Loss of ANCO1 Expression Regulates Chromatin Accessibility and Drives Progression of Early-Stage Triple-Negative Breast Cancer
Abstract
1. Introduction
2. Results
2.1. High ANCO1 Expression Is a Positive Prognostic Indicator in Breast Cancer Patients
2.2. Reduction in ANCO1 Causes Abnormal Cell Morphology and Leads to Aneuploidy and Senescence
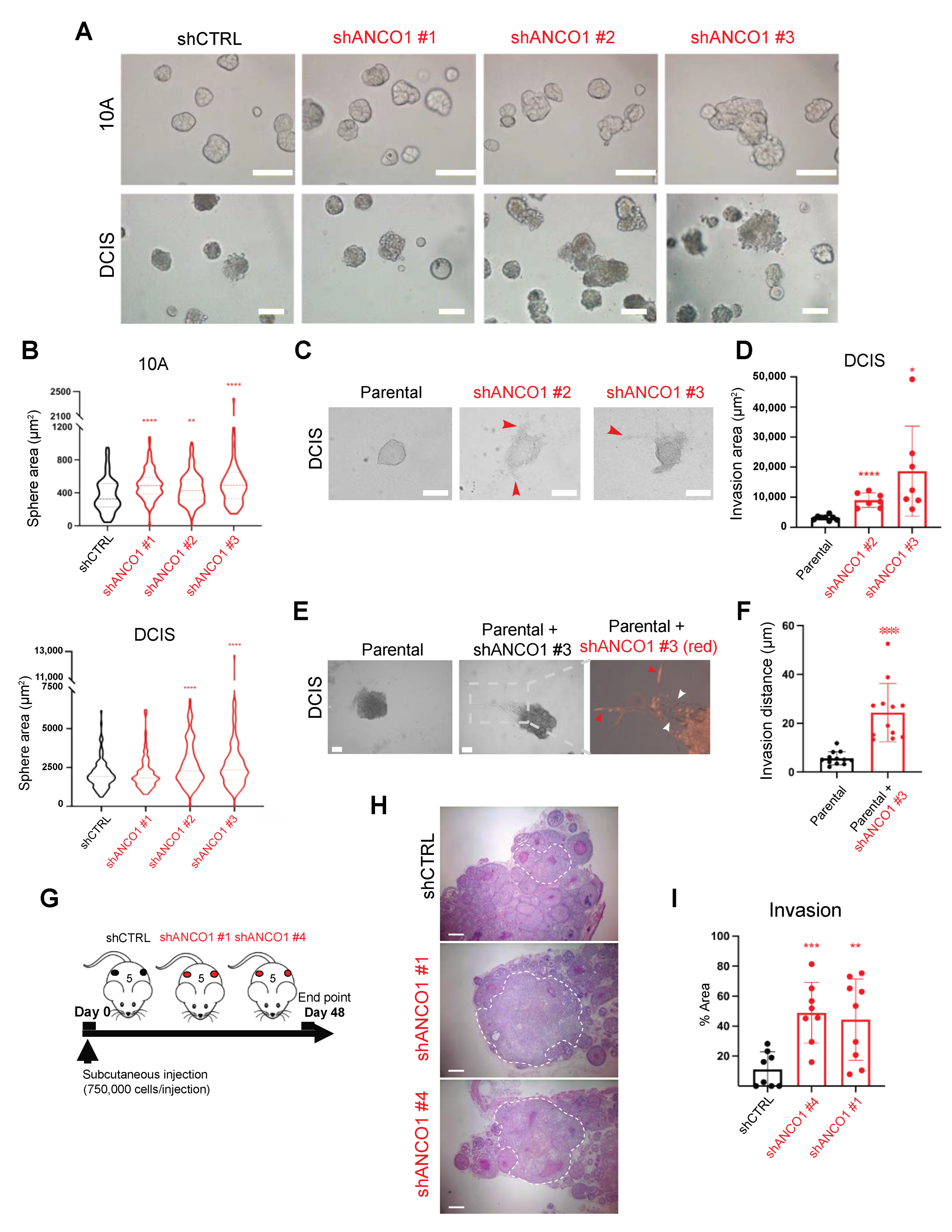
2.3. Reduction in ANCO1 Expression Leads to Aberrant Sphere Formation and Enables Invasion In Vitro
2.4. Loss of ANCO1 Expression Enables Collective Invasion in 3D Spheres
2.5. Loss of ANCO1 Expression Enhances Invasion In Vivo
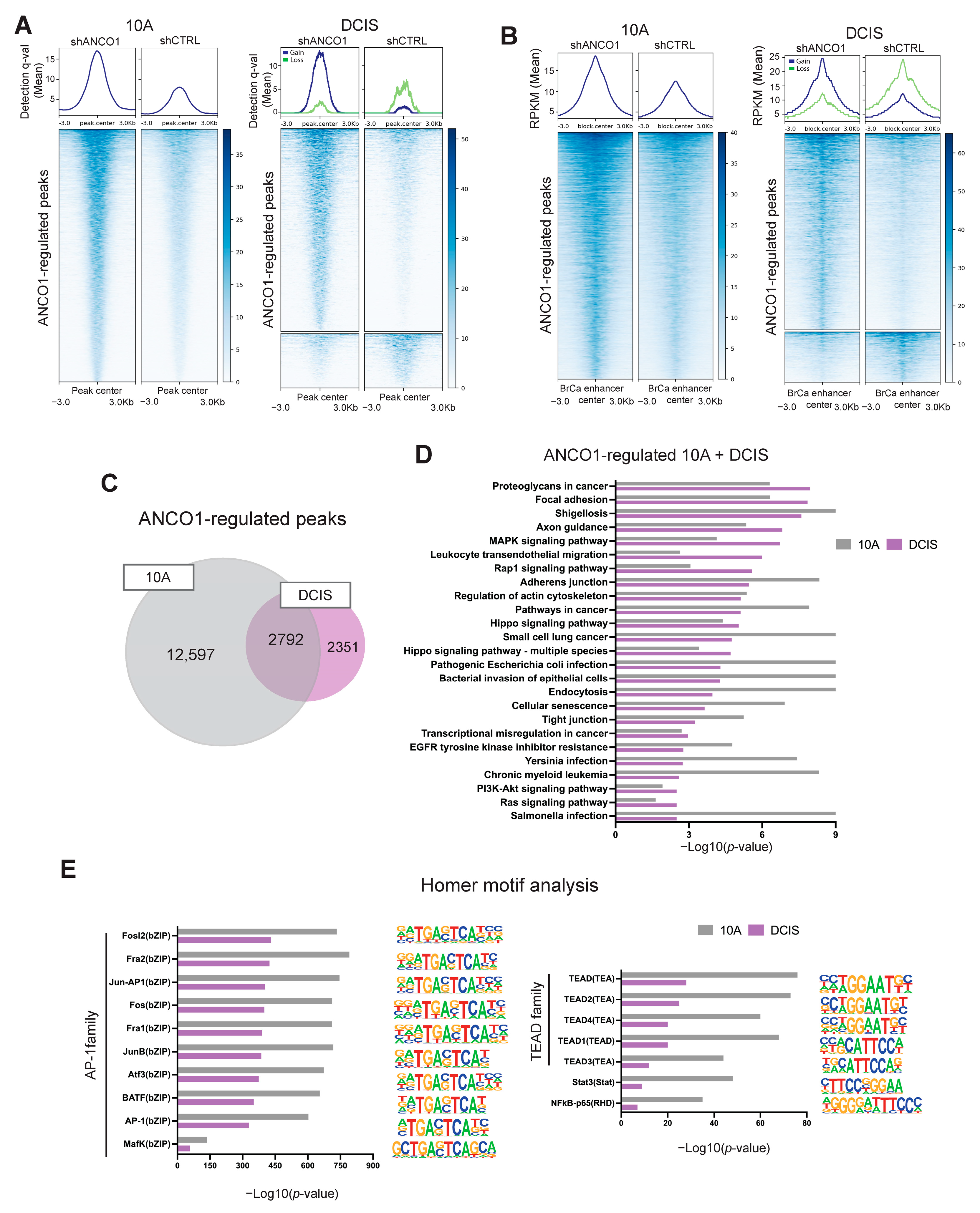
2.6. ANCO1 Reduction Increases Chromatin Accessibility and Promotes H3K27Ac Binding at Known Breast Cancer Enhancer Regions
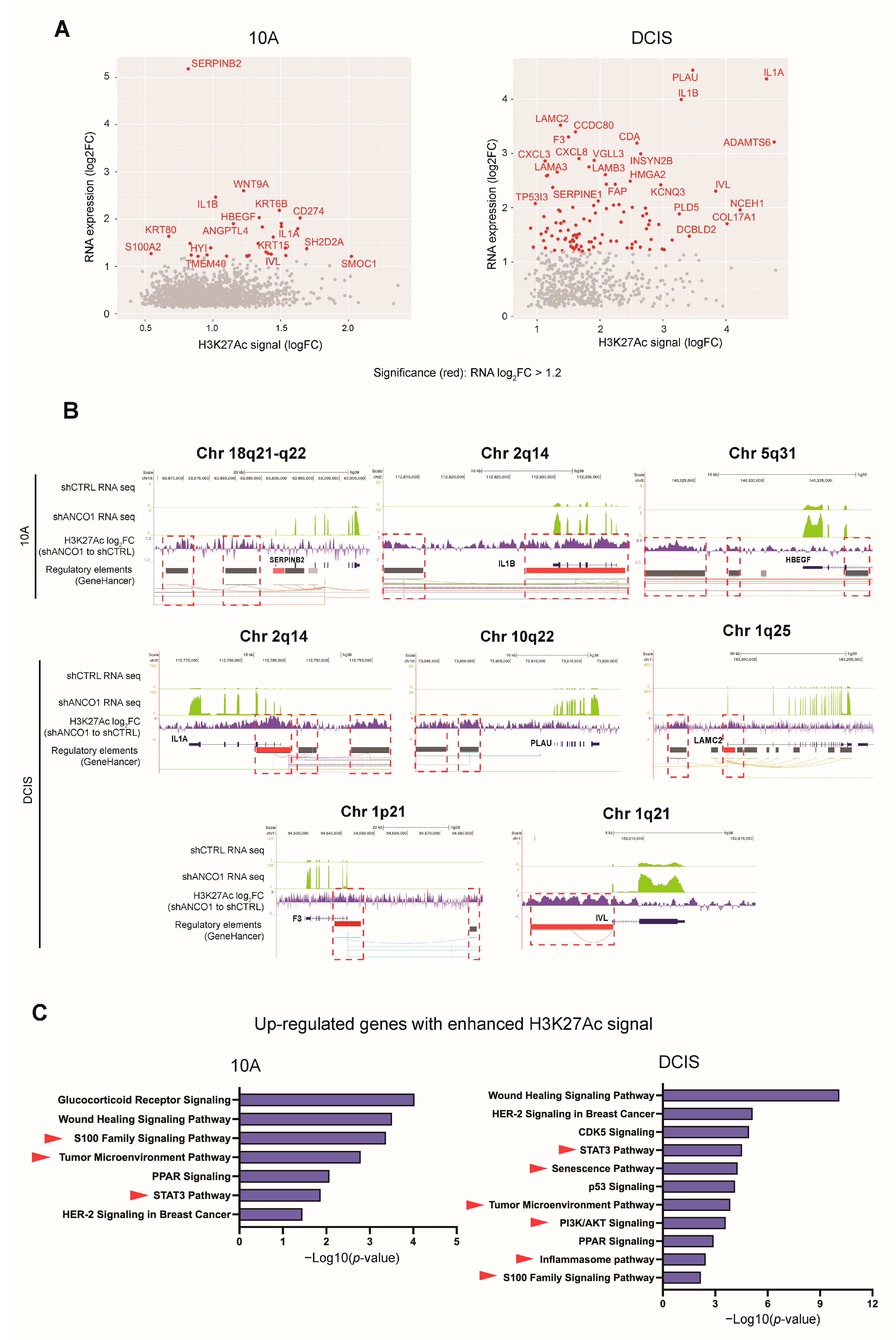
2.7. Loss of ANCO1 Expression Leads to Activation of Breast Cancer Progression Genes
2.8. ANCO1 Reduction Promotes Transcriptional Activation through Chromatin Remodeling
3. Discussion
4. Materials and Methods
4.1. Approval of Studies Involving Humans and Patient Informed Consent
4.2. TMA Staining
4.3. TMA Analysis Using Vectra3
4.4. Gene Expression Analysis
4.5. Cell Culture
4.6. Cell Transfection and Infection
4.7. Real-Time Quantitative PCR (RT-qPCR)
4.8. Western Blotting
4.9. IF Staining
4.10. FISH Analysis
4.11. SA β-Galactosidase Analysis
4.12. Apoptotic Analysis
4.13. Sphere Formation Assay
4.14. Sphere Invasion Assay
4.15. Animal Experiments
4.16. RNA-seq
4.17. ChIP-seq
4.18. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- American Cancer Society. Triple-Negative Breast Cancer. Available online: https://www.cancer.org/cancer/breast-cancer/about/types-of-breast-cancer/triple-negative.html (accessed on 27 April 2023).
- Sirmaci, A.; Spiliopoulos, M.; Brancati, F.; Powell, E.; Duman, D.; Abrams, A.; Bademci, G.; Agolini, E.; Guo, S.; Konuk, B. Mutations in ANKRD11 cause KBG syndrome, characterized by intellectual disability, skeletal malformations, and macrodontia. Am. J. Hum. Genet. 2011, 89, 289–294. [Google Scholar] [CrossRef] [PubMed]
- Kline, A.D.; Moss, J.F.; Selicorni, A.; Bisgaard, A.-M.; Deardorff, M.A.; Gillett, P.M.; Ishman, S.L.; Kerr, L.M.; Levin, A.V.; Mulder, P.A.; et al. Diagnosis and management of Cornelia de Lange syndrome: First international consensus statement. Nat. Rev. Genet. 2018, 19, 649–666. [Google Scholar] [CrossRef] [PubMed]
- Powell, J.A.; Gardner, A.E.; Bais, A.J.; Hinze, S.J.; Baker, E.; Whitmore, S.; Crawford, J.; Kochetkova, M.; Spendlove, H.E.; Doggett, N.A.; et al. Sequencing, transcript identification, and quantitative gene expression profiling in the breast cancer loss of heterozygosity region 16q24.3 reveal three potential tumor-suppressor genes. Genomics 2002, 80, 303–310. [Google Scholar] [CrossRef] [PubMed]
- Miller, B.J.; Wang, D.; Krahe, R.; Wright, F.A. Pooled analysis of loss of heterozygosity in breast cancer: A genome scan provides comparative evidence for multiple tumor suppressors and identifies novel candidate regions. Am. J. Hum. Genet. 2003, 73, 748–767. [Google Scholar] [CrossRef]
- Kushner, M.H.; Ory, V.; Graham, G.T.; Sharif, G.M.; Kietzman, W.B.; Thevissen, S.; Yuan, M.; Schmidt, M.O.; Wellstein, A.; Riegel, A.T. Loss of ANCO1 repression at AIB1/YAP targets drives breast cancer progression. EMBO Rep. 2020, 21, e48741. [Google Scholar] [CrossRef]
- Jiang, G.; Zhang, S.; Yazdanparast, A.; Li, M.; Pawar, A.V.; Liu, Y.; Inavolu, S.M.; Cheng, L. Comprehensive comparison of molecular portraits between cell lines and tumors in breast cancer. BMC Genom. 2016, 17, 525. [Google Scholar] [CrossRef]
- Lim, S.P.; Wong, N.C.; Suetani, R.J.; Ho, K.; Ng, J.L.; Neilsen, P.M.; Gill, P.G.; Kumar, R.; Callen, D.F. Specific-site methylation of tumour suppressor ANKRD11 in breast cancer. Eur. J. Cancer 2012, 48, 3300–3309. [Google Scholar] [CrossRef]
- Schachter, N.F.; Adams, J.R.; Skowron, P.; Kozma, K.J.; Lee, C.A.; Raghuram, N.; Yang, J.; Loch, A.J.; Wang, W.; Kucharczuk, A.; et al. Single allele loss-of-function mutations select and sculpt conditional cooperative networks in breast cancer. Nat. Commun. 2021, 12, 5238. [Google Scholar] [CrossRef]
- Gallagher, D.; Voronova, A.; Zander, M.A.; Cancino, G.I.; Bramall, A.; Krause, M.P.; Abad, C.; Tekin, M.; Neilsen, P.M.; Callen, D.F.; et al. Ankrd11 is a chromatin regulator involved in autism that is essential for neural development. Dev. Cell 2015, 32, 31–42. [Google Scholar] [CrossRef]
- Zhang, A.; Yeung, P.L.; Li, C.-W.; Tsai, S.-C.; Dinh, G.K.; Wu, X.; Li, H.; Chen, J.D. Identification of a novel family of ankyrin repeats containing cofactors for p160 nuclear receptor coactivators. J. Biol. Chem. 2004, 279, 33799–33805. [Google Scholar] [CrossRef]
- Li, C.-W.; Dinh, G.K.; Zhang, A.; Chen, J.D. Ankyrin repeats-containing cofactors interact with ADA3 and modulate its co-activator function. Biochem. J. 2008, 413, 349–357. [Google Scholar] [CrossRef] [PubMed]
- Garee, J.P.; Chien, C.D.; Li, J.V.; Wellstein, A.; Riegel, A.T. Regulation of HER2 oncogene transcription by a multifunctional coactivator/corepressor complex. Mol. Endocrinol. 2014, 28, 846–859. [Google Scholar] [CrossRef] [PubMed]
- Neilsen, P.M.; Cheney, K.M.; Li, C.-W.; Chen, J.D.; Cawrse, J.E.; Schulz, R.B.; Powell, J.A.; Kumar, R.; Callen, D.F. Identification of ANKRD11 as a p53 coactivator. J. Cell Sci. 2008, 121, 3541–3552. [Google Scholar] [CrossRef]
- Goh, J.Y.; Feng, M.; Wang, W.; Oguz, G.; Yatim, S.M.J.M.; Lee, P.L.; Bao, Y.; Lim, T.H.; Wang, P.; Tam, W.L.; et al. Chromosome 1q21.3 amplification is a trackable biomarker and actionable target for breast cancer recurrence. Nat. Med. 2017, 23, 1319–1330. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature 2012, 490, 61–70. [Google Scholar] [CrossRef] [PubMed]
- Curtis, C.; Shah, S.P.; Chin, S.-F.; Turashvili, G.; Rueda, O.M.; Dunning, M.J.; Speed, D.; Lynch, A.G.; Samarajiwa, S.; Yuan, Y.; et al. The genomic and transcriptomic architecture of 2000 breast tumours reveals novel subgroups. Nature 2012, 486, 346–352. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, A.I.; Geng, X.; Chaldekas, K.; Harris, B.; Duttargi, A.; Berry, V.L.; Berry, D.L.; Mahajan, A.; Cavalli, L.R.; Győrffy, B.; et al. The orphan nuclear receptor estrogen-related receptor beta (ERRβ) in triple-negative breast cancer. Breast Cancer Res. Treat. 2020, 179, 585–604. [Google Scholar] [CrossRef]
- Soule, H.D.; Maloney, T.M.; Wolman, S.R.; Peterson, W.D.; Brenz, R.; McGrath, C.M.; Russo, J.; Pauley, R.J.; Jones, R.F.; Brooks, S.C. Isolation and characterization of a spontaneously immortalized human breast epithelial cell line, MCF-10. Cancer Res. 1990, 50, 6075–6086. [Google Scholar]
- Miller, F.R.; Santner, S.J.; Tait, L.; Dawson, P.J. MCF10DCIS.com xenograft model of human comedo ductal carcinoma in situ. JNCI J. Natl. Cancer Inst. 2000, 92, 1185–1186. [Google Scholar] [CrossRef]
- Masood, S.; Bui, M.M.; Yung, J.F.; Mark, H.F.; Wong, E.Y.; Birkmeier, J.M.; Yang, S.J.; Hsu, P. Reproducibility of LSI HER-2/neu SpectrumOrange and CEP 17 SpectrumGreen Dual Color deoxyribonucleic acid probe kit. For enumeration of gene amplification in paraffin-embedded specimens: A multicenter clinical validation study. Ann. Clin. Lab. Sci. 1998, 28, 215–223. [Google Scholar]
- González-Gualda, E.; Baker, A.G.; Fruk, L.; Muñoz-Espín, D. A guide to assessing cellular senescence in vitro and in vivo. FEBS J. 2021, 288, 56–80. [Google Scholar] [CrossRef] [PubMed]
- Dimri, G.P.; Lee, X.; Basile, G.; Acosta, M.; Scott, G.; Roskelley, C.; Medrano, E.E.; Linskens, M.; Rubelj, I.; Pereira-Smith, O.; et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc. Natl. Acad. Sci. USA 1995, 92, 9363–9367. [Google Scholar] [CrossRef]
- Debnath, J.; Muthuswamy, S.K.; Brugge, J.S. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods 2003, 30, 256–268. [Google Scholar] [CrossRef]
- O’Brien, L.E.; Zegers, M.M.P.; Mostov, K.E. Building epithelial architecture: Insights from three-dimensional culture models. Nat. Rev. Mol. Cell Biol. 2002, 3, 531–537. [Google Scholar] [CrossRef] [PubMed]
- Wellings, S.R.; Jensen, H.M. On the origin and progression of ductal carcinoma in the human breast. J. Natl. Cancer Inst. 1973, 50, 1111–1118. [Google Scholar] [CrossRef] [PubMed]
- Chien, C.D.; Kirilyuk, A.; Li, J.V.; Zhang, W.; Lahusen, T.; Schmidt, M.O.; Oh, A.S.; Wellstein, A.; Riegel, A.T. Role of the nuclear receptor coactivator AIB1-Δ4 splice variant in the control of gene transcription. J. Biol. Chem. 2011, 286, 26813–26827. [Google Scholar] [CrossRef]
- Sharif, G.M.; Campbell, M.J.; Nasir, A.; Sengupta, S.; Graham, G.T.; Kushner, M.H.; Kietzman, W.B.; Schmidt, M.O.; Pearson, G.W.; Loudig, O.; et al. An AIB1 isoform alters enhancer access and enables progression of early-stage triple-negative breast cancer. Cancer Res. 2021, 81, 4230–4241. [Google Scholar] [CrossRef]
- Hu, M.; Yao, J.; Carroll, D.K.; Weremowicz, S.; Chen, H.; Carrasco, D.; Richardson, A.; Violette, S.; Nikolskaya, T.; Nikolsky, Y.; et al. Regulation of in situ to invasive breast carcinoma transition. Cancer Cell 2008, 13, 394–406. [Google Scholar] [CrossRef]
- Ory, V.; Tassi, E.; Cavalli, L.; Sharif, G.; Saenz, F.; Baker, T.; O Schmidt, M.; Mueller, S.C.; A Furth, P.; Wellstein, A.; et al. The nuclear coactivator Amplified in Breast Cancer 1 maintains tumor initiating cells during development of Ductal Carcinoma In Situ. Oncogene 2014, 33, 3033–3042. [Google Scholar] [CrossRef]
- Creyghton, M.P.; Cheng, A.W.; Welstead, G.G.; Kooistra, T.; Carey, B.W.; Steine, E.J.; Hanna, J.; Lodato, M.A.; Frampton, G.M.; Sharp, P.A.; et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. USA 2010, 107, 21931–21936. [Google Scholar] [CrossRef]
- Patten, D.K.; Corleone, G.; Győrffy, B.; Perone, Y.; Slaven, N.; Barozzi, I.; Erdős, E.; Saiakhova, A.; Goddard, K.; Vingiani, A.; et al. Enhancer mapping uncovers phenotypic heterogeneity and evolution in patients with luminal breast cancer. Nat. Med. 2018, 24, 1469–1480. [Google Scholar] [CrossRef]
- Fingar, D.C.; Salama, S.; Tsou, C.; Harlow, E.; Blenis, J. Mammalian cell size is controlled by mTOR and its downstream targets S6K1 and 4EBP1/eIF4E. Gene. Dev. 2002, 16, 1472–1487. [Google Scholar] [CrossRef]
- Xu, W.; Yang, Z.; Lu, N. A new role for the PI3K/Akt signaling pathway in the epithelial-mesenchymal transition. Cell Adhes. Migr. 2015, 9, 317–324. [Google Scholar] [CrossRef]
- Shaulian, E.; Karin, M. AP-1 in cell proliferation and survival. Oncogene 2001, 20, 2390–2400. [Google Scholar] [CrossRef] [PubMed]
- Ozanne, B.W.; Spence, H.J.; McGarry, L.C.; Hennigan, R.F. Transcription factors control invasion: AP-1 the first among equals. Oncogene 2007, 26, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Lin, K.C.; Park, H.W.; Guan, K.-L. Regulation of the Hippo pathway transcription factor TEAD. Trends Biochem. Sci. 2017, 42, 862–872. [Google Scholar] [CrossRef]
- Lamar, J.M.; Stern, P.; Liu, H.; Schindler, J.W.; Jiang, Z.-G.; Hynes, R.O. The Hippo pathway target, YAP, promotes metastasis through its TEAD-interaction domain. Proc. Natl. Acad. Sci. USA 2012, 109, E2441–E2450. [Google Scholar] [CrossRef]
- Nguyen, C.D.K.; Yi, C. YAP/TAZ signaling and resistance to cancer therapy. Trends Cancer 2019, 5, 283–296. [Google Scholar] [CrossRef] [PubMed]
- Zanconato, F.; Forcato, M.; Battilana, G.; Azzolin, L.; Quaranta, E.; Bodega, B.; Rosato, A.; Bicciato, S.; Cordenonsi, M.; Piccolo, S. Genome-wide association between YAP/TAZ/TEAD and AP-1 at enhancers drives oncogenic growth. Nat. Cell Biol. 2015, 17, 1218–1227. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Pardoll, D.; Jove, R. STATs in cancer inflammation and immunity: A leading role for STAT3. Nat. Rev. Cancer 2009, 9, 798–809. [Google Scholar] [CrossRef]
- Taniguchi, K.; Karin, M. NF-κB, inflammation, immunity and cancer: Coming of age. Nat. Rev. Immunol. 2018, 18, 309–324. [Google Scholar] [CrossRef]
- Jeon, H.-M.; Kim, J.-Y.; Lee, W.J.; Nguyen, D.; Kim, S.; Park, J.B.; Park, M.J.; Nam, D.H.; Lee, J. Tissue factor promotes oncogenic senescence signaling and radiation resistance in glioblastoma. Brain Tumor Res. Treat. 2022, 10, S44-1. [Google Scholar]
- Simmen, F.A.; Alhallak, I.; Simmen, R.C.M. Malic enzyme 1 (ME1) in the biology of cancer: It is not just intermediary metabolism. J. Mol. Endocrinol. 2020, 65, R77–R90. [Google Scholar] [CrossRef]
- Jiang, P.; Du, W.; Mancuso, A.; Wellen, K.E.; Yang, X. Reciprocal regulation of p53 and malic enzymes modulates metabolism and senescence. Nature 2013, 493, 689–693. [Google Scholar] [CrossRef] [PubMed]
- Chou, C.-K.; Fan, C.-C.; Lin, P.-S.; Liao, P.-Y.; Tung, J.-C.; Hsieh, C.-H.; Hung, M.-C.; Chen, C.-H.; Chang, W.-C. Sciellin mediates mesenchymal-to-epithelial transition in colorectal cancer hepatic metastasis. Oncotarget 2016, 7, 25742–25754. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Abraham, A.D.; Li, L.; Babalmorad, A.; Bagby, S.; Arcaroli, J.J.; Hansen, R.J.; A Valeriote, F.; Gustafson, D.L.; Schaack, J.; et al. Topoisomerase IIα mediates TCF-dependent epithelial–mesenchymal transition in colon cancer. Oncogene 2016, 35, 4990–4999. [Google Scholar] [CrossRef] [PubMed]
- Stoll, S.W.; Rittié, L.; Johnson, J.L.; Elder, J.T. Heparin-binding EGF-like growth factor promotes epithelial-mesenchymal transition in human keratinocytes. J. Investig. Dermatol. 2012, 132, 2148–2157. [Google Scholar] [CrossRef]
- Moquet-Torcy, G.; Tolza, C.; Piechaczyk, M.; Jariel-Encontre, I. Transcriptional complexity and roles of Fra-1/AP-1 at the uPA/Plau locus in aggressive breast cancer. Nucleic Acids Res. 2014, 42, 11011–11024. [Google Scholar] [CrossRef]
- Pei, Y.-F.; Liu, J.; Cheng, J.; Wu, W.-D.; Liu, X.-Q. Silencing of LAMC2 reverses epithelial-mesenchymal transition and inhibits angiogenesis in cholangiocarcinoma via inactivation of the epidermal growth factor receptor signaling pathway. Am. J. Pathol. 2019, 189, 1637–1653. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Bakhoum, S.F.; Cantley, L.C. The multifaceted role of chromosomal instability in cancer and its microenvironment. Cell 2018, 174, 1347–1360. [Google Scholar] [CrossRef] [PubMed]
- Ippolito, M.R.; Martis, V.; Martin, S.; Tijhuis, A.E.; Hong, C.; Wardenaar, R.; Dumont, M.; Zerbib, J.; Spierings, D.C.; Fachinetti, D.; et al. Gene copy-number changes and chromosomal instability induced by aneuploidy confer resistance to chemotherapy. Dev. Cell 2021, 56, 2440–2454.e6. [Google Scholar] [CrossRef] [PubMed]
- Piché, J.; Vliet, P.P.V.; Pucéat, M.; Andelfinger, G. The expanding phenotypes of cohesinopathies: One ring to rule them all! Cell Cycle 2019, 18, 2828–2848. [Google Scholar] [CrossRef] [PubMed]
- Parenti, I.; Gervasini, C.; Pozojevic, J.; Graul-Neumann, L.; Azzollini, J.; Braunholz, D.; Watrin, E.; Wendt, K.; Cereda, A.; Cittaro, D.; et al. Broadening of cohesinopathies: Exome sequencing identifies mutations in ANKRD11 in two patients with Cornelia de Lange-overlapping phenotype. Clin. Genet. 2016, 89, 74–81. [Google Scholar] [CrossRef]
- Walz, K.; Cohen, D.; Neilsen, P.M.; Foster, J.; Brancati, F.; Demir, K.; Fisher, R.; Moffat, M.; Verbeek, N.E.; Bjørgo, K.; et al. Characterization of ANKRD11 mutations in humans and mice related to KBG syndrome. Hum. Genet. 2015, 134, 181–190. [Google Scholar] [CrossRef]
- Noll, J.E.; Jeffery, J.; Al-Ejeh, F.; Kumar, R.; Khanna, K.K.; Callen, D.F.; Neilsen, P.M. Mutant p53 drives multinucleation and invasion through a process that is suppressed by ANKRD11. Oncogene 2012, 31, 2836–2848. [Google Scholar] [CrossRef]
- Rodier, F.; Coppé, J.-P.; Patil, C.K.; Hoeijmakers, W.A.M.; Muñoz, D.P.; Raza, S.R.; Freund, A.; Campeau, E.; Davalos, A.R.; Campisi, J. Persistent DNA damage signalling triggers senescence-associated inflammatory cytokine secretion. Nat. Cell Biol. 2009, 11, 973–979. [Google Scholar] [CrossRef]
- Laberge, R.-M.; Sun, Y.; Orjalo, A.V.; Patil, C.K.; Freund, A.; Zhou, L.; Curran, S.C.; Davalos, A.R.; Wilson-Edell, K.A.; Liu, S.; et al. MTOR regulates the pro-tumorigenic senescence-associated secretory phenotype by promoting IL1A translation. Nat. Cell Biol. 2015, 17, 1049–1061. [Google Scholar] [CrossRef]
- Sur, I.; Taipale, J. The role of enhancers in cancer. Nat. Rev. Cancer 2016, 16, 483–493. [Google Scholar] [CrossRef]
- Tumaneng, K.; Schlegelmilch, K.; Russell, R.C.; Yimlamai, D.; Basnet, H.; Mahadevan, N.; Fitamant, J.; Bardeesy, N.; Camargo, F.D.; Guan, K.-L. YAP mediates crosstalk between the Hippo and PI(3)K–TOR pathways by suppressing PTEN via miR-29. Nat. Cell Biol. 2012, 14, 1322–1329. [Google Scholar] [CrossRef]
- Tilli, M.T.; Reiter, R.; Oh, A.S.; Henke, R.T.; McDonnell, K.; Gallicano, G.I.; Furth, P.A.; Riegel, A.T. Overexpression of an N-terminally truncated isoform of the nuclear receptor coactivator amplified in breast cancer 1 leads to altered proliferation of mammary epithelial cells in transgenic mice. Mol. Endocrinol. 2005, 19, 644–656. [Google Scholar] [CrossRef] [PubMed]
- Kassambara, A.; Kosinski, M.; Biecek, P. Survminer: Drawing Survival Curves Using ‘ggplot2’. R Package Version 0.4.9. 2021. Available online: https://cran.r-project.org/web/packages/survminer/index.html (accessed on 9 July 2023).
- Wickham, H.; François, R.; Henry, L.; Müller, K.; Vaughan, D. dplyr: A Grammar of Data Manipulation. 2023. Available online: https://dplyr.tidyverse.org (accessed on 9 July 2023).
- Pinkel, D.; Straume, T.; Gray, J.W. Cytogenetic analysis using quantitative, high-sensitivity, fluorescence hybridization. Proc. Natl. Acad. Sci. USA 1986, 83, 2934–2938. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.-N.; Nasir, A.; Camacho, S.; Berry, D.L.; Schmidt, M.O.; Pearson, G.W.; Riegel, A.T.; Wellstein, A. Monitoring cancer cell invasion and T-Cell cytotoxicity in 3D culture. J. Vis. Exp. 2020, e61392. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- McCarthy, D.J.; Chen, Y.; Smyth, G.K. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012, 40, 4288–4297. [Google Scholar] [CrossRef]
- Chen, Y.; Lun, A.T.L.; Smyth, G.K. From reads to genes to pathways: Differential expression analysis of RNA-Seq experiments using Rsubread and the edgeR quasi-likelihood pipeline. F1000research 2016, 5, 1438. [Google Scholar]
- Ploner, A. heatplus: Heatmaps with Row and/or Column Covariates and Colored Clusters. R Package Version 3.6.0. 2022. Available online: https://github.com/alexploner/Heatplus (accessed on 9 July 2023).
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016; ISBN 978-3-319-24277-4. Available online: https://ggplot2.tidyverse.org (accessed on 9 July 2023).
- Slowikowski, K. ggrepel: Automatically Position Non-Overlapping Text Labels with ‘ggplot2’. R Package Version 0.9.2. 2022. Available online: https://github.com/slowkow/ggrepel (accessed on 9 July 2023).
- Kent, W.J.; Sugnet, C.W.; Furey, T.S.; Roskin, K.M.; Pringle, T.H.; Zahler, A.M.; Haussler, D. The human genome browser at UCSC. Genome Res. 2002, 12, 996–1006. [Google Scholar] [CrossRef]
- Lun, A.T.L.; Smyth, G.K. De novo detection of differentially bound regions for ChIP-seq data using peaks and windows: Controlling error rates correctly. Nucleic Acids Res. 2014, 42, e95. [Google Scholar] [CrossRef]
- Lun, A.T.L.; Smyth, G.K. csaw: A Bioconductor package for differential binding analysis of ChIP-seq data using sliding windows. Nucleic Acids Res. 2016, 44, e45. [Google Scholar] [CrossRef]
- Zhu, L.J.; Gazin, C.; Lawson, N.D.; Pagès, H.; Lin, S.M.; Lapointe, D.S.; Green, M.R. ChIPpeakAnno: A Bioconductor package to annotate ChIP-seq and ChIP-chip data. BMC Bioinform. 2010, 11, 237. [Google Scholar] [CrossRef]
- Zhu, L.J. Integrative analysis of ChIP-chip and ChIP-seq dataset. Methods Mol. Biol. 2013, 1067, 105–124. [Google Scholar] [PubMed]
- Zhang, Y.; Liu, T.; Meyer, C.A.; Eeckhoute, J.; Johnson, D.S.; Bernstein, B.E.; Nusbaum, C.; Myers, R.M.; Brown, M.; Li, W.; et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008, 9, R137. [Google Scholar] [CrossRef] [PubMed]
- Ramírez, F.; Ryan, D.P.; Grüning, B.; Bhardwaj, V.; Kilpert, F.; Richter, A.S.; Heyne, S.; Dündar, F.; Manke, T. deepTools2: A next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016, 44, W160–W165. [Google Scholar] [CrossRef] [PubMed]
- Heinz, S.; Benner, C.; Spann, N.; Bertolino, E.; Lin, Y.C.; Laslo, P.; Cheng, J.X.; Murre, C.; Singh, H.; Glass, C.K. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 2010, 38, 576–589. [Google Scholar] [CrossRef] [PubMed]


| Plasmid | Manufacturer | Category Number |
|---|---|---|
| pLKO.1 ANCO1 shRNA #1 | Sigma-Aldrich | TRCN0000140052 |
| pLKO.1 ANCO1 shRNA #2 | Sigma-Aldrich | TRCN0000144209 |
| pLKO.1 ANCO1 shRNA #3 | Sigma-Aldrich | TRCN0000140474 |
| pLKO.1 ANCO1 shRNA #4 | Sigma-Aldrich | TRCN0000344174 |
| pLKO.1 GFP shRNA | A gift from David Sabatini (Addgene) | 30323 |
| pVSV-G | A gift from Robert Weinberg (Addgene) | 8454 |
| pPACKF1 FIV | System Biosciences | LV100A-1 |
| pCDH-EF1-Luc2-P2A-tdTomato | Addgene | 72486 |
| Plasmid | Forward | Reverse |
|---|---|---|
| ANCO1/ANKRD11 (human) | TTGATGAGGACGACGAGCAG | TGACAGGATACGATGGGACG |
| ACTIN (human) | CCTGGCACCCAGCACAAT | GCCGATCCACACGGAGTACT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yuan, M.; Barefoot, M.E.; Peterson, K.; Campbell, M.J.; Blancato, J.K.; Chen, M.; Schmidt, M.O.; Kiliti, A.J.; Fang, H.-B.; Wellstein, A.; et al. Loss of ANCO1 Expression Regulates Chromatin Accessibility and Drives Progression of Early-Stage Triple-Negative Breast Cancer. Int. J. Mol. Sci. 2023, 24, 11505. https://doi.org/10.3390/ijms241411505
Yuan M, Barefoot ME, Peterson K, Campbell MJ, Blancato JK, Chen M, Schmidt MO, Kiliti AJ, Fang H-B, Wellstein A, et al. Loss of ANCO1 Expression Regulates Chromatin Accessibility and Drives Progression of Early-Stage Triple-Negative Breast Cancer. International Journal of Molecular Sciences. 2023; 24(14):11505. https://doi.org/10.3390/ijms241411505
Chicago/Turabian StyleYuan, Meng, Megan E. Barefoot, Kendell Peterson, Moray J. Campbell, Jan K. Blancato, Manjing Chen, Marcel O. Schmidt, Amber J. Kiliti, Hong-Bin Fang, Anton Wellstein, and et al. 2023. "Loss of ANCO1 Expression Regulates Chromatin Accessibility and Drives Progression of Early-Stage Triple-Negative Breast Cancer" International Journal of Molecular Sciences 24, no. 14: 11505. https://doi.org/10.3390/ijms241411505
APA StyleYuan, M., Barefoot, M. E., Peterson, K., Campbell, M. J., Blancato, J. K., Chen, M., Schmidt, M. O., Kiliti, A. J., Fang, H.-B., Wellstein, A., Riegel, A. T., & Sharif, G. M. (2023). Loss of ANCO1 Expression Regulates Chromatin Accessibility and Drives Progression of Early-Stage Triple-Negative Breast Cancer. International Journal of Molecular Sciences, 24(14), 11505. https://doi.org/10.3390/ijms241411505







