Systematic Review, Meta-Analysis and Radiomics Quality Score Assessment of CT Radiomics-Based Models Predicting Tumor EGFR Mutation Status in Patients with Non-Small-Cell Lung Cancer
Abstract
1. Introduction
2. Materials and Methods
2.1. Search Strategy
2.2. Inclusion and Exclusion Criteria
2.3. Data Extraction
2.4. Quality Analysis
2.5. Statistical Analysis
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Imyanitov, E.N.; Iyevleva, A.G.; Levchenko, E.V. Molecular testing and targeted therapy for non-small cell lung cancer: Current status and perspectives. Crit. Rev. Oncol. Hematol. 2021, 157, 103194. [Google Scholar] [CrossRef] [PubMed]
- Lindeman, N.I.; Cagle, P.T.; Aisner, D.L.; Arcila, M.E.; Beasley, M.B.; Bernicker, E.H.; Colasacco, C.; Dacic, S.; Hirsch, F.R.; Kerr, K.; et al. Updated Molecular Testing Guideline for the Selection of Lung Cancer Patients for Treatment With Targeted Tyrosine Kinase Inhibitors. J. Thorac. Oncol. 2018, 13, 323–358. [Google Scholar] [CrossRef]
- Dang, Y.; Wang, R.; Qian, K.; Lu, J.; Zhang, H.; Zhang, Y. Clinical and radiological predictors of epidermal growth factor receptor mutation in nonsmall cell lung cancer. J. Appl. Clin. Med. Phys. 2021, 22, 271–280. [Google Scholar] [CrossRef]
- Metro, G.; Finocchiaro, G.; Toschi, L.; Bartolini, S.; Magrini, E.; Cancellieri, A.; Trisolini, R.; Castaldini, L.; Tallini, G.; Crino, L. Epidermal Growth Factor Receptor (EGFR) Targeted Therapies in Non-Small Cell Lung Cancer (NSCLC). Rev. Recent. Clin. Trials 2006, 1, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Zito Marino, F.; Bianco, R.; Accardo, M.; Ronchi, A.; Cozzolino, I.; Morgillo, F.; Rossi, G.; Franco, R. Molecular heterogeneity in lung cancer: From mechanisms of origin to clinical implications. Int. J. Med. Sci. 2019, 16, 981–989. [Google Scholar] [CrossRef] [PubMed]
- Di Capua, D.; Bracken-Clarke, D.; Ronan, K.; Baird, A.M.; Finn, S. The Liquid Biopsy for Lung Cancer: State of the Art, Limitations and Future Developments. Cancers 2021, 13, 3923. [Google Scholar] [CrossRef]
- Lambin, P.; Leijenaar, R.T.H.; Deist, T.M.; Peerlings, J.; de Jong, E.E.C.; van Timmeren, J.; Sanduleanu, S.; Larue, R.T.H.M.; Even, A.J.G.; Jochems, A.; et al. Radiomics: The bridge between medical imaging and personalized medicine. Nat. Rev. Clin. Oncol. 2017, 14, 749–762. [Google Scholar] [CrossRef]
- Lambin, P.; Rios-Velazquez, E.; Leijenaar, R.; Carvalho, S.; van Stiphout, R.G.; Granton, P.; Zegers, C.M.; Gillies, R.; Boellard, R.; Dekker, A.; et al. Radiomics: Extracting more information from medical images using advanced feature analysis. Eur. J. Cancer 2012, 48, 441–446. [Google Scholar] [CrossRef]
- Wang, C.; Ma, J.; Shao, J.; Zhang, S.; Liu, Z.; Yu, Y.; Li, W. Predicting EGFR and PD-L1 Status in NSCLC Patients Using Multitask AI System Based on CT Images. Front. Immunol. 2022, 13, 813072. [Google Scholar] [CrossRef]
- Yang, L.; Xu, P.; Li, M.; Wang, M.; Peng, M.; Zhang, Y.; Wu, T.; Chu, W.; Wang, K.; Meng, H.; et al. PET/CT Radiomic Features: A Potential Biomarker for EGFR Mutation Status and Survival Outcome Prediction in NSCLC Patients Treated With TKIs. Front. Oncol. 2022, 12, 894323. [Google Scholar] [CrossRef]
- Nair, J.K.R.; Saeed, U.A.; McDougall, C.C.; Sabri, A.; Kovacina, B.; Raidu, B.V.S.; Khokhar, R.A.; Probst, S.; Hirsh, V.; Chankowsky, J.; et al. Radiogenomic Models Using Machine Learning Techniques to Predict EGFR Mutations in Non-Small Cell Lung Cancer. Can. Assoc. Radiol. J. 2021, 72, 109–119. [Google Scholar] [CrossRef] [PubMed]
- Jiang, M.; Zhang, Y.; Xu, J.; Ji, M.; Guo, Y.; Guo, Y.; Xiao, J.; Yao, X.; Shi, H.; Zeng, M. Assessing EGFR gene mutation status in non-small cell lung cancer with imaging features from PET/CT. Nucl. Med. Commun. 2019, 40, 842–849. [Google Scholar] [CrossRef] [PubMed]
- McInnes, M.D.F.; Moher, D.; Thombs, B.D.; McGrath, T.A.; Bossuyt, P.M.; The PRISMA-DTA Group; Clifford, T.; Cohen, J.F.; Deeks, J.J.; Gatsonis, C.; et al. Preferred Reporting Items for a Systematic Review and Meta-analysis of Diagnostic Test Accuracy Studies. JAMA 2018, 319, 388. [Google Scholar] [CrossRef]
- Spadarella, G.; Stanzione, A.; Akinci D’Antonoli, T.; Andreychenko, A.; Fanni, S.C.; Ugga, L.; Kotter, E.; Cuocolo, R. Systematic review of the radiomics quality score applications: An EuSoMII Radiomics Auditing Group Initiative. Eur. Radiol. Mar. 2023, 33, 1884–1894. [Google Scholar] [CrossRef]
- Agazzi, G.M.; Ravanelli, M.; Roca, E.; Medicina, D.; Balzarini, P.; Pessina, C.; Vermi, W.; Berruti, A.; Maroldi, R.; Farina, D. CT texture analysis for prediction of EGFR mutational status and ALK rearrangement in patients with non-small cell lung cancer. Radiol. Med. 2021, 126, 786–794. [Google Scholar] [CrossRef]
- Digumarthy, S.R.; Padole, A.M.; Gullo, R.L.; Sequist, L.V.; Kalra, M.K. Can CT radiomic analysis in NSCLC predict histology and EGFR mutation status? Medicine 2019, 98, e13963. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Jiang, Z.; Li, C.; Dong, S.; Zhang, S.; Lv, Y.; Sun, F.; Liu, S. Development and validation of novel radiomics-based nomograms for the prediction of EGFR mutations and Ki-67 proliferation index in non-small cell lung cancer. Quant. Imaging Med. Surg. 2022, 12, 2658–2671. [Google Scholar] [CrossRef] [PubMed]
- He, R.; Yang, X.; Li, T.; He, Y.; Xie, X.; Chen, Q.; Zhang, Z.; Cheng, T. A Machine Learning-Based Predictive Model of Epidermal Growth Factor Mutations in Lung Adenocarcinomas. Cancers 2022, 14, 4664. [Google Scholar] [CrossRef]
- Hong, D.; Xu, K.; Zhang, L.; Wan, X.; Guo, Y. Radiomics Signature as a Predictive Factor for EGFR Mutations in Advanced Lung Adenocarcinoma. Front. Oncol. 2020, 10, 28. [Google Scholar] [CrossRef]
- Jia, T.Y.; Xiong, J.F.; Li, X.Y.; Yu, W.; Xu, Z.Y.; Cai, X.W.; Ma, J.C.; Ren, Y.C.; Larsson, R.; Zhang, J.; et al. Identifying EGFR mutations in lung adenocarcinoma by noninvasive imaging using radiomics features and random forest modeling. Eur. Radiol. 2019, 29, 4742–4750. [Google Scholar] [CrossRef] [PubMed]
- Koyasu, S.; Nishio, M.; Isoda, H.; Nakamoto, Y.; Togashi, K. Usefulness of gradient tree boosting for predicting histological subtype and EGFR mutation status of non-small cell lung cancer on 18F FDG-PET/CT. Ann. Nucl. Med. 2020, 34, 49–57. [Google Scholar] [CrossRef]
- Le, N.Q.K.; Kha, Q.H.; Nguyen, V.H.; Chen, Y.C.; Cheng, S.J.; Chen, C.Y. Machine Learning-Based Radiomics Signatures for EGFR and KRAS Mutations Prediction in Non-Small-Cell Lung Cancer. Int. J. Mol. Sci. 2021, 22, 9254. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Ding, C.; Zhang, H.; Song, J.; Wu, L. Radiomics for the prediction of EGFR mutation subtypes in non-small cell lung cancer. Med. Phys. 2019, 46, 4545–4552. [Google Scholar] [CrossRef] [PubMed]
- Li, X.Y.; Xiong, J.F.; Jia, T.Y.; Shen, T.L.; Hou, R.P.; Zhao, J.; Fu, X.L. Detection of epithelial growth factor receptor (EGFR) mutations on CT images of patients with lung adenocarcinoma using radiomics and/or multi-level residual convolutionary neural networks. J. Thorac. Dis. 2018, 10, 6624–6635. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Kim, J.; Balagurunathan, Y.; Li, Q.; Garcia, A.L.; Stringfield, O.; Ye, Z.; Gillies, R.J. Radiomic Features Are Associated with EGFR Mutation Status in Lung Adenocarcinomas. Clin. Lung Cancer 2016, 17, 441–448.e6. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhou, J.; Wu, J.; Wang, W.; Wang, X.; Guo, J.; Wang, Q.; Zhang, X.; Li, D.; Xie, J.; et al. Development and Validation of Machine Learning Models to Predict Epidermal Growth Factor Receptor Mutation in Non-Small Cell Lung Cancer: A Multi-Center Retrospective Radiomics Study. Cancer Control 2022, 29, 107327482210929. [Google Scholar] [CrossRef]
- Lu, L.; Sun, S.H.; Yang, H.; E, L.; Guo, P.; Schwartz, L.H.; Zhao, B. Radiomics Prediction of EGFR Status in Lung Cancer—Our Experience in Using Multiple Feature Extractors and The Cancer Imaging Archive Data. Tomography 2020, 6, 223–230. [Google Scholar] [CrossRef]
- Ninomiya, K.; Arimura, H.; Chan, W.Y.; Tanaka, K.; Mizuno, S.; Muhammad Gowdh, N.F.; Yaakup, N.A.; Liam, C.K.; Chai, C.S.; Ng, K.H. Robust radiogenomics approach to the identification of EGFR mutations among patients with NSCLC from three different countries using topologically invariant Betti numbers. PLoS ONE 2021, 16, e0244354. [Google Scholar] [CrossRef]
- Rossi, G.; Barabino, E.; Fedeli, A.; Ficarra, G.; Coco, S.; Russo, A.; Adamo, V.; Buemi, F.; Zullo, L.; Dono, M.; et al. Radiomic Detection of EGFR Mutations in NSCLC. Cancer Res. 2021, 81, 724–731. [Google Scholar] [CrossRef]
- Shiri, I.; Amini, M.; Nazari, M.; Hajianfar, G.; Haddadi Avval, A.; Abdollahi, H.; Oveisi, M.; Arabi, H.; Rahmim, A.; Zaidi, H. Impact of feature harmonization on radiogenomics analysis: Prediction of EGFR and KRAS mutations from non-small cell lung cancer PET/CT images. Comput. Biol. Med. 2022, 142, 105230. [Google Scholar] [CrossRef] [PubMed]
- Tu, W.; Sun, G.; Fan, L.; Wang, Y.; Xia, Y.; Guan, Y.; Li, Q.; Zhang, D.; Liu, S.; Li, Z. Radiomics signature: A potential and incremental predictor for EGFR mutation status in NSCLC patients, comparison with CT morphology. Lung Cancer 2019, 132, 28–35. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Yin, G.; Zhang, Y.; Dai, D.; Liu, J.; Chen, P.; Zhu, L.; Ma, W.; Xu, W. Predictive Power of a Radiomic Signature Based on 18F-FDG PET/CT Images for EGFR Mutational Status in NSCLC. Front. Oncol. 2019, 9, 1062. [Google Scholar] [CrossRef]
- Li, Y.; Lu, L.; Xiao, M.; Dercle, L.; Huang, Y.; Zhang, Z.; Schwartz, L.H.; Li, D.; Zhao, B. CT Slice Thickness and Convolution Kernel Affect Performance of a Radiomic Model for Predicting EGFR Status in Non-Small Cell Lung Cancer: A Preliminary Study. Sci. Rep. 2018, 8, 17913. [Google Scholar] [CrossRef]
- Yang, C.; Chen, W.; Gong, G.; Li, Z.; Qiu, Q.; Yin, Y. Application of CT radiomics features to predict the EGFR mutation status and therapeutic sensitivity to TKIs of advanced lung adenocarcinoma. Transl. Cancer Res. 2020, 9, 6683–6690. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Liu, M.; Ren, Y.; Chen, H.; Yu, P.; Wang, S.; Zhang, R.; Dai, H.; Wang, C. Using contrast-enhanced CT and non-contrast-enhanced CT to predict EGFR mutation status in NSCLC patients—A radiomics nomogram analysis. Eur. Radiol. 2022, 32, 2693–2703. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhao, X.; Zhao, Y.; Zhang, J.; Zhang, Z.; Wang, J.; Wang, Y.; Dai, M.; Han, J. Value of pre-therapy 18F-FDG PET/CT radiomics in predicting EGFR mutation status in patients with non-small cell lung cancer. Eur. J. Nucl. Med. Mol. Imaging 2020, 47, 1137–1146. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Chen, B.; Liu, X.; Song, J.; Fang, M.; Hu, C.; Dong, D.; Li, W.; Tian, J. Quantitative Biomarkers for Prediction of Epidermal Growth Factor Receptor Mutation in Non-Small Cell Lung Cancer. Transl. Oncol. 2018, 11, 94–101. [Google Scholar] [CrossRef] [PubMed]
- Abdurixiti, M.; Nijiati, M.; Shen, R.; Ya, Q.; Abuduxiku, N.; Nijiati, M. Current progress and quality of radiomic studies for predicting EGFR mutation in patients with non-small cell lung cancer using PET/CT images: A systematic review. Br. J. Radiol. 2021, 94, 20201272. [Google Scholar] [CrossRef]
- Azour, L.; Liu, S.; Washer, S.L.; Moore, W.H. Percutaneous Transthoracic Lung Biopsy: Optimizing Yield and Mitigating Risk. J. Comput. Assist. Tomogr. 2021, 45, 765–775. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Mayer, A.T.; Li, R. Integrated imaging and molecular analysis to decipher tumor microenvironment in the era of immunotherapy. Semin. Cancer Biol. 2022, 84, 310–328. [Google Scholar] [CrossRef]
- Binczyk, F.; Prazuch, W.; Bozek, P.; Polanska, J. Radiomics and artificial intelligence in lung cancer screening. Transl. Lung Cancer Res. 2021, 10, 1186–1199. [Google Scholar] [CrossRef] [PubMed]
- Mali, S.A.; Ibrahim, A.; Woodruff, H.C.; Andrearczyk, V.; Müller, H.; Primakov, S.; Salahuddin, Z.; Chatterjee, A.; Lambin, P. Making Radiomics More Reproducible across Scanner and Imaging Protocol Variations: A Review of Harmonization Methods. J. Pers. Med. 2021, 11, 842. [Google Scholar] [CrossRef] [PubMed]
- Horng, H.; Singh, A.; Yousefi, B.; Cohen, E.A.; Haghighi, B.; Katz, S.; Noël, P.B.; Shinohara, R.T.; Kontos, D. Generalized ComBat harmonization methods for radiomic features with multi-modal distributions and multiple batch effects. Sci. Rep. 2022, 12, 4493. [Google Scholar] [CrossRef] [PubMed]

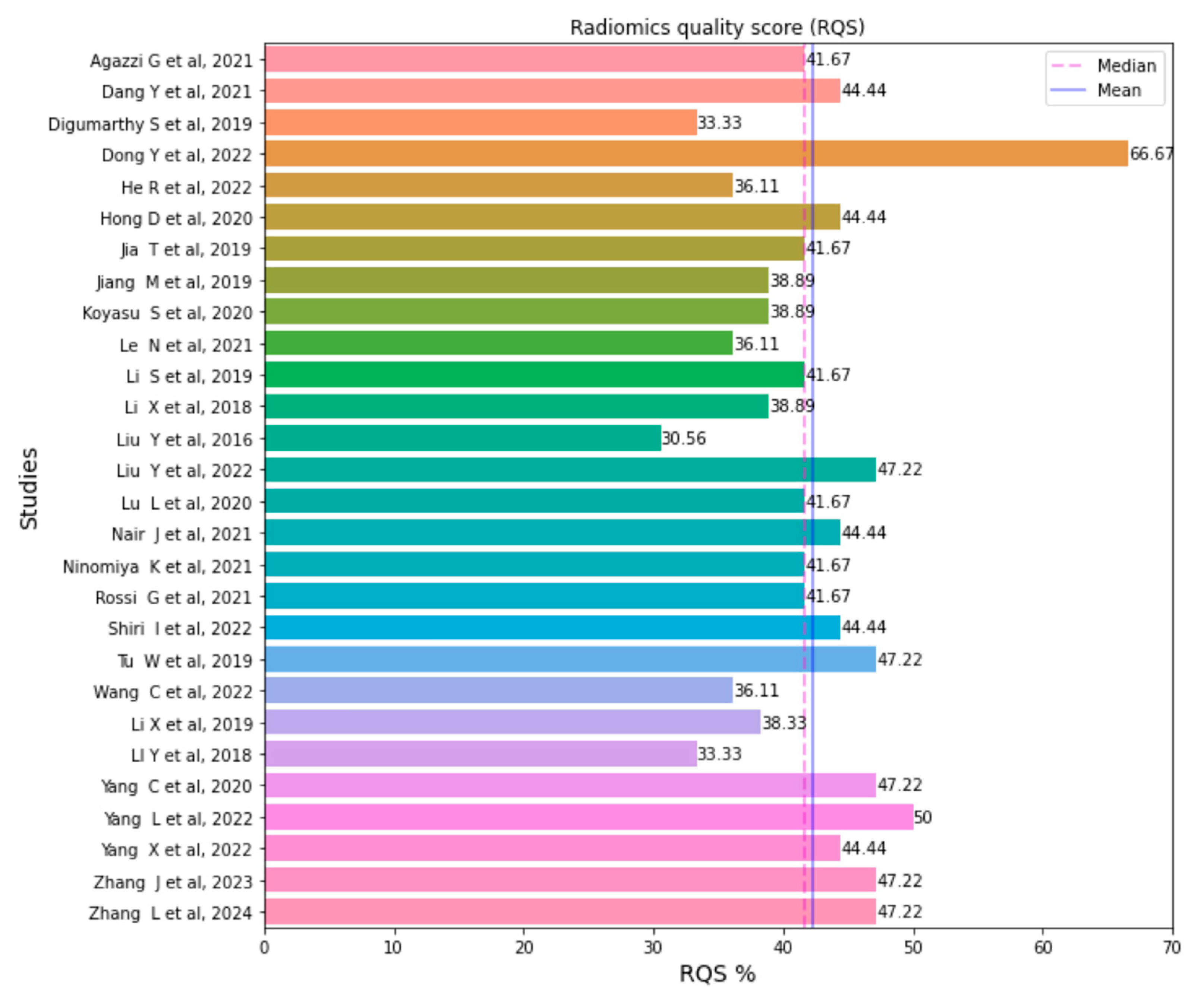

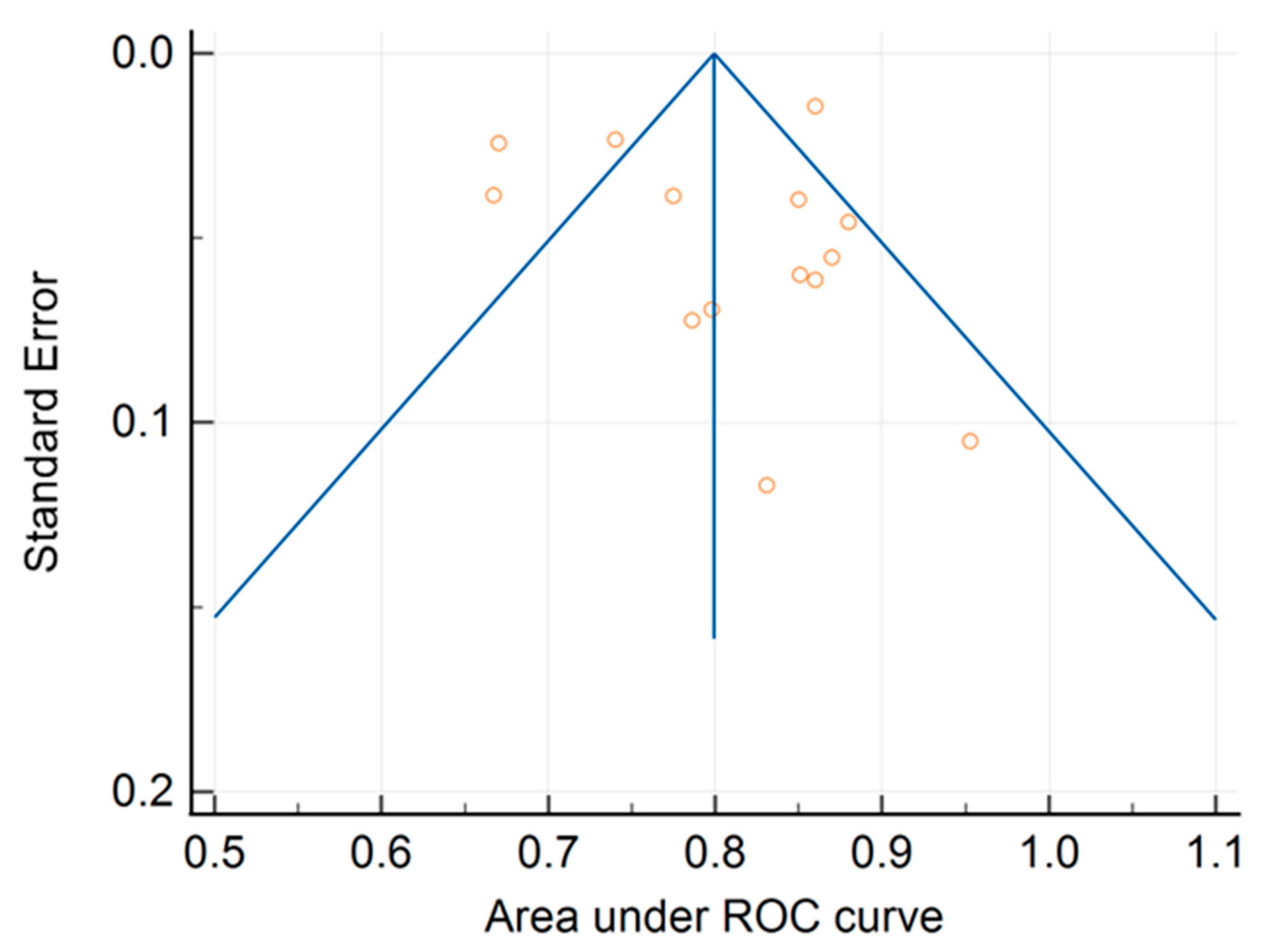
| Study | ROC Curve Area | Standard Error | 95% CI | z-Score | p-Value | Weight | (%) | Sample Size | Study Centers |
|---|---|---|---|---|---|---|---|---|---|
| Fixed | Random | ||||||||
| Dong et al., 2022 [17] | 0.798 | 0.0694 | 0.662 to 0.934 | 1.68 | 5.47 | 132 | Single center | ||
| Dang et al., 2021 [3] | 0.786 | 0.0723 | 0.644 to 0.928 | 1.55 | 5.24 | 118 | Multicenter | ||
| He et al., 2022 [18] | 0.670 | 0.0243 | 0.622 to 0.718 | 13.67 | 9.99 | 758 | Multicenter | ||
| Hong et al., 2020 [19] | 0.851 | 0.0600 | 0.733 to 0.969 | 2.25 | 6.29 | 449 | Single center | ||
| Jiang et al., 2019 [12] | 0.953 | 0.105 | 0.747 to 1.000 | 0.73 | 3.28 | 80 | Single center | ||
| Li et al., 2018 [24] | 0.740 | 0.0233 | 0.694 to 0.786 | 14.91 | 10.09 | 810 | Single center | ||
| Liu et al., 2016 [25] | 0.667 | 0.0384 | 0.592 to 0.742 | 5.50 | 8.52 | 397 | Single center | ||
| Liu et al., 2022 [26] | 0.880 | 0.0456 | 0.791 to 0.969 | 3.90 | 7.74 | 211 | Single center | ||
| Nair et al., 2021 [11] | 0.831 | 0.117 | 0.600 to 1.000 | 0.59 | 2.79 | 80 | Single center | ||
| Ninomiya et al., 2021 [28] | 0.860 | 0.0614 | 0.740 to 0.980 | 2.15 | 6.16 | 194 | Multicenter | ||
| Tu et al., 2019 [31] | 0.775 | 0.0386 | 0.699 to 0.851 | 5.45 | 8.50 | 404 | Single center | ||
| Wang et al., 2022 [9] | 0.860 | 0.0143 | 0.832 to 0.888 | 39.82 | 10.81 | 3629 | Single center | ||
| Yang et al., 2022 [10] | 0.850 | 0.0396 | 0.772 to 0.928 | 5.16 | 8.39 | 1028 | Single center | ||
| Zhang et al., 2020 [36] | 0.870 | 0.0552 | 0.762 to 0.978 | 2.65 | 6.74 | 728 | Single center | ||
| Total (fixed effects) | 0.800 | 0.00900 | 0.782 to 0.817 | 88.858 | <0.001 | 100.00 | 100.00 | ||
| Total (random effects) | 0.801 | 0.0226 | 0.757 to 0.845 | 35.459 | <0.001 | 100.00 | 100.00 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Felfli, M.; Liu, Y.; Zerka, F.; Voyton, C.; Thinnes, A.; Jacques, S.; Iannessi, A.; Bodard, S. Systematic Review, Meta-Analysis and Radiomics Quality Score Assessment of CT Radiomics-Based Models Predicting Tumor EGFR Mutation Status in Patients with Non-Small-Cell Lung Cancer. Int. J. Mol. Sci. 2023, 24, 11433. https://doi.org/10.3390/ijms241411433
Felfli M, Liu Y, Zerka F, Voyton C, Thinnes A, Jacques S, Iannessi A, Bodard S. Systematic Review, Meta-Analysis and Radiomics Quality Score Assessment of CT Radiomics-Based Models Predicting Tumor EGFR Mutation Status in Patients with Non-Small-Cell Lung Cancer. International Journal of Molecular Sciences. 2023; 24(14):11433. https://doi.org/10.3390/ijms241411433
Chicago/Turabian StyleFelfli, Mehdi, Yan Liu, Fadila Zerka, Charles Voyton, Alexandre Thinnes, Sebastien Jacques, Antoine Iannessi, and Sylvain Bodard. 2023. "Systematic Review, Meta-Analysis and Radiomics Quality Score Assessment of CT Radiomics-Based Models Predicting Tumor EGFR Mutation Status in Patients with Non-Small-Cell Lung Cancer" International Journal of Molecular Sciences 24, no. 14: 11433. https://doi.org/10.3390/ijms241411433
APA StyleFelfli, M., Liu, Y., Zerka, F., Voyton, C., Thinnes, A., Jacques, S., Iannessi, A., & Bodard, S. (2023). Systematic Review, Meta-Analysis and Radiomics Quality Score Assessment of CT Radiomics-Based Models Predicting Tumor EGFR Mutation Status in Patients with Non-Small-Cell Lung Cancer. International Journal of Molecular Sciences, 24(14), 11433. https://doi.org/10.3390/ijms241411433






