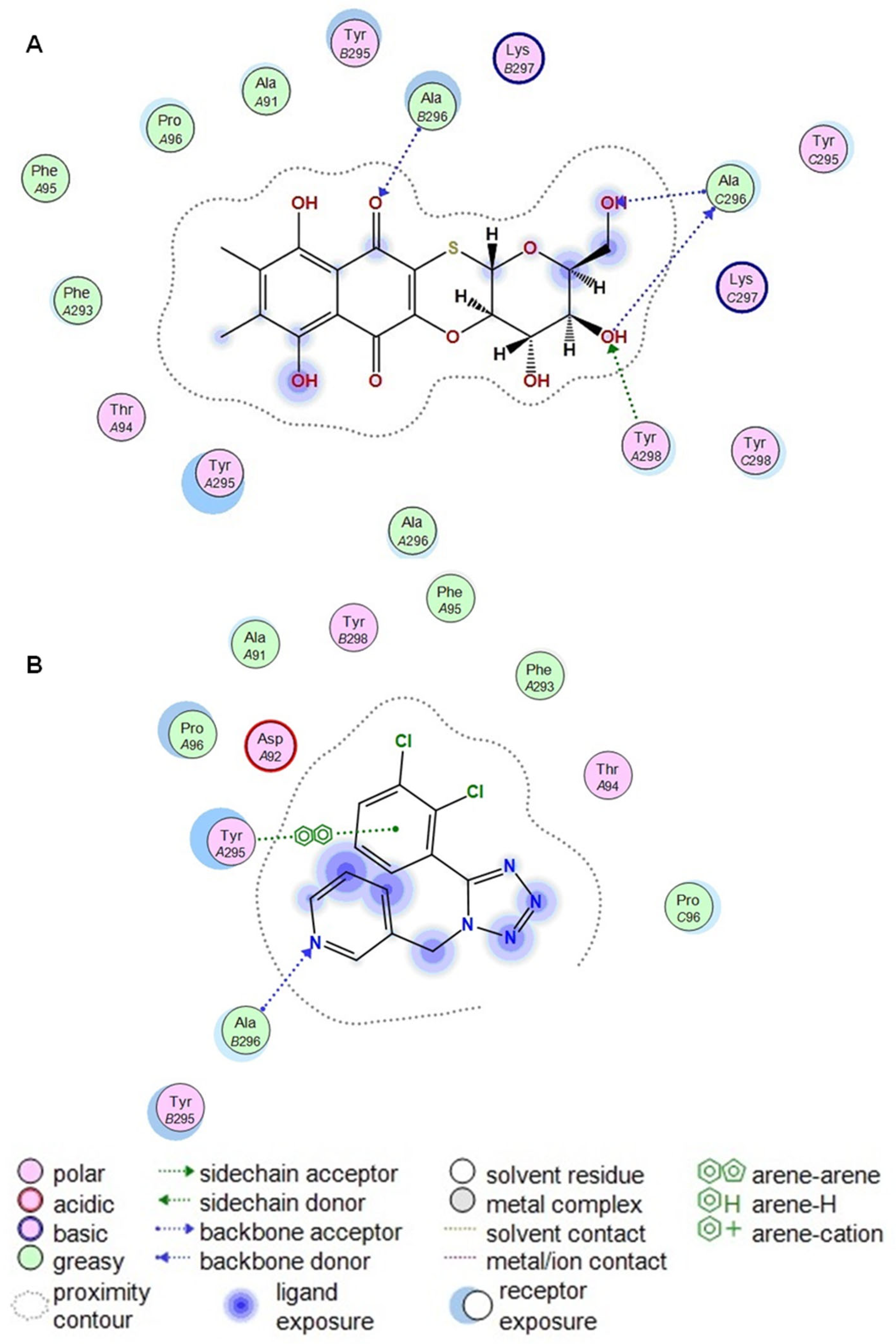
We showed that quinone-thioglucoside conjugate
U-556 can penetrate the allosteric binding site and interact with the inner part of the pocket. The contacts between the quinone-thioglucoside conjugate core and Ala296_C and Ala296_B amino acid residues seem to be the most substantial contributors to this reciprocity. Computer modeling predicted that
U-556 binds the P2X7R allosteric binding site, topographically similar to that of the specific A438079 blocker. The failure in an additive effect of the inhibitory action of
U-556 and A438079 when used in combination (
Figure 2E) is the further verification of a single allosteric binding site existence for these two compounds to the P2X7 receptor.
At present, the crystal structure of the complex of A438079 with P2X7R has not been established. Work [
19] showed that antagonists with different structures bind to the same P2X7R allosteric site. Depending on the structure of antagonists, various contacts are possible at the allosteric site. At the same time, there are papers in which A438079 was docked to the allo- and orthosteric P2X7R sites [
20]. The allosteric site has been shown to be the most beneficial.
Amino acid residues Phe88, Ile310, Met105, Phe103, Phe293, Tyr295, Tyr298, Asp92, Glu73, Lys110, and Lys297 are the residues that form the allosteric site of the P2X7 receptor. The set of these residues is the same both in article [
12] and in this study, there are no differences. However, not all these residues form H-donor and H-acceptor contacts with
U-556. Previously, we noted that
U-556 forms contacts with Asp92 and Glu73. At the same time, in this study, we have shown that such amino acid residues are Ala296_C and Ala296_B. Previously, we performed docking using the A740003 antagonist as a template in the allosteric site. In this work, the A438079 blocker was docked to the allosteric site, and then the A438079 was used as a template for docking
U-556. This resulted in
U-556 making contact with Ala296_B, as did A438079, and additionally with Ala296_C.
In our study, we found the partial inhibition of P2X7R by
U-556 and A438079. The incomplete blocking of the receptor by A438079 has been described in a number of papers. For instance, in adenocarcinoma cells, this selective blocker had an unimpressive effect on ATP-induced P2X7R pore formation and cell death [
21]. Partial blocking of receptor function by the investigated 1,4-naphthoquinone
U-556 may be of a similar nature to the A438079 blocker since both of these compounds bind to the same allosteric site of the receptor.
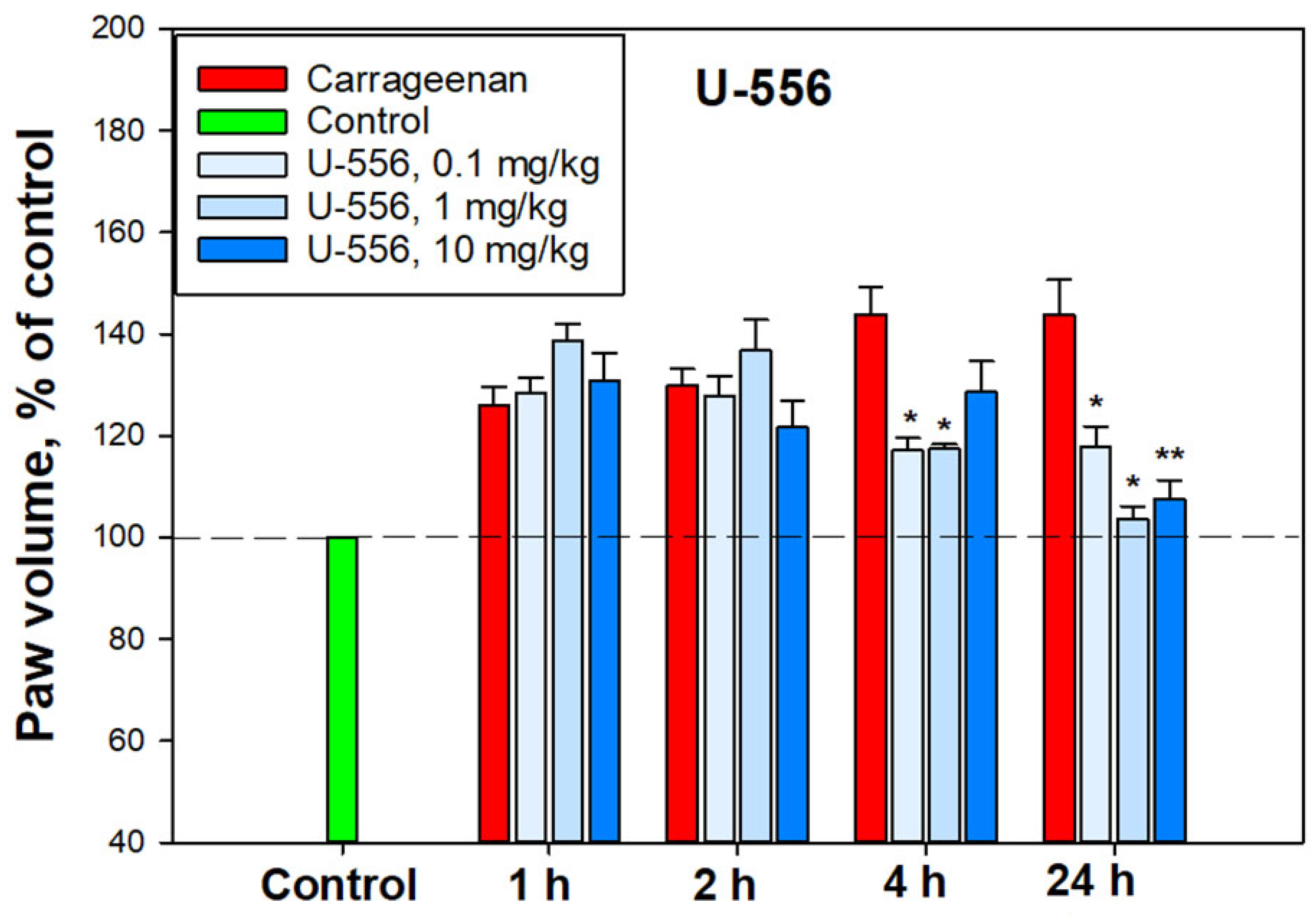
The process of carrageenan-induced paws edema develops over time and is a biphasic process. In the early phase, histamine, serotonin, and bradykinin are the first mediators involved, whereas prostaglandins, COX-2, various cytokines, such as IL-1β, IL-6, IL-10, and TNF-α, reactive oxygen species, and hydroxyl radicals are implicated in the second phase [
22]. Notably, the maturation and implementation of the pro-inflammatory cytokines depend on the canonical activation of the NLRP3 inflammasome through Pannexin-1/P2X7/K
+ efflux signaling. Blocking P2X7R by specific ligands or using macrophages of knockout
P2rx7−/− mice resulted in a significant decrease in the carrageenan effect [
23]. In our experiments, acute inflammation induced by carrageenan showed significant mitigation by
U-556 in the second phase after 4 and especially 24 h. Apparently, this is a consequence of the conjugate
U-556 impact on P2X7R followed by inhibition of pro-inflammatory cytokine release and ROS generation responsible for paws edema. COX-2 inhibition by the studied compound described for some 1,4-NQs in our previous investigation [
13] may also contribute to diminishing the edema rate in the second phase.
4. Materials and Methods
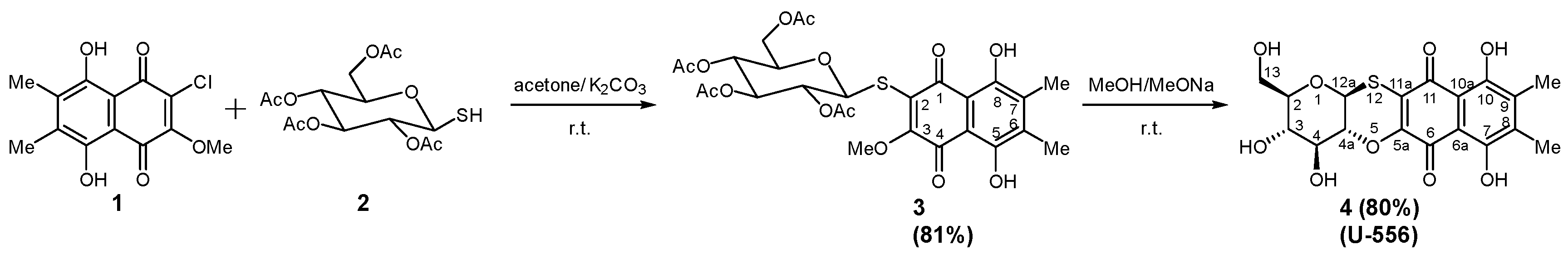
4.1. Synthesis of Tetracyclic Conjugate of 1,4-Naphthoquinone Thioglucoside (U-556)
Synthesis of 3-(tetra-O-acetyl-β-D-glucopyranosyl-1-thio)-5,8-dihydroxy-2-methoxy-6,7-dimethylnaphthalene-1,4-dione
3 was carried out by condensation of 2-chloro-5,8-dihydroxy-3-methoxy-6,7-dimethylnaphtalene-1,4-dione
1 with tetra-O-acetyl-1-thio-β-D-glucose
2 according [
12]. Acetylated thioglucoside
3 was characterized as red solid with R
f 0.43 (hexane-benzene-acetone, 2:1:1 v/v, silufol plates), yield 81%, mp 169–171 °C.
1 H NMR (700 MHz, CDCl
3): δ 1.95 (s, 3H), 2.01 (s, 3H), 2.02 (s, 3H), 2.08 (s, 3H), 2.26 (s, 3H), 2.27 (s, 3H), 3.70 (ddd, 1H,
J 2.4, 5.1, 10.0 Hz), 4.06 (dd, 1H,
J 2.4, 12.3 Hz), 4.15 (m, 1H,
J 5.1, 12.3 Hz), 4.21 (s, 3H), 5.09 (dd, 1H,
J 9.3, 10.0 Hz), 5.11 (dd, 1H,
J 9.3 10.0 Hz), 5.26 (dd, 1H,
J 9.3, 9.4 Hz), 5.50 (d, 1H,
J 10.0 Hz), 12.06 (s, 1H), 13.41 (s, 1H).
13C NMR (CDCl
3, 176 MHz): δ 12.33, 12.57, 20.48, 20.55, 20.58, 20.67, 61.91, 62.06, 68.30, 71.23, 73.99, 75.94, 82.06, 107.95, 109.70, 126.98, 139.62, 140.81, 159.52, 165.29, 166.25, 169.34, 169.41, 170.17, 170.49, 173.24, 178.13.
Tetracyclic conjugate
4 (
U-556) was prepared according to [
12] by treatment
3 with MeONa solution in dry MeOH. The red precipitate of
4 was filtered off, dried, and yielded (2R,3S,4S,4aR,12aS)-3,4,7,10-tetrahydroxy- 2-hydroxymethyl-3,4,4a,12a-tetrahydro-2H-naphtho[2,3-b]pyrano[2,3-e][
1,
4]oxathiine-6,11-dione (
4); R
f 0.47 (benzene-ethylacetate- methanol, 7:4:2 v/v, silufol plates), yield 80%, mp 314–316 °C [
21].
1H NMR (500 MHz, DMSO-
d6): δ 2.18 (s, 3H), 2.19 (s, 3H), 3.32 (m, 1H), 3.50 (m, 2H), 3.60 (m, 2H), 3.75 (m, 1H), 4.81 (m, 1H), 5.01 (m, 1H), 5.49 (d, 1H,
J 5.9 Hz), 5.76 (m, 1H), 12.64 (s, 1H), 12.94. (s, 1H).
13C NMR (DMSO-
d6, 125 MHz): δ 12.1, 12.2, 60.7, 70.4, 73.4, 73.9, 79.3, 82.3, 117.3, 107.4, 123.5, 137.8, 137.9, 150.4, 156.3, 157.3, 177.7, 183.0 [
14].
The total scheme of compound
U-556 synthesis is shown in
Figure 1.
Compound U-556 was dissolved in 100% DMSO at a concentration of 10 mM and stored as a stock solution. Intermediate stock solutions were then prepared at a concentration of 100 μM in 10% DMSO. For experiments, U-556 solution was added in a volume of 20 µL per well of 96-well plates to 180 µL of cell medium so that the final concentration of DMSO in the incubation medium did not exceed 1%.
4.2. Cell Line
Mouse macrophage cell line RAW 264.7 TIB-71™ was purchased from ATCC (American Type Culture Collection, Manassas, VA, USA). Cells were grown in DMEM medium (Biolot, St. Petersburg, Russia) supplemented with 10% fetal bovine serum (Biolot, St. Petersburg, Russia) and 1% penicillin/streptomycin (Biolot, St. Petersburg, Russia) in a CO2 incubator at 37 °C and 5% CO2.
4.3. Ca2+ Influx Measurement
RAW 264.7 cells were seeded in 96-well plates (4 × 104 cells per well) in a DMEM cultural medium and incubated overnight at 37 °C, 5% CO2. Then, the cells were washed once with HBSS saline (140 mM NaCl, 5 mM KCl, 0.8 mM MgCl2, 2 mM CaCl2, 10 mM glucose, 10 mM HEPES, pH 7.4) and loaded with 5 μM Fluo-8 AM (Abcam, Cambridge, UK) and 0.05% (w/v) Pluronic F-127 (Sigma-Aldrich, Burlington, MA, USA) in the same buffer solution. Cells were incubated for 40 min (37 °C, 5%) and then were washed with HBSS buffer without the fluorescent dye and treated by studied compounds for 20 min at RT in the dark. ATP (Sigma, Burlington, MA, USA) was used as an agonist of P2X7R. Competitive P2X7R antagonist A438079 (10 μM, Sigma, Burlington, MA, USA) were used as inhibitory controls. Ionomycin (Sigma-Aldrich, St. Louis, MO, USA) was used to generate a generic calcium signal in cells. Fluo-8 was excited at 485 nm, and the emission at 520 nm was measured with a PHERAstar FS plate reader (BMG LABTECH, Ortenberg, Germany). The data were processed by MARS Data Analysis v. 3.01R2 (BMG Labtech, Ortenberg, Germany). Mean values and standard error of peak height calcium were calculated.
4.4. YO-PRO-1 Uptake Assay
RAW 264.7 cells were seeded in 96-well plates at a density of 4 × 104 cells/well and incubated in DMEM for 24 h, 37 °C, 5% CO2. The cells were then washed twice with HBSS, and the medium was changed to the same buffer containing YO-PRO-1 dye (Sigma, Burlington, MA, USA, 2.5 μM final concentration). Further, the studied compound was added to the cells at concentrations of 5, 1, and 0.1 μM. P2X7 receptor blocker A438079 at a concentration of 10 μM was used as a comparison control. Two mM ATP (Sigma, Burlington, MA, USA) or 200 μM 2,3-O-(4-benzoylbenzoyl)ATP (BzATP, Sigma-Aldrich, St. Louis, MO, USA) were used as an agonist of P2X7R. In some experiments, cells were treated with 350 μM oxidized ATP (OxATP, Sigma-Aldrich, St. Louis, MO, USA) for 2 h. The YO-PRO-1 fluorescence was measured continuously every 100 sec for 800 sec after adding ATP or BzATP with a PHERAstar FS plate reader (BMG Labtech, Ortenberg, Germany) at λex = 480 nm and λem = 520 nm, ATP 2 mM or BzATP 200 μM were added after baseline recording and HBSS (20 μL) was added as a negative control.
4.5. ROS Formation Assay
RAW 264.7 cells were seeded in 96-well plates at a density of 4 × 104 cells per well for 24 h. The medium was replaced with fresh DMEM and treated with LPS (100 ng/mL) for 3 h at 37 °C. After that, the cells were treated with the studied compound (5.0, 1.0, 0.1 μM final concentrations) for an additional 30 min. P2X7R blocker A438079 (10 μM) was used as inhibitory control. Then ATP (300 μM final concentration) was added to each well for 30 min. For fluorescence measurement, the cells were washed once, and the cell medium was replaced by HBSS, containing 10 μM of fluorescent dye 2,7-dichlorodihydrofluorescein diacetate (H2DCFDA) (Sigma, Darmstadt, Germany). Cells were incubated for 30 min at 37 °C, washed twice with HBSS without fluorescent probe, and additionally incubated for 20 min at RT in the dark. The measurement of fluorescence intensity was carried out using PHERAstar FS plate reader (BMG Labtech, Ortenberg, Germany) at λex = 485 nm and λem = 520 nm.
4.6. Cytoprotection Activity Assay
4.6.1. Detection of Lactate Dehydrogenase (LDH) Release
RAW 264.7 cells (4 × 104 cells/well of a 96-well plate) were seeded and incubated for 24 h at 37 °C, 5% CO2 in an incubator. Then, the cells were treated with U-556 at final concentrations of 5.0, 1.0, and 0.1 μM and incubated for 1 h. A438079 (10 μM) and apyrase (30 U/mL) were used as inhibition control. Next, LPS (100 ng/mL, Sigma, St. Louis, MO, USA) was added to the cells, and the cells were incubated for 4 h. Thereafter, ATP (2 mM final concentration) was added to the cells, and the plates were incubated for an additional 1 or 24 h. Cells incubated without ATP and LPS were used as negative controls. The plate was then centrifuged at 250× g, and 100 μL of the supernatant from each well was transferred to the relevant wells of an optically clean 96-well plate. An equal volume of the reaction mixture (100 μL) from the LDH Cytotoxicity Assay Kit (Abcam, Cambridge, UK) was added to each well and incubated for 30 min at room temperature. The absorbance was measured at λ = 490 nm using a Multiscan FC spectrophotometer (Thermo Scientific, Nummela, Finland).
4.6.2. FDA Cell Viability Assay
A stock solution of fluorescein diacetate (FDA) (Sigma-Aldrich, St. Louis, MO, USA) in DMSO (1 mg/mL) was prepared. RAW 264.7 cells were seeded and treated as described above. Further, FDA solution (50 μg/mL) was added to each well, and the plate was incubated in the dark at 37 °C for 15 min. Cells were washed, and fluorescence intensity corresponding to nonspecific esterase activity was measured using a Fluoroscan plate reader (Thermo Labsystems, Helsinki, Finland) at λex = 485 nm and λem = 518 nm. Cell viability was expressed as a percentage of control.
4.6.3. MTT Assay
RAW 264.7 cells (4 × 104 cells/well) were seeded and incubated in 96-well plates for 24 h at 37 °C and 5% CO2 in an incubator. Compound U-556 was added to the wells at different concentrations, and the plates were incubated for an additional 1 h. A438079 (10 μM) and apyrase (30 U/mL, Sigma, Burlington, MA, USA) were also added to cells as standard blockers of ATP action. Then ATP (2 mM, final concentration) and LPS (100 ng/mL, final concentration) were added to the cells. Cells were incubated for 24 h, after which cell viability was determined using the MTT reagent (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide, Sigma-Aldrich, Saint-Louis, MO, USA). For this purpose, 10 μL of MTT stock solution (5 mg/mL) was added to each well, and the microplate was incubated for 4 h at 37 °C. After that, 100 μL of SDS-HCl (1 g SDS/10 mL dH2O/17 μL 6 N HCl) were added to each well, followed by incubation for 4–18 h. The absorbance of the converted dye formazan was measured using Multiskan FC microplate photometer (Thermo Scientific, Waltham, MA, USA) at a wavelength of 570 nm. The results were presented as percentage of control data. The tested compound was added to the wells of the plates in a volume of 20 μL dissolved in PBS (DMSO concentration > 1%).
4.6.4. Trypan Blue Exclusion Cell Viability Test
RAW 264.7 cells (4 × 104 cells/well of a 96-well plate) were seeded and incubated for 24 h at 37 °C, 5% CO2 in an incubator. Then, the cells were treated with LPS (100 ng/mL) and inhibitors, as described above. After 24 h of incubation with ATP (2 mM), the cells were resuspended in the medium of each well. Ten µL of the cell medium containing the cell suspension was mixed with 10 µL of 0.4% trypan blue. The mixture was loaded onto counting slides, and cell viability was assessed using the TC20 automated cell counter (Bio-Rad, Hercules, CA, USA).
4.7. Radical Scavenging Assay
DPPH radical scavenging activity of compounds was tested as described [
24] with minor modifications. The compounds were dissolved in MeOH, and the solutions of conjugate
U-556, ascorbic acid (Vekton, St. Petersburg, Russia), or quercetin (Sigma-Aldrich, Steinheim, Germany) as a positive control (120 µL) were dispensed into wells of a 96-well microplate. In all of them, 30 µL of the DPPH (Sigma-Aldrich, Steinheim, Germany) solution in MeOH (0.75 mM) was added to each well. The concentrations of compounds and ascorbic acid in the mixture were 0.01–100.0 µM. The plates were incubated in the dark at room temperature for 30 min, and then the absorbance was measured at 517 nm with a Multiskan FC microplate photometer (Thermo Scientific, Waltham, MA, USA). The negative control contained no test compound. The final results were reported as DPPH radical scavenging (%) in the reaction solution.
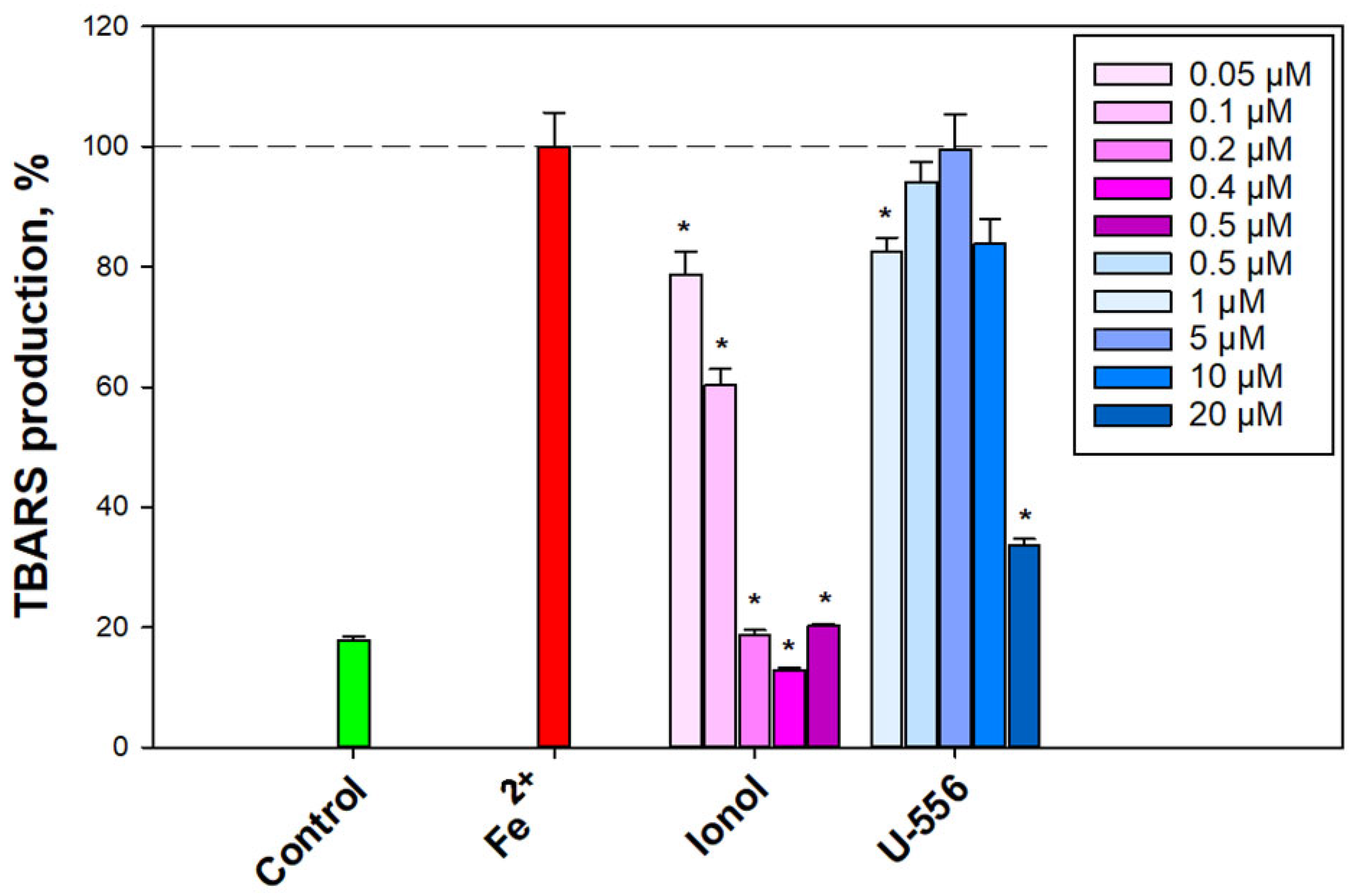
4.8. Determination of Antioxidant Activity in Brain Homogenate
The procedure was performed according to the protocol described in [
25]. The brain of female C57BL/6 mouse was washed with cold buffer (140 mM KCl, 10 mM K
2HPO
4, pH 7.4) and then homogenized using a Potter–Elehjem tissue homogenizer, adding the buffer to the brain in a weight ratio of 1:4. 10 mL of buffer was added to the resulting homogenate and centrifuged for 10 min at 900 rpm in a Labofuge 44R centrifuge (Thermo Scientific, Karlsruhe, Germany). The supernatant was transferred to a volumetric flask and adjusted to 25 mL with a buffer. The Lowry protein concentration in homogenate was in the range of 0.27–0.3 mg/mL. To determine the antioxidant activity, the solution of the tested compound was added to the brain homogenate, then 2 mM FeSO
4 solution was added, and the reaction was stopped by adding of 20% trichloroacetic acid (TCA) (Reachim, Moscow, Russia). The homogenate was centrifuged, supernatant was collected, and 0.7% solution of thiobarbituric acid (TBA) (Sigma, Saint-Louis, MO, USA) in 50% glacial acetic acid was added to reveal the products reacting with TBA. The content of TBA-reacting substances (TBARS) was determined spectrophotometrically using PHERAstar FS plate reader (BMG Labtech, Ortenberg, Germany) by measurement of fluorescence intensity at 480/520 nm (λex/λem).
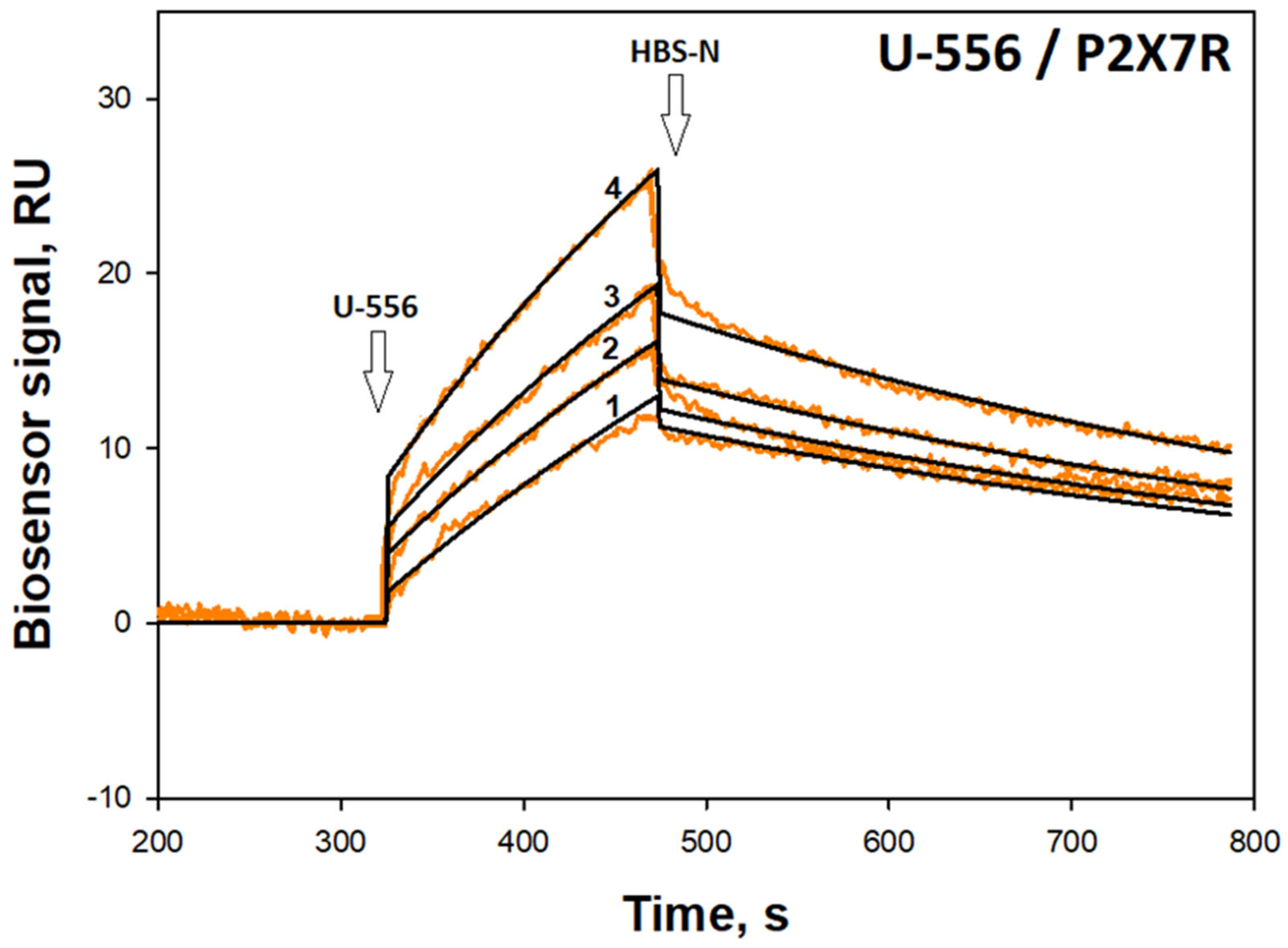
4.9. Surface Plasmon Resonance
SPR analysis was performed at 25 °C using a Biacore® 3000 optical biosensor and CM5 sensor chips (GE Healthcare, Chicago, IL USA). Buffer solution HBS-N (10 mM HEPES, 150 mM NaCl, pH 7.4) (Cytiva, Chicago, IL, USA) was used as a working buffer. Protein hP2X7 (47–334 a.a., extracellular domain, 36.8 kDa) (abx167020 Abbexa LTD, Cambridge, UK) was covalently immobilized on the surface of the chip by forming amide bonds between dextran carboxyl groups and free amino groups of the protein. The carboxyl groups of the chip were activated for 5 min by injecting a mixture 1:1 v/v of 0.2 M 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride (EDC) and 0.05 M N-hydroxysuccinimide (NHS) at a rate of 5 μL/min followed by a 1 min wash with HBS-N buffer at the same flow rate. Next, hP2X7 (15 μg/mL) in 10 mM sodium acetate (pH 5.0) was injected into the working channel of the biosensor for 10 min at a flow rate of 5 μL/min. The final level of immobilization was 12,300 RU (12.3 ng of protein). The control channel without immobilized P2X7R was used to correct the effects of the nonspecific binding of analytes to the chip surface. Compound U-556 was prepared as 1 mM stock solutions in DMSO and then was dissolved in HBS-N buffer (1:100 v/v). The samples were injected through the channels of the biosensor (working and reference) at a flow rate of 10 μL/min for 6 min. The dissociation of the formed 1,4-NQ/P2X7 complexes was recorded for at least 6 min at the same flow rate after the injection of the sample. After each cycle of measurement, the bound analyte was removed by double injection of the regenerating solution (2 M NaCl, 1% CHAPS) at a flow rate of 30 μL/min for 30 s.
SPR sensorgrams were processed in Biacore 3000 Evaluation Software v.1.0 (GE Healthcare, Chicago, IL, USA) and BIAevaluation Software v. 4.1.1 (GE Healthcare, Chicago, IL, USA) using “1:1 binding (Langmuir)” model. The 1:1 (Langmuir) binding model is a 1:1 interaction model between a compound (C) and an immobilized protein (P) and is equivalent to the Langmuir isotherm for surface adsorption: C + P ↔ CP. The final kinetic parameters were obtained from the fits that best agree with the experimental curves for the minimum of the obtained value of chi2. Chi2 is a measure of the average squared residual between the experimental data and the fitted curve. The equation describing the interaction model used is as follows: (1) 1:1 (Langmuir) binding:
where Kd is the equilibrium dissociation constant, koff is the dissociation rate constant, kon is the association rate constant.
4.10. Molecular Modeling
The homology model of the mouse P2X7R trimer (UniProt ID Q9Z1M0) in the closed form was built with the Homology module of the Molecular Operating Environment (MOE) program [
26], using as a template structure of the rat P2X7R closed form (PDB ID 6U9V). Energy minimization of the model mP2X7R trimer with force field Amber10:EHT and 3D-protonation were carried out using MOE 2020 program. The 3D structure of
U-556 was generated with the Build module of MOE program using MMFF94 potential for structure optimization. The molecular docking of compounds to the model of mP2X7R trimer was carried out using SiteFinder and Dock modules of the MOE program. Template docking of A438079 was carried out using P2X7R antagonist A740003 binding site as template and refinement 5 poses with Induced Fit and Score GBVI/WSA dG. Template docking of
U-556 was carried out using P2X7R antagonist A438079 binding site as a template and Score GBVI/WSA dG. Analysis of ligands contacts was carried out using Ligand Interaction module of the MOE program.
4.11. Carrageenan-Induced Paw Edema
Mouse paw edema, an acute inflammatory model, was used to investigate the antagonistic action of U-556. The female C57BL/6 mice were divided into groups of 6 individuals, each mouse was measured by the control volume of the paw using a plethysmometer (Ugo basile 37140, Milan, Italy). Each group then received tested 1,4-naphthoquinone diluted in sterile H2O from a stock solution in DMSO (10 mM) at different dosages. The final concentration of DMSO was 1%. In these experiments, 1,4-naphthoquinone at dosages of 0.1, 1.0, and 10.0 mg/kg was injected intraperitoneally at 60 min prior to carrageenan-σ (Sigma, Saint-Louis, MO, USA) administration. Control mice received an intraperitoneal injection of the solvent. Carrageenan in saline solution at a concentration of 1.5 mg/mL in the volume of 20 μL was injected into the back paw. After 1, 2, 4, 6, and 24 h, paw edema was measured using a plethysmometer. The volume of the paw before the start of the experiment was taken as 100%.
4.12. Animals
Female C57BL/6 mice (two months of age, 20–22 g) purchased from the Russian National Center for Genetic Resources of Laboratory Animals on the basis of the SPF vivarium (ICiG SB RAS, Novosibirsk, Russia) were used for experiments. All mice were raised in an environment with a 12-h light/dark cycle at a temperature of 22 ± 1 °C with available food and water. All experiments were conducted according to the International Recommendations for Biomedical Research Using Animals, adopted by the International Council of Medical Scientific Societies (CIOMS) and the Order of the Ministry of Health and Social Development of Russia dated 23.08.2010 No. 708n “On Approval of Laboratory Practice Rules”.
4.13. Statistics
All data were obtained in independent replicates, and calculated values were expressed as mean ± standard error of the mean (SEM). The Student’s t-test was performed using SigmaPlot 14.0 (Systat Software Inc., San Jose, CA, USA) to determine statistical significance.