Genetic Modulation of the Erythrocyte Phenotype Associated with Retinopathy of Prematurity—A Multicenter Portuguese Cohort Study
Abstract
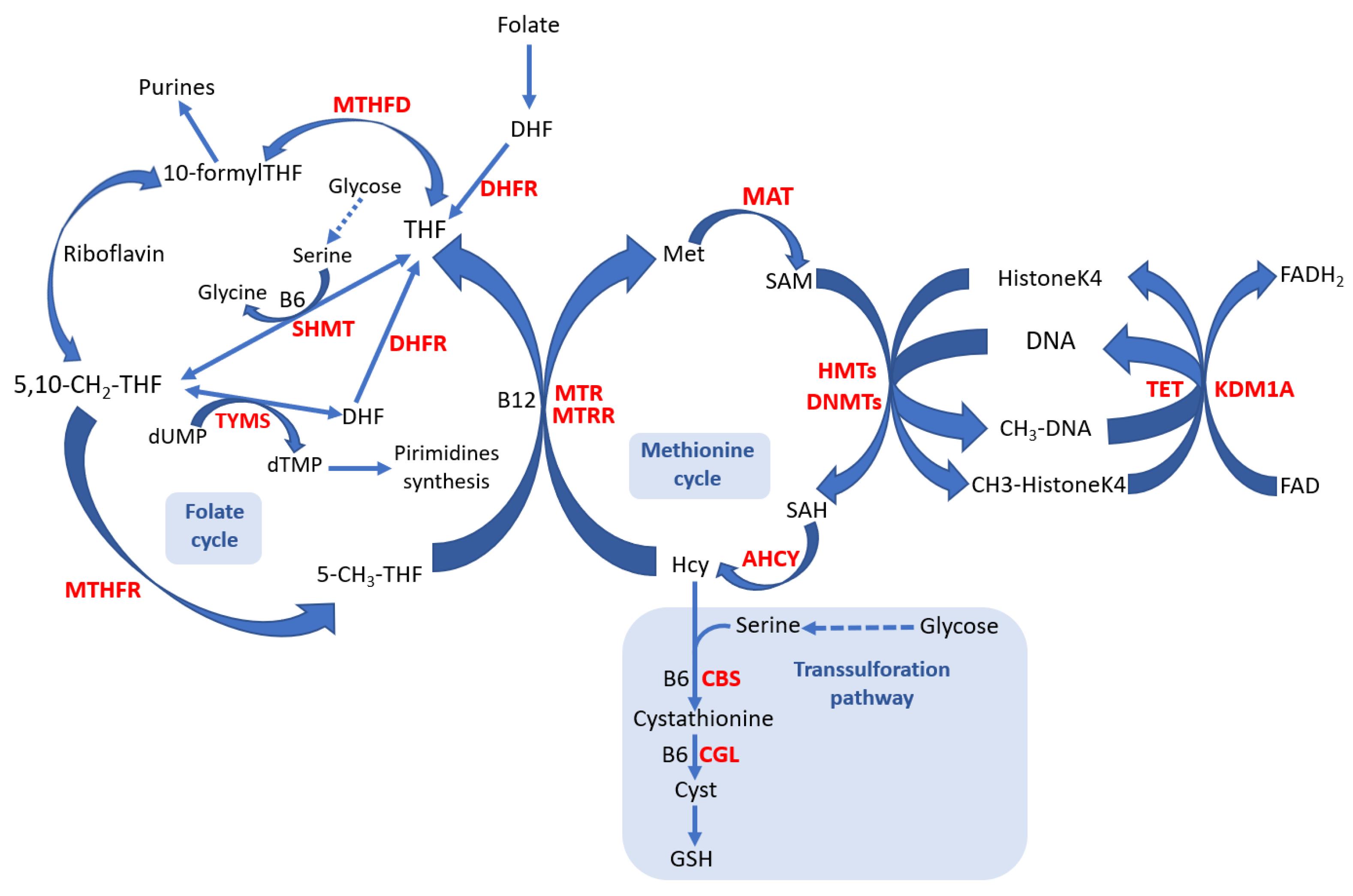
1. Introduction
2. Results and Discussion
2.1. Clinical Characteristics
2.2. Anemia
2.3. DNMT3B, KDM1A, and MTHFR Polymorphism and ROP
2.4. Epistatic Relations and ROP
2.5. DNMT3B, KDM1A, and MTHFR Polymorphism and Hematological Parameters
2.6. Epistatic Relations and Hematological Parameters
2.7. DNMT3B, KDM1A, and MTHFR Polymorphism, Hematological Parameters, and ROP
2.8. Discussion
3. Materials and Methods
3.1. Population
3.2. ROP Screening and Ophthalmological Data Collection
3.3. Genetic Polymorphism Identification
3.4. Demographic and Clinical Data
3.5. Statistical Analysis
3.6. Ethics Approval
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Graziosi, A.; Perrotta, M.; Russo, D.; Gasparroni, G.; D’egidio, C.; Marinelli, B.; Di Marzio, G.; Falconio, G.; Mastropasqua, L.; Li Volti, G.; et al. Oxidative Stress Markers and the Retinopathy of Prematurity. J. Clin. Med. 2020, 9, 2711. [Google Scholar] [CrossRef] [PubMed]
- Bai, W.H.; Gu, D.F.; Dai, Y.; Chen, Y.H.; Yang, Z.M.; Lu, L.J. The Relationship between Probiotics and Retinopathy of Prematurity in Preterm Infants: A Population-Based Retrospective Study in China. Front. Pediatr. 2023, 11, 1055992. [Google Scholar] [CrossRef] [PubMed]
- Joyal, J.S.; Omri, S.; Sitaras, N.; Rivera, J.C.; Sapieha, P.; Chemtob, S. Neovascularization in Retinopathy of Prematurity: Opposing Actions of Neuronal Factors GPR91 and Semaphorins 3A. Acta Paediatr. 2012, 101, 819–826. [Google Scholar] [CrossRef] [PubMed]
- Fevereiro-Martins, M.; Marques-Neves, C.; Guimarães, H.; Bicho, M. Retinopathy of Prematurity: A Review of Pathophysiology and Signaling Pathways. Surv. Ophthalmol. 2023, 68, 175–210. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.J.; Port, A.D.; Swan, R.; Campbell, J.P.; Chan, R.V.P.; Chiang, M.F. Retinopathy of Prematurity: A Review of Risk Factors and Their Clinical Significance. Surv. Ophthalmol. 2018, 63, 618–637. [Google Scholar] [CrossRef]
- Hartnett, M.E.; Cotten, C.M. Genomics in the Neonatal Nursery: Focus on ROP. Semin. Perinatol. 2015, 39, 604–610. [Google Scholar] [CrossRef]
- Lundgren, P.; Hellgren, G.; Pivodic, A.; Sävman, K.; Smith, L.E.H.; Hellström, A. Erythropoietin Serum Levels, versus Anaemia as Risk Factors for Severe Retinopathy of Prematurity. Pediatr. Res. 2019, 86, 276–282. [Google Scholar] [CrossRef]
- Pheng, E.; Di Lim, Z.; Tai Li Min, E.; Van Rostenberghe, H.; Shatriah, I. Haemoglobin Levels in Early Life among Infants with and without Retinopathy of Prematurity. Int. J. Environ. Res. Public Health 2021, 18, 7054. [Google Scholar] [CrossRef]
- Perez, M.; Robbins, M.E.; Revhaug, C.; Saugstad, O.D. Oxygen Radical Disease in the Newborn, Revisited: Oxidative Stress and Disease in the Newborn Period. Free Radic. Biol. Med. 2019, 142, 61. [Google Scholar] [CrossRef]
- Kalteren, W.S.; Bos, A.F.; Bergman, K.A.; van Oeveren, W.; Hulscher, J.B.F.; Kooi, E.M.W. The Short-Term Effects of RBC Transfusions on Intestinal Injury in Preterm Infants. Pediatr. Res. 2023, 93, 1307–1313. [Google Scholar] [CrossRef]
- Krug, A.W.; Tille, E.; Sun, B.; Pojoga, L.; Williams, J.; Chamarthi, B.; Lichtman, A.H.; Hopkins, P.N.; Adler, G.K.; Williams, G.H. Lysine-Specific Demethylase-1 Modifies the Age Effect on Blood Pressure Sensitivity to Dietary Salt Intake. Age 2013, 35, 1809–1820. [Google Scholar] [CrossRef]
- Riancho, J.; del Real, A.; Riancho, J.A. How to Interpret Epigenetic Association Studies: A Guide for Clinicians. Bonekey Rep. 2016, 5, 797. [Google Scholar] [CrossRef] [PubMed]
- Raghubeer, S.; Matsha, T.E.; Mccaddon, A.; Miller, J.W. Methylenetetrahydrofolate (MTHFR), the One-Carbon Cycle, and Cardiovascular Risks. Nutrients 2021, 13, 4562. [Google Scholar] [CrossRef] [PubMed]
- Majstorović, D.; Barišić, A.; Božović, I.B.; Čače, I.B.; Čače, N.; Štifanić, M.; Vraneković, J. DNMT3B Rs2424913 as a Risk Factor for Congenital Heart Defects in Down Syndrome. Genes 2023, 14, 576. [Google Scholar] [CrossRef]
- Saradalekshmi, K.R.; Neetha, N.V.; Sathyan, S.; Nair, I.V.; Nair, C.M.; Banerjee, M. DNA Methyl Transferase (DNMT) Gene Polymorphisms Could Be a Primary Event in Epigenetic Susceptibility to Schizophrenia. PLoS ONE 2014, 9, e98182. [Google Scholar] [CrossRef]
- Lorente-Pozo, S.; Parra-Llorca, A.; Lara-Cantón, I.; Solaz, A.; García-Jiménez, J.L.; Pallardó, F.V.; Vento, M. Oxygen in the Neonatal Period: Oxidative Stress, Oxygen Load and Epigenetic Changes. Semin. Fetal Neonatal Med. 2020, 25, 101090. [Google Scholar] [CrossRef] [PubMed]
- Coppede, F.; Teresa Zitarosa, M.; Migheli, F.; Lo Gerfo, A.; Bagnoli, S.; Dardano, A.; Nacmias, B.; Mancuso, M.; Monzani, F.; Siciliano, G.; et al. DNMT3B Promoter Polymorphisms and Risk of Late Onset Alzheimer’s Disease. Curr. Alzheimer Res. 2012, 9, 550–554. [Google Scholar] [CrossRef]
- Barišic, A.; Kolak, M.; Peterlin, A.; Tul, N.; Krpina, M.G.; Ostojic, S.; Peterlin, B.; Pereza, N. DNMT3B Rs1569686 and Rs2424913 Gene Polymorphisms Are Associated with Positive Family History of Preterm Birth and Smoking Status. Croat. Med. J. 2020, 61, 8–17. [Google Scholar] [CrossRef]
- Singh, R.K.; Mallela, R.K.; Hayes, A.; Dunham, N.R.; Hedden, M.E.; Enke, R.A.; Fariss, R.N.; Sternberg, H.; West, M.D.; Nasonkin, I.O. Dnmt1, Dnmt3a and Dnmt3b Cooperate in Photoreceptor and Outer Plexiform Layer Development in the Mammalian Retina. Exp. Eye Res. 2017, 159, 132–146. [Google Scholar] [CrossRef]
- Haggarty, P.; Hoad, G.; Horgan, G.W.; Campbell, D.M. DNA Methyltransferase Candidate Polymorphisms, Imprinting Methylation, and Birth Outcome. PLoS ONE 2013, 8, e68896. [Google Scholar] [CrossRef]
- Mabhida, S.E.; Muhamed, B.; Sharma, J.R.; Apalata, T.; Nomatshila, S.; Mabasa, L.; Benjeddou, M.; Masilela, C.; Ziqubu, K.; Shabalala, S.; et al. Methylenetetrahydrofolate Reductase Polymorphism (Rs1801133) and the Risk of Hypertension among African Populations: A Narrative Synthesis of Literature. Genes 2022, 13, 631. [Google Scholar] [CrossRef] [PubMed]
- Nan, Y.; Li, H. MTHFR Genetic Polymorphism Increases the Risk of Preterm Delivery. Int. J. Clin. Exp. Pathol. 2015, 8, 7397–7402. [Google Scholar] [PubMed]
- Da Silva Dias, L.N.; Coêlho, M.D.C.; Persuhn, D.C.; Arrais Ribeiro, I.L.; Medeiros Freire, E.A.; De Oliveira, N.F.P.; De Aquino, S.G. DNMT3B (Rs2424913) Polymorphism Is Associated with Systemic Lupus Erythematosus Alone and with Co-Existing Periodontitis in a Brazilian Population. J. Appl. Oral Sci. 2022, 30, e20210567. [Google Scholar] [CrossRef]
- Malagraba, G.; Yarmohammadi, M.; Javed, A.; Barceló, C.; Rubio-Tomás, T. The Role of LSD1 and LSD2 in Cancers of the Gastrointestinal System: An Update. Biomolecules 2022, 12, 462. [Google Scholar] [CrossRef]
- Patel, D.; Shimomura, A.; Majumdar, S.; Holley, M.C.; Hashino, E. The Histone Demethylase LSD1 Regulates Inner Ear Progenitor Differentiation through Interactions with Pax2 and the NuRD Repressor Complex. PLoS ONE 2018, 13, e0191689. [Google Scholar] [CrossRef]
- Wojtala, M.; Dąbek, A.; Rybaczek, D.; Śliwińska, A.; Świderska, E.; Słapek, K.; El-Osta, A.; Balcerczyk, A. Silencing Lysine-Specific Histone Demethylase 1 (LSD1) Causes Increased HP1-Positive Chromatin, Stimulation of DNA Repair Processes, and Dysregulation of Proliferation by Chk1 Phosphorylation in Human Endothelial Cells. Cells 2019, 8, 1212. [Google Scholar] [CrossRef]
- Zhao, A.; Zhou, H.; Yang, J.; Li, M.; Niu, T. Epigenetic Regulation in Hematopoiesis and Its Implications in the Targeted Therapy of Hematologic Malignancies. Signal Transduct. Target. Ther. 2023, 8, 71. [Google Scholar] [CrossRef]
- Hino, S.; Kohrogi, K.; Nakao, M. Histone Demethylase LSD1 Controls the Phenotypic Plasticity of Cancer Cells. Cancer Sci. 2016, 107, 1187–1192. [Google Scholar] [CrossRef] [PubMed]
- Domingo, J.; Baeza-Centurion, P.; Lehner, B. The Causes and Consequences of Genetic Interactions (Epistasis). Annu. Rev. Genom. Hum. Genet. 2019, 20, 433–460. [Google Scholar] [CrossRef] [PubMed]
- Jia, C.H.; Feng, Z.S.; Lin, X.J.; Cui, Q.L.; Han, S.S.; Jin, Y.; Liu, G.S.; Yang, C.Z.; Ye, X.T.; Dai, Y.H.; et al. Short Term Outcomes of Extremely Low Birth Weight Infants from a Multicenter Cohort Study in Guangdong of China. Sci. Rep. 2022, 12, 11119. [Google Scholar] [CrossRef]
- Gilfillan, M.; Bhandari, A.; Bhandari, V. Diagnosis and Management of Bronchopulmonary Dysplasia. BMJ 2021, 375, n1974. [Google Scholar] [CrossRef] [PubMed]
- Good, M.; Chu, T.; Shaw, P.; Nolan, L.S.; Mcclain, L.; Chamberlain, A.; Castro, C.; Gong, Q.; Cooksey, K.; Linneman, L.; et al. Neonatal Necrotizing Enterocolitis-Associated DNA Methylation Signatures in the Colon Are Evident in Stool Samples of Affected Individuals. Epigenomics 2021, 13, 829–844. [Google Scholar] [CrossRef] [PubMed]
- Wood, E.H.; Chang, E.Y.; Beck, K.; Hadfield, B.R.; Quinn, A.R.; Harper, C.A. 80 Years of Vision: Preventing Blindness from Retinopathy of Prematurity. J. Perinatol. 2021, 41, 1216. [Google Scholar] [CrossRef] [PubMed]
- Teofili, L.; Papacci, P.; Bartolo, M.; Molisso, A.; Orlando, N.; Pane, L.; Giannantonio, C.; Serrao, F.; Bianchi, M.; Valentini, C.G.; et al. Transfusion-Free Survival Predicts Severe Retinopathy in Preterm Neonates. Front. Pediatr. 2022, 10, 814194. [Google Scholar] [CrossRef]
- Lundgren, P.; Athikarisamy, S.E.; Patole, S.; Lam, G.C.; Smith, L.E.; Simmer, K. Duration of Anaemia during the First Week of Life Is an Independent Risk Factor for Retinopathy of Prematurity. Acta Paediatr. 2018, 107, 759–766. [Google Scholar] [CrossRef]
- Gire, C.; Fournier, N.; Pirrello, J.; Marret, S.; Patural, H.; Flamant, C.; Pierrat, V.; Kaminski, M.; Ancel, P.-Y.; Tosello, B.; et al. Impact of Early Hemoglobin Levels on Neurodevelopment Outcomes of Two-Year-Olds in Very Preterm Children. Children 2023, 10, 209. [Google Scholar] [CrossRef] [PubMed]
- Asada, N. Tubular Immaturity Causes Erythropoietin-Deficiency Anemia of Prematurity in Preterm Neonates. Sci. Rep. 2018, 8, 4448. [Google Scholar] [CrossRef]
- Wang, X.; Jing, J.; Huang, S.; He, X.; Gao, P.; Li, H.; Lin, Z.; Sangild, P.T.; Zhu, Y. Relationship of Early Anemia with Neurodevelopment and Brain Injury in Very Low Birth Weight Preterm Infants—A Prospective Cohort Study. Nutrients 2022, 14, 4931. [Google Scholar] [CrossRef]
- Banerjee, J.; Asamoah, F.K.; Singhvi, D.; Kwan, A.W.; Morris, J.K.; Aladangady, N. Haemoglobin Level at Birth Is Associated with Short Term Outcomes and Mortality in Preterm Infants. BMC Med. 2013, 13, 16. [Google Scholar] [CrossRef]
- Song, J.; Dong, H.; Xu, F.; Wang, Y.; Li, W.; Jue, Z.; Wei, L.; Yue, Y.; Zhu, C. The Association of Severe Anemia, Red Blood Cell Transfusion and Necrotizing Enterocolitis in Neonates. PLoS ONE 2021, 16, e0254810. [Google Scholar] [CrossRef]
- Meyer, M.P.; O’Connor, K.L.; Meyer, J.H. Thresholds for Blood Transfusion in Extremely Preterm Infants: A Review of the Latest Evidence from Two Large Clinical Trials. Front. Pediatr. 2022, 10, 957585. [Google Scholar] [CrossRef] [PubMed]
- Scrivens, A.; Johanna Reibel, N.; Heeger, L.; Stanworth, S.; Lopriore, E.; New, H.V.; Dame, C.; Fijnvandraat, K.; Deschmann, E.; Aguar, M.; et al. Survey of Transfusion Practices in Preterm Infants in Europe On Behalf of the Neonatal Transfusion Network. Arch. Dis. Child. Fetal Neonatal Ed. 2023, 108, 360–366. [Google Scholar] [CrossRef]
- Fevereiro-Martins, M.; Santos, A.C.; Marques-Neves, C.; Guimarães, H.; Bicho, M.; Afonso, C.; Ferreira, J.; Santo, R.E.; Teixeira, F.; Rosa, R.; et al. Complete Blood Count Parameters as Biomarkers of Retinopathy of Prematurity: A Portuguese Multicenter Study. Graefes Arch. Clin. Exp. Ophthalmol. 2023. [Google Scholar] [CrossRef]
- Lust, C.; Vesoulis, Z.; Jackups, R.; Liao, S.; Rao, R.; Mathur, A.M. Early Red Cell Transfusion Is Associated with Development of Severe Retinopathy of Prematurity. J. Perinatol. 2019, 39, 393–400. [Google Scholar] [CrossRef]
- Hellström, W.; Martinsson, T.; Morsing, E.; Gränse, L.; Ley, D.; Hellström, A. Low Fraction of Fetal Haemoglobin Is Associated with Retinopathy of Prematurity in the Very Preterm Infant. Br. J. Ophthalmol. 2022, 106, 970–974. [Google Scholar] [CrossRef]
- Podraza, W.; Podraza, H.; Jezierska, K.; Szwed, J.; Modrzejewska, M.; Rudnicki, J.; Kordek, A.; Domek, H. The Role of Hemoglobin Variant Replacement in Retinopathy of Prematurity. Indian J. Pediatr. 2011, 78, 1498–1502. [Google Scholar] [CrossRef]
- Stutchfield, C.J.; Jain, A.; Odd, D.; Williams, C.; Markham, R. Foetal Haemoglobin, Blood Transfusion, and Retinopathy of Prematurity in Very Preterm Infants: A Pilot Prospective Cohort Study. Eye 2017, 31, 1451–1455. [Google Scholar] [CrossRef] [PubMed]
- Jiramongkolchai, K.; Repka, M.X.; Tian, J.; Aucott, S.W.; Shepard, J.; Collins, M.; Clemens, J.; Feller, M.; Burd, I.; Roizenblatt, M.; et al. Effects of Fetal Haemoglobin on Systemic Oxygenation in Preterm Infants and the Development of Retinopathy of Prematurity PacIFiHER Report No. 2. Br. J. Ophthalmol. 2023, 107, 380–383. [Google Scholar] [CrossRef]
- Scaglione, F.; Panzavolta, G. Folate, Folic Acid and 5-Methyltetrahydrofolate Are Not the Same Thing. Xenobiotica 2014, 44, 480–488. [Google Scholar] [CrossRef] [PubMed]
- McKay, J.A.; Groom, A.; Potter, C.; Coneyworth, L.J.; Ford, D.; Mathers, J.C.; Relton, C.L. Genetic and Non-Genetic Influences during Pregnancy on Infant Global and Site Specific DNA Methylation: Role for Folate Gene Variants and Vitamin B12. PLoS ONE 2012, 7, e33290. [Google Scholar] [CrossRef]
- Bi, H.; Hou, Y.; Wang, J.; Xia, Z.; Wang, D.; Liu, Y.; Bao, H.; Han, X.; Ren, K.; Li, E.; et al. Chromatin Reconstruction during Mouse Terminal Erythropoiesis. iScience 2022, 25, 105554. [Google Scholar] [CrossRef] [PubMed]
- Wells, M.; Steiner, L. Epigenetic and Transcriptional Control of Erythropoiesis. Front. Genet. 2022, 13, 805265. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Yu, L.; Engel, J.D.; Singh, S.A. Epigenetic Activities in Erythroid Cell Gene Regulation. Semin. Hematol. 2021, 58, 4–9. [Google Scholar] [CrossRef] [PubMed]
- Ryzhkova, A.; Battulin, N. Genome Reorganization during Erythroid Differentiation. Genes 2021, 12, 1012. [Google Scholar] [CrossRef]
- Gu, X.; Qiao, X.; Yu, S.; Song, X.; Wang, L.; Song, L. Histone Lysine-Specific Demethylase 1 Regulates the Proliferation of Hemocytes in the Oyster Crassostrea Gigas. Front. Immunol. 2022, 13, 1088149. [Google Scholar] [CrossRef]
- Yu, L.; Myers, G.; Ku, C.J.; Schneider, E.; Wang, Y.; Singh, S.A.; Jearawiriyapaisarn, N.; White, A.; Moriguchi, T.; Khoriaty, R.; et al. An Erythroid-to-Myeloid Cell Fate Conversion Is Elicited by LSD1 Inactivation. Blood 2021, 138, 1691–1704. [Google Scholar] [CrossRef]
- Çömez, A.; Yurttutan, S.; Seringec Akkececi, N.; Bozkaya, A.; Köküsarı, G.; Evgin, İ.; İpek, S. Red Cell Distribution Width and Its Association with Retinopathy of Prematurity. Int. Ophthalmol. 2021, 41, 699–706. [Google Scholar] [CrossRef]
- Akyüz Ünsal, A.İ.; Key, Ö.; Güler, D.; Omurlu, İ.K.; Anık, A.; Demirci, B.; Dündar, S. Can Complete Blood Count Parameters Predict Retinopathy of Prematurity? Turkish J. Ophthalmol. 2020, 50, 87–93. [Google Scholar] [CrossRef]
- Fava, C.; Cattazzo, F.; Hu, Z.-D.; Lippi, G.; Montagnana, M. The Role of Red Blood Cell Distribution Width (RDW) in Cardiovascular Risk Assessment: Useful or Hype? Ann. Transl. Med. 2019, 7, 581. [Google Scholar] [CrossRef]
- Xanthopoulos, A.; Giamouzis, G.; Dimos, A.; Skoularigki, E.; Starling, R.C.; Skoularigis, J.; Triposkiadis, F. Clinical Medicine Red Blood Cell Distribution Width in Heart Failure: Pathophysiology, Prognostic Role, Controversies and Dilemmas. J. Clin. Med. 2022, 11, 1951. [Google Scholar] [CrossRef]
- Said, A.S.; Spinella, P.C.; Hartman, M.E.; Steffen, K.M.; Jackups, R.; Holubkov, R.; Wallendorf, M.; Doctor, A. Red Blood Cell Distribution Width: Biomarker for Red Cell Dysfunction and Critical Illness Outcome? Pediatr. Crit. Care Med. 2017, 18, 134–142. [Google Scholar] [CrossRef] [PubMed]
- Yuan, H.; Fu, M.; Yang, X.; Huang, K.; Ren, X. Single Nucleotide Polymorphism of MTHFR Rs1801133 Associated with Elevated Hcy Levels Affects Susceptibility to Cerebral Small Vessel Disease. PeerJ 2020, 8, e8627. [Google Scholar] [CrossRef] [PubMed]
- Lubetzky, R.; Stolovitch, C.; Dollberg, S.; Mimouni, F.B.; Salomon, M.; Mandel, D. Nucleated Red Blood Cells in Preterm Infants with Retinopathy of Prematurity. Pediatrics 2005, 116, e619–e622. [Google Scholar] [CrossRef] [PubMed]
- Niranjan, H.S.; Bharath Kumar Reddy, K.R.; Benakappa, N.; Murthy, K.; Shivananda, S.; Veeranna, V. Role of Hematological Parameters in Predicting Retinopathy of Prematurity (ROP) in Preterm Neonates. Indian J. Pediatr. 2013, 80, 726–730. [Google Scholar] [CrossRef] [PubMed]
- García-Giménez, J.L.; Pallardó, F.V. Maintenance of Glutathione Levels and Its Importance in Epigenetic Regulation. Front. Pharmacol. 2014, 5, 88. [Google Scholar] [CrossRef] [PubMed]
- Tsantes, A.E.; Bonovas, S.; Travlou, A.; Sitaras, N.M. Redox Imbalance, Macrocytosis, and RBC Homeostasis. Antioxid. Redox Signal. 2006, 8, 1205–1216. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Gao, S.; Dong, M.; Luo, J.; Xu, C.; Wen, W.; Huang, Y.; Wu, Y.; Zhou, J.; Yuan, Z. Relationship between Red Blood Cell Indices (MCV, MCH, and MCHC) and Major Adverse Cardiovascular Events in Anemic and Nonanemic Patients with Acute Coronary Syndrome. Dis. Markers 2022, 2022, 2193343. [Google Scholar] [CrossRef]
- SPN. Consenso Clínico Retinopatia da Prematuridade. Available online: https://www.spneonatologia.pt/wp-content/uploads/2016/11/2014-ROP.pdf (accessed on 12 February 2023).
- An International Classification of Retinopathy of Prematurity. The Committee for the Classification of Retinopathy of Prematurity. Arch. Ophthalmol. 1984, 102, 1130–1134. [CrossRef]
- Quinn, G.E. The International Classification of Retinopathy of Prematurity Revisited. Arch. Ophthalmol. 2005, 123, 991–999. [Google Scholar] [CrossRef]
- Good, W.V.; Hardy, R.J.; Dobson, V.; Palmer, E.A.; Phelps, D.L.; Quintos, M.; Tung, B. Revised Indications for the Treatment of Retinopathy of Prematurity: Results of the Early Treatment for Retinopathy of Prematurity Randomized Trial. Arch. Ophthalmol. 2003, 121, 1684–1696. [Google Scholar] [CrossRef]
| Clinical Characteristics n (%) or n; Median, Interquartile Range | Total | No ROP | ROP | p-Value | p-Value * |
|---|---|---|---|---|---|
| Number of individuals n (%) | 396 | 238 (60.1) | 158 (39.9) | n.a. | n.a. |
| Gestational age (weeks) | 396; 29.5, 3.1 | 238; 30.4, 2.3 | 158; 28.0, 2.9 | <0.001 | n.a. |
| Birth weight (g) | 396; 1187.5, 444.3 | 238; 1300.0, 381.0 | 158; 980.0, 434.3 | <0.001 | 0.016 |
| Gender | |||||
| Male | 198 (50.0) | 126 (52.9) | 72 (45.6) | 0.182 | 0.139 |
| Female | 198 (50.0) | 112 (47.1) | 86 (54.4) | ||
| Apgar at minute 5 | |||||
| ≥7 | 353 (90.5) | 223 (94.5) | 130 (84.4) | 0.001 | 0.298 |
| <7 | 37 (9.5) | 13 (5.5) | 24 (15,6) | ||
| Assisted reproductive technologies (yes) | 48 (14.3) | 26 (54.2) | 22 (45.8) | 0.753 | 0.724 |
| Twin or multiple births (yes) | 112 (28.4) | 77 (68.8) | 35 (31.3) | 0.04 | 0.240 |
| Type of delivery (Cesarean) | 269 (68.3) | 165 (61.3) | 104 (38.7) | 0.538 | 0.705 |
| Maternal chorioamnionitis (yes) | 43 (11.0) | 23 (53.5) | 20 (46.5) | 0.328 | 0.999 |
| Prenatal steroids (yes) | 366 (92.7) | 223 (60.9) | 143 (39.1) | 0.332 | 0.169 |
| Prenatal magnesium (yes) | 147 (63.4) | 81 (55.1) | 66 (44.9) | 0.168 | 0.322 |
| Days of oxygen ventilation | 394; 16.5, 43 | 236; 7.0, 24 | 158; 42.5, 51 | <0.001 | 0.438 |
| Days with glycemia ≥125 mg/dL | 396; 1.0, 3 | 238, 1.0, 2 | 158; 2.5, 5 | <0.001 | 0.400 |
| Days of red cell transfusions | 396; 0.0, 1 | 238; 0.0, 0 | 158; 1.0, 4 | <0.001 | n.a. |
| Days of platelets transfusions | 396; 0.2, 0 | 238; 0.1, 0 | 158; 0.4, 0 | <0.001 | 0.082 |
| Bronchopulmonary dysplasia (yes) | 81 (20.7) | 22 (27.2) | 59 (72.8) | <0.001 | 0.978 |
| Necrotizing enterocolitis (yes) | 23 (5.9) | 9 (39.1) | 14 (60.9) | 0.045 | 0.847 |
| Periventricular/intraventricular hemorrhage (yes) | 47 (12.1) | 16 (34.0) | 31 (66.0) | <0.001 | 0.478 |
| Patent ductus arteriosus (yes) | 55 (14.1) | 15 (27.3) | 40 (72.7) | <0.001 | 0.218 |
| Periventricular leukomalacia (yes) | 12 (3.1) | 4 (33.3) | 8 (66.7) | 0.071 | 0.656 |
| Hematological Parameters n (%) or n; Median, Interquartile Range | Total | No ROP | ROP | p-Value | p-Value * |
|---|---|---|---|---|---|
| Hemoglobin (g/dL) | 343 | 197; 16.1, 3.1 | 146; 14.8, 3.4 | <0.001 | 0.240 |
| Red cell distribution width (%) | 325 | 184; 16.5, 2.2 | 141; 17.1, 2.9 | 0.001 | 0.031 |
| Erythroblasts (× 109/L) | 310 | 176; 7.8, 15.9 | 134; 14.8, 32.2 | <0.001 | 0.014 |
| Mean corpuscular hemoglobin (pg) | 326 | 185; 37.7, 3.3 | 141; 37.3, 4.0 | 0.146 | 0.579 |
| Mean corpuscular hemoglobin concentration (pg) | 328 | 186; 35.2, 1.6 | 142; 33.9, 2.7 | <0.001 | <0.001 |
| Mean corpuscular volume (fl) | 324 | 184; 107.4, 9.6 | 140; 108.6, 12.2 | 0.022 | 0.026 |
| (a) | ||||||
| Without ROP N (%) | With ROP N (%) | OR [CI 95%] | p-Value | p-Value * | p-Value # | |
| Hemoglobin ≥ 14.5 g/dL | 148 (63.8) | 55 (35.0) | 1 | <0.001 | 0.157 | 0.063 |
| Hemoglobin < 14.5 g/dL | 84 (36.2) | 102 (65.0) | 3.23 [2.14–4.99] | |||
| (b) | ||||||
| DNMT3B N (%) | Hemoglobin ≥ 14.5 g/dL | Hemoglobin < 14.5 g/dL | OR [CI 95%] | p-value | p-value * | p-value # |
| CC | 65 (33.0) | 38 (20.7) | 1 | 0.008 | 0.073 | 0.043 |
| Allele T | 132 (67.0) | 146 (79.3) | 1.89 [1.19–3.01] | |||
| TT | 46 (23.4) | 46 (25.0) | n.a | 0.721 | 0.696 | 0.664 |
| Allele C | 151 (76.6) | 138 (75.0) | n.a | |||
| CC + TT | 111 (56.3) | 84 (45.7) | 1 | 0.041 | 0.183 | 0.144 |
| CT | 86 (43.7) | 100 (54.3) | 1.54 [1.03–2.30] | |||
| Genetic Polymorphisms | ||||||
|---|---|---|---|---|---|---|
| Polymorphism | Genotype | Without ROP N (%) | With ROP N (%) | OR [CI 95%] | p-Value | p-Value * |
| DNMT3B | TT | 52 (22.2) | 41 (26.6) | n.a. | 0.333 | 0.412 |
| Allele C | 182 (77.8) | 113 (73.4) | n.a. | |||
| CC | 74 (31.6) | 33 (21.4) | 1 | 0.029 | 0.193 | |
| Allele T | 160 (68.4) | 121 (78.6) | 2.16 [1.06–2.72] | |||
| CT | 108 (46.2) | 80 (51.9) | n.a. | 0.299 | 0.695 | |
| CC + TT | 126 (53.8) | 74 (48.1) | n.a. | |||
| KDM1A | AA | 32 (13.5) | 24 (15.2) | n.a. | 0.661 | 0.406 |
| Allele T | 205 (86.5) | 134 (84.8) | n.a. | |||
| TT | 98 (41.4) | 65 (41.1) | n.a. | 0.967 | 0.393 | |
| Allele A | 139 (58.6) | 93 (58.9) | n.a. | |||
| TA | 107 (45.1) | 69 (43.7) | n.a. | 0.836 | 0.151 | |
| AA + TT | 130 (54.9) | 89 (56.3) | n.a. | |||
| MTHFR | TT | 18 (7.7) | 14 (8.9) | n.a. | 0.709 | 0.679 |
| Allele C | 216 (92.3) | 143 (91.1) | n.a. | |||
| CC | 128 (54.7) | 73 (46.5) | n.a. | 0.122 | 0.554 | |
| Allele T | 106 (45.3) | 84 (53.5) | n.a. | |||
| CT | 88 (37.6) | 70 (44.6) | n.a. | 0.173 | 0.700 | |
| CC + TT | 146 (62.4) | 87 (55.4) | n.a. | |||
| Epistatic Relations between Polymorphisms and ROP | |||||
|---|---|---|---|---|---|
| Epistatic Relations | No ROP N (%) | ROP N (%) | OR [CI 95%] | p-Value | p-Value * |
| DNMT3B (Allele T) + MTHFR (CC) + KDM1A (AA) | 10 (4.3) | 13 (8.5) | n.a. | 0.123 | n.a. |
| Other genotypes | 221 (95.7) | 140 (91.5) | n.a. | ||
| DNMT3B (Allele T) + KDM1A (AA) | 19 (8.1) | 19 (12.4) | n.a. | 0.168 | n.a. |
| Other genotypes | 215 (91.9) | 134 (87.6) | n.a. | ||
| DNMT3B (Allele T) + MTHFR (CC) | 88 (37.9) | 61 (39.6) | n.a. | 0.750 | n.a. |
| Other genotypes | 144 (62.1) | 93 (60.4) | n.a. | n.a. | |
| KDM1A (AA) + MTHFR (CC) | 18 (7.7) | 15 (9.6) | n.a. | 0.579 | n.a. |
| Other genotypes | 215 (92.3) | 142 (90.4) | n.a. | n.a. | |
| Polymorphism | Genotype | RDW (%) n; Median, Interquartile Range | p-Value | p-Value * | Erythroblasts (× 109/L) n; Median, Interquartile Range | p-Value | p-Value * | MCH (pg) n; Median, Interquartile Range | p-Value | p-Value * | MCHC (pg) n; Median, Interquartile Range | p-Value | p-Value * |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DNMT3B | TT | 80; 16.0, 1.6 | 0.036 | 0.025 | 75; 10.1, 19.4 | 0.867 | 0.307 | 80; 37.1, 4.6 | 0.274 | 0.232 | 83; 34.8, 2.3 | 0.895 | 0.965 |
| Allele C | 237; 16.7, 2.3 | 228; 10.8, 22.8 | 238; 37.5, 3.3 | 237; 34.9, 2.4 | |||||||||
| CC | 86; 16.4, 2.2 | 0.592 | 0.414 | 82; 10.6, 15.7 | 0.221 | 0.564 | 86; 37.4, 2.8 | 0.279 | 0.291 | 86; 35.2, 2.3 | 0.044 | 0.153 | |
| Allele T | 231; 16.1, 1.9 | 221; 10.4, 27.1 | 232; 37.5, 4.0 | 234; 34.8, 2.3 | |||||||||
| CT | 151; 17.0, 2.9 | 0.179 | 0.196 | 146; 11.0, 31.6 | 0.218 | 0.665 | 166; 37.5, 3.9 | 0.989 | 0.912 | 169; 34.8, 2.3 | 0.095 | 0.216 | |
| CC + TT | 166; 16.6, 2.1 | 157; 10.2, 16.6 | 152; 37.3, 3.6 | 151; 35.1, 2.2 | |||||||||
| KDM1A | AA | 41; 17.1, 2.8 | 0.037 | 0.039 | 40; 18.5, 34.9 | 0.019 | 0.044 | 41; 38.2, 4.4 | 0.569 | 0.583 | 42; 34.8, 2.1 | 0.728 | 0.624 |
| Allele T | 283; 16.2, 1.9 | 269; 9.4, 34.9 | 28437.5, 3.4 | 285; 34.9, 2.4 | |||||||||
| TT | 135; 16.4, 1.8 | 0.517 | 0.454 | 130; 9.5, 24.5 | 0.772 | 0.555 | 135; 37.6, 3.7 | 0.581 | 0.430 | 137; 34.9, 2.5 | 0.845 | 0.409 | |
| Allele A | 189; 16.2, 2.2 | 179; 10.9, 22.0 | 190; 37.4, 3.7 | 190; 34.9, 2.2 | |||||||||
| TA | 148; 16.6, 2.2 | 0.452 | 0.510 | 139; 9.4, 17.3 | 0.193 | 0.057 | 149; 37.3, 3.3 | 0.206 | 0.253 | 148; 34.9, 2.3 | 0.194 | 0.627 | |
| AA + TT | 176; 16.8, 2.6 | 170; 12.6, 28.9 | 176; 37.6, 3.9 | 179; 34.9, 2.5 | |||||||||
| MTHFR | TT | 25; 15.8, 1.2 | 0.154 | 0.103 | 24; 5.6, 17.9 | 0.131 | 0.197 | 25; 38.1, 4.7 | 0.473 | 0.535 | 25; 34.9, 2.0 | 0.565 | 0.414 |
| Allele C | 295; 16.3, 2.1 | 282; 11.2, 23.1 | 296; 37.4, 3.7 | 298; 34.9, 2.4 | |||||||||
| CC | 162; 16.6, 2.6 | 0.004 | 0.002 | 157; 23.5, 27.8 | 0.004 | 0.090 | 162; 37.3, 3.4 | 0.115 | 0.095 | 165; 34.8, 2.5 | 0.132 | 0.124 | |
| Allele T | 158; 16.0, 2.0 | 149; 8.2, 17.3 | 159; 37.7, 3.8 | 158; 34.9, 2.0 | |||||||||
| CT | 133; 16.6, 2.3 | 0.031 | 0.023 | 125; 8.3, 18.1 | 0.038 | 0.295 | 134; 37.6, 3.6 | 0.227 | 0.173 | 133; 34.9, 2.0 | 0.224 | 0.064 | |
| CC + TT | 187; 16.8, 2.7 | 181; 13.4, 26.7 | 187; 37.4, 3.5 | 190; 34.8, 2.5 |
| Epistatic Relationships | RDW (%) n; Median, Interquartile Range | p-Value | p-Value * | Erythroblasts (× 109/L) n; Median, Interquartile Range | p-Value | p-Value * | MCH (pg) n; Median, Interquartile Range | p-Value | p-Value * | MCHC (pg) n; Median, Interquartile Range | p-Value | p-Value * |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DNMT3B (Allele T) + MTHFR (CC) | 121; 17.0, 2.8 | 0.062 | 0.543 | 118; 13.8, 32.9 | 0.005 | 0.073 | 121; 37.1, 3.6 | 0.031 | 0.035 | 124; 34.4, 2.4 | 0.032 | 0.027 |
| Other genotypes | 194; 16.6, 2.4 | 183; 9.0, 17.6 | 195; 37.6, 3.5 | 194; 35.0, 2.1 | ||||||||
| KDM1A (AA) + MTHFR (CC) | 23; 17.8, 5.0 | 0.019 | 0.019 | 21; 20.8, 13.4 | 0.006 | 0.006 | 23; 38.5, 4.6 | 0.349 | 0.334 | 24; 34.8, 2.4 | 0.507 | 0.498 |
| Other genotypes | 296; 16.7, 2.3 | 284; 9.6, 22.1 | 297; 37.4, 3.5 | 298; 34.9, 2.4 | ||||||||
| DNMT3B (Allele T) + KDM1A (AA) | 28; 17.7, 2.3 | 0.006 | 0.009 | 28; 26.0, 35.8 | 0.001 | 0.019 | 28; 38.0, 4.5 | 0.944 | 0.100 | 29; 34.9, 2.4 | 0.219 | 0.309 |
| Other genotypes | 288; 16.7, 2.3 | 274; 9.4, 19.4 | 289; 37.5, 3.5 | 290; 34.5, 2.4 | ||||||||
| DNMT3B (Allele T) + MTHFR (CC) + KDM1A (AA) | 16; 17.9, 4.8 | 0.040 | 0.009 | 16; 26,7, 22.7 | 0.002 | 0.003 | 16; 38.9, 4.9 | 0.049 | 0.233 | 17; 34.2, 2.6 | 0.134 | 0.854 |
| Other genotypes | 297; 16.7, 2.3 | 283; 9.7, 21.4 | 298; 37.4, 3.5 | 299; 34.9, 2.4 |
| (a) Genetic Polymorphisms in the Group without ROP | |||||||||||||
| Polymorphism | Genotype | RDW (%) n; Median, Interquartile Range | p-Value | p-Value * | Erythroblasts (× 109/L) n; Median, Interquartile Range | p-Value | p-Value * | MCH (pg) n; Median, Interquartile Range | p-Value | p-Value * | MCV (fl) n; Median, Interquartile Range | p-Value | p-Value * |
| DNMT3B | TT | 42; 16.5, 1.9 | 0.585 | 0.355 | 40; 6.6, 17.9 | 0.911 | 0.515 | 42; 37.7, 4.2 | 0.487 | 0.400 | 42; 107.1, 9.8 | 0.547 | 0.469 |
| Allele C | 138; 16.6, 2.4 | 133; 8.3, 16.2 | 139; 37.7, 3.2 | 138; 107.6, 9.7 | |||||||||
| CC | 57; 16.6, 2.2 | 0.359 | 0.338 | 54; 8.0, 14.9 | 0.503 | 0.561 | 57; 37.5, 3.5 | 0.372 | 0.304 | 57; 107.7, 10.2 | 0.599 | 0.511 | |
| Allele T | 123; 16.5, 2.1 | 119; 8.0, 16.6 | 124; 37.7, 3.6 | 123; 107.2, 9.6 | |||||||||
| CT | 81; 16.4, 2.4 | 0.695 | 0.894 | 79; 8.3, 16.6 | 0.473 | 0.311 | 82; 37.7, 3.3 | 0.807 | 0.776 | 81; 107.3, 9.6 | 0.984 | 0.966 | |
| CC + TT | 99; 16.6, 1.9 | 94; 7.2, 16.4 | 99; 37.6, 3.5 | 99; 107.6, 9.7 | |||||||||
| KDM1A | AA | 23; 16.5, 2.5 | 0.997 | 0.989 | 22; 7.9, 23.8 | 0.676 | 0.789 | 23; 38.2, 3.3 | 0.702 | 0.560 | 23; 105.9, 6.1 | 0.110 | 0.117 |
| Allele T | 160; 16.6, 2.0 | 153; 7.6, 15.5 | 161; 37.7, 3.3 | 160; 107.9, 9.9 | |||||||||
| TT | 77; 16.7, 1.8 | 0.869 | 0.575 | 74; 7.8, 16.1 | 0.945 | 0.796 | 77; 37.8, 3.0 | 0.801 | 0.639 | 76; 108.1, 9.0 | 0.203 | 0.210 | |
| Allele A | 106; 16.5, 2.4 | 101; 7.6, 15.1 | 107; 37.7, 3.6 | 107; 107.1, 9.4 | |||||||||
| TA | 83; 16.4, 2.4 | 0.868 | 0.589 | 79; 7.6, 12.3 | 0.728 | 0.946 | 84; 37.6, 3.6 | 0.997 | 0.925 | 84; 107.5, 10.1 | 0.845 | 0.829 | |
| AA + TT | 100; 16.7, 2.0 | 96; 7.8, 16.7 | 100; 37.8, 3.0 | 99; 107.6, 8.5 | |||||||||
| MTHFR | TT | 13; 16.2, 1.4 | 0.041 | 0.046 | 13; 4.4, 5.7 | 0.068 | 0.385 | 13; 38.9, 3.5 | 0.011 | 0.004 | 13; 110.3, 7.0 | 0.060 | 0.082 |
| Allele C | 167; 16.7, 2.2 | 160; 8.3, 16.7 | 168; 37.5, 3.4 | 167; 107.1, 9.4 | |||||||||
| CC | 97; 16.7, 2.5 | 0.079 | 0.057 | 93; 10.4, 19.3 | 0.022 | 0.075 | 97; 37.2, 2.7 | 0.002 | 0.002 | 97; 106.7, 9.0 | 0.033 | 0.020 | |
| Allele T | 83; 16.5, 2.1 | 80; 6.3, 9.4 | 84; 38.4, 3.6 | 83; 108.3, 9.5 | |||||||||
| CT | 70; 16.5, 2.1 | 0.475 | 0.222 | 67; 7.0, 9.6 | 0.175 | 0.112 | 71; 38.2, 3.8 | 0.070 | 0.069 | 70; 108.1, 10.1 | 0.239 | 0.155 | |
| TT + CC | 110; 16.5, 2.2 | 106; 9.2, 17.5 | 110; 37.4, 2.8 | 110; 107.1, 9.3 | |||||||||
| (b) Genetic polymorphisms in the group with ROP | |||||||||||||
| Polymorphism | Genotype | RDW (%) n; median, interquartile range | p-value | p-value * | Erythroblasts (× 109/L) n; median, interquartile range | p-value | p-value * | MCH (pg) n; median, interquartile range | p-value | p-value * | MCV (fl) n; median, interquartile range | p-value | p-value * |
| DNMT3B | TT | 38; 16.4, 2.1 | 0.010 | 0.015 | 35; 14.2, 20.5 | 0.551 | 0.297 | 38; 36.7, 4.2 | 0.428 | 0.417 | 37; 108.1, 13.3 | 0.383 | 0.312 |
| Allele C | 99; 17.6, 2.9 | 95; 14.9, 41.9 | 99; 37.4, 3.7 | 99; 109.2, 12.1 | |||||||||
| CC | 29; 17.0, 2.9 | 0.782 | 0.446 | 28; 14.3, 18.2 | 0.558 | 0.261 | 29; 37.1, 1.9 | 0.602 | 0.739 | 29; 108.4, 8.0 | 0.960 | 0.827 | |
| Allele T | 108; 17.2, 2.8 | 102; 15.2, 33.6 | 108; 37.2, 4.5 | 107; 109.2, 15.0 | |||||||||
| CT | 70; 17.6, 3.0 | 0.037 | 0.104 | 63; 18.5, 44.5 | 0.312 | 0.983 | 67; 37.4, 4.4 | 0.776 | 0.622 | 66; 109.4, 15.3 | 0.413 | 0.261 | |
| CC + TT | 67; 16.8, 2.4 | 67; 14.2, 19.4 | 70; 37.0, 3.6 | 70;108.2, 9.6 | |||||||||
| KDM1A | AA | 18; 19.1, 3.1 | 0.002 | 0.006 | 18; 30.9, 213.2 | 0.002 | 0.064 | 18; 38.9, 4.9 | 0.232 | 0.197 | 18; 112.5, 15.6 | 0.112 | 0.075 |
| Allele T | 123; 16.9, 2.7 | 116; 14.0, 30.2 | 123; 37.1, 3.6 | 122; 108.1, 10.4 | |||||||||
| TT | 58; 16.8, 2.5 | 0.483 | 0.659 | 56; 14.6, 42.8 | 0.740 | 0.473 | 58; 37.3, 5.2 | 0.695 | 0.542 | 58; 109.6, 13.7 | 0.533 | 0.759 | |
| Allele A | 83; 17.5, 3.1 | 78; 14.9, 27.8 | 83; 37.1, 3.8 | 82; 108.1, 10.3 | |||||||||
| TA | 65; 17.0, 2.6 | 0.166 | 0.132 | 60; 12.9, 20.1 | 0.071 | 0.053 | 65; 37.4, 5.1 | 0.235 | 0.147 | 64; 107.5, 9.3 | 0.092 | 0.135 | |
| AA + TT | 76; 17.4, 3.0 | 74; 18.2, 58.8 | 76; 37.0, 3.4 | 76; 109.6, 13.7 | |||||||||
| MTHFR | TT | 12; 16.8, 1.8 | 0.864 | 0.767 | 11; 14.6, 30.2 | 0.636 | 0.298 | 12; 36.0, 5.2 | 0.124 | 0.111 | 12; 103.2, 14.6 | 0.073 | 0.095 |
| Allele C | 128; 17.2, 3.0 | 122; 14.9, 33.9 | 128; 37.3, 4.3 | 127; 109.2, 13.0 | |||||||||
| CC | 65; 17.6, 2.7 | 0.008 | 0.019 | 64; 17.9, 62.7 | 0.031 | 0.218 | 65;37.6, 4.7 | 0.472 | 0.461 | 64; 109.6, 15.0 | 0.079 | 0.059 | |
| Allele T | 75; 16.7, 2.8 | 69; 13.8, 22.8 | 75; 37.1, 3.5 | 75; 108.1, 10.1 | |||||||||
| CT | 63; 16.6, 3.1 | 0.011 | 0.031 | 58; 13.3, 22.4 | 0.057 | 0.508 | 63; 37.1, 3.2 | 0.885 | 0.877 | 63; 108.1, 8.9 | 0.455 | 0.296 | |
| CC + TT | 77; 17.6, 2.6 | 133; 17.8, 42.5 | 77; 37.4, 4.8 | 76; 109.4, 15.8 | |||||||||
| (a) Epistatic Relationships in the Group without ROP | ||||||||||||
| Epistatic Relationships | RDW (%) n; Median, Interquartile Range | p-Value | p-Value * | Erythroblasts (× 109/L) n; Median, Interquartile Range | p-Value | p-Value * | MCH (pg) n; Median, Interquartile Range | p-Value | p-Value * | MCV (fl) n; Median, Interquartile Range | p-Value | p-Value * |
| DNMT3B (Allele T) + MTHFR (CC) | 66; 16.6, 2.2 | 0.750 | 0.497 | 64; 9.9, 21.9 | 0.092 | 0.068 | 66; 37.0, 3.0 | <0.001 | 0.001 | 66; 106.3, 9.2 | 0.006 | 0.003 |
| Other genotypes | 112; 16.5, 2.2 | 107; 7.0 12.4 | 113; 38.2, 3.6 | 112; 108.2, 9.6 | ||||||||
| KDM1A (AA) + MTHFR (CC) | 12; 16.3, 4.6 | 0.598 | 0.324 | 10; 10.3, 16.4 | 0.470 | 0.950 | 12; 38.2, 4.4 | 0.328 | 0.140 | 12; 105.6, 11.6 | 0.144 | 0.141 |
| Other genotypes | 167; 16.6, 2.1 | 162; 7.3, 16.2 | 168; 37.7, 3.3 | 167; 107.7, 9.5 | ||||||||
| DNMT3B (Allele C) + KDM1A (AA) | 13; 17.4, 2.5 | 0.270 | 0.249 | 13; 12.0, 34.3 | 0.080 | 0.369 | 13; 37.2, 3.0 | 0.302 | 0.406 | 13; 107.0, 8.2 | 0.309 | 0.317 |
| Other genotypes | 167; 16.6, 2.1 | 160; 7.3, 16.7 | 168; 37.7, 3.3 | 167; 107.7, 9.8 | ||||||||
| DNMT3B (Allele C) + MTHFR (CC) + KDM1A (AA) | 6; 16.6, 4.7 | 0.448 | 0.096 | 6; 10.3, 25.6 | 0.325 | 0.770 | 6; 36.8, 4.2 | 0.371 | 0.327 | 6; 104.1, 12.3 | 0.384 | 0.431 |
| Other genotypes | 171; 16.6, 2.2 | 164; 7.5, 16.7 | 172; 37.7, 3.3 | 171; 107.6, 9.7 | ||||||||
| (b) Epistatic relationships in the group with ROP | ||||||||||||
| Epistatic relationships | RDW (%) n; median, interquartile range | p-value | p-value * | Erythroblasts (× 109/L) n; median, interquartile range | p-value | p-value * | MCH (pg) n; median, interquartile range | p-value | p-value * | MCV (fl) n; median, interquartile range | p-value | p-value * |
| DNMT3B (Allele C) + MTHFR (CC) | 55; 17.6, 2.8 | 0.076 | 0.084 | 54; 18.2, 8.5 | 0.034 | 0.265 | 55; 37.4, 5.0 | 0.642 | 0.664 | 54; 109.6, 15.2 | 0.103 | 0.100 |
| Other genotypes | 82; 16.8, 2.8 | 76; 13.9, 21.7 | 82; 37.1, 3.2 | 82; 108.1, 10.0 | ||||||||
| KDM1A (AA) + MTHFR (CC) | 11; 19.5, 4.8 | 0.037 | 0.021 | 11; 60.0, 35.9 | 0.001 | 0.013 | 11; 39.7, 3.0 | 0.008 | 0.014 | 11; 119.2, 13.9 | 0.002 | 0.003 |
| Other genotypes | 129; 17.0, 2.7 | 122; 14.6, 39.6 | 129; 37.4, 3.5 | 128; 108.1, 10.5 | ||||||||
| DNMT3B (Allele C) + KDM1A (AA) | 15; 19.2, 3.2 | 0.011 | 0.028 | 15; 26.4, 21.5 | 0.006 | 0.062 | 15; 39.0, 4.7 | 0.350 | 0.270 | 15; 115.6, 13.1 | 0.106 | 0.071 |
| Other genotypes | 121; 17.0, 2.7 | 114; 14.4, 30.8 | 121; 37.0, 3.6 | 120; 108.1, 10.8 | ||||||||
| DNMT3B (Allele C) + MTHFR (CC) + KDM1A (AA) | 10; 18.7, 4.5 | 0.066 | 0.063 | 10; 132.6, 25.1 | 0.003 | 0.412 | 10; 39.4, 3.1 | 0.020 | 0.038 | 10; 118.8, 14.9 | 0.005 | 0.007 |
| Other genotypes | 126; 17.0, 2.7 | 119; 14.6, 29.7 | 126; 37.0, 3.5 | 125; 108.1, 10.9 | ||||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fevereiro-Martins, M.; Santos, A.C.; Marques-Neves, C.; Guimarães, H.; Bicho, M.; on behalf of the GenE-ROP Study Group. Genetic Modulation of the Erythrocyte Phenotype Associated with Retinopathy of Prematurity—A Multicenter Portuguese Cohort Study. Int. J. Mol. Sci. 2023, 24, 11817. https://doi.org/10.3390/ijms241411817
Fevereiro-Martins M, Santos AC, Marques-Neves C, Guimarães H, Bicho M, on behalf of the GenE-ROP Study Group. Genetic Modulation of the Erythrocyte Phenotype Associated with Retinopathy of Prematurity—A Multicenter Portuguese Cohort Study. International Journal of Molecular Sciences. 2023; 24(14):11817. https://doi.org/10.3390/ijms241411817
Chicago/Turabian StyleFevereiro-Martins, Mariza, Ana Carolina Santos, Carlos Marques-Neves, Hercília Guimarães, Manuel Bicho, and on behalf of the GenE-ROP Study Group. 2023. "Genetic Modulation of the Erythrocyte Phenotype Associated with Retinopathy of Prematurity—A Multicenter Portuguese Cohort Study" International Journal of Molecular Sciences 24, no. 14: 11817. https://doi.org/10.3390/ijms241411817
APA StyleFevereiro-Martins, M., Santos, A. C., Marques-Neves, C., Guimarães, H., Bicho, M., & on behalf of the GenE-ROP Study Group. (2023). Genetic Modulation of the Erythrocyte Phenotype Associated with Retinopathy of Prematurity—A Multicenter Portuguese Cohort Study. International Journal of Molecular Sciences, 24(14), 11817. https://doi.org/10.3390/ijms241411817









