Investigating the Crime Scene—Molecular Signatures in Inflammatory Bowel Disease
Abstract
1. Introduction
- Diet, gut microbes and patient immune factors are essential factors for IBD initiation and progression
- Nearly every second case of IBD can be prevented by a healthy lifestyle including a healthy diet
- The enormous diverse nature of patients poses challenges in disease management
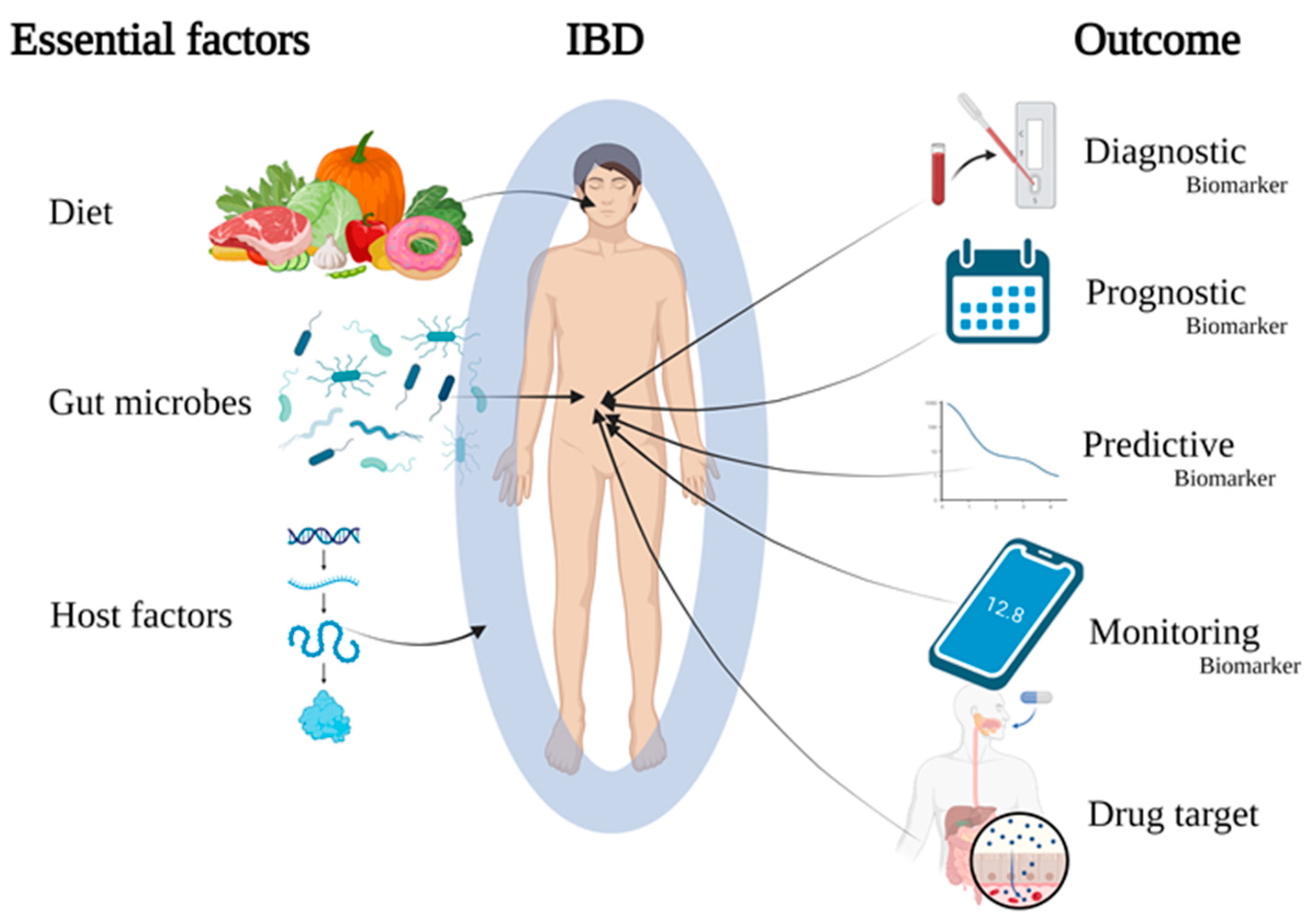
- An update on cutting-edge research identifying molecular profiles to reflect patient diversity based on interactions between diet, gut microbes and the patient’s immune system (Figure 1)
- Patient diet interferes with gut inflammation depending on the patient immune status, genetics and gut microbiome
- Certain faecal metabolome profiles, specifically short-chain fatty acids, are better indicators of IBD phenotypes compared to faecal metagenome or metatranscriptome.
- Potential biomarkers derived from the gut mucosa (e.g., cytokines, gasdermins, neutrophil extracellular traps, Faecalibacterium prausnitzii) and circulating biomarkers (e.g., redox status, N6-methyladenosine modification) are discussed
- Better understanding of specific microbiota–patient interactions by characterising mucosa-associated microbiota and the accompanying immune responses
- Better molecular characterising of IBD sub-phenotypes such as patients with specific disease courses, complications and other immune-related diseases
- Better molecular characterisation of specific IBD phenotypes improving from specific dietary and drug interventions
- Better molecular understanding of patient diversity based on careful phenotypic patient stratification
- Combining specific clinical information for IBD phenotypes with omics data using data integration as a way forward to identify clinically useful biomarkers
- Prospective longitudinal observational studies of biomarkers considered for clinical translation to validate potential biomarkers
- Replication and validation of promising biomarkers in patient cohorts from different geographic regions (e.g., Asia) ultimately leading to clinical translation
2. Identifying Biomarkers Based on Diet–Gut Microbes–Patient Interactions
3. Essential Factors Involved in IBD
3.1. Diet and IBD
3.2. The Gut Microbiota and IBD
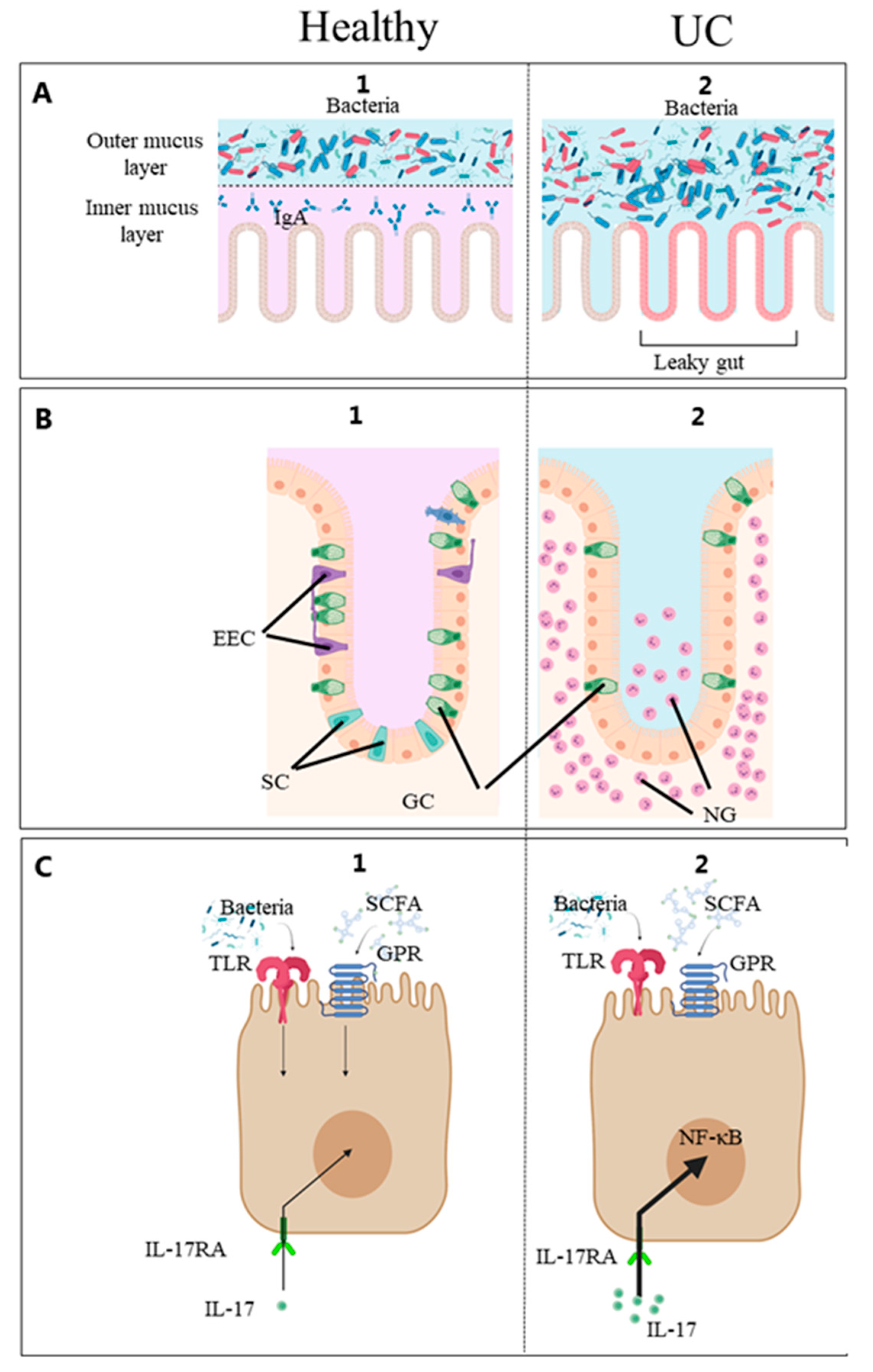
3.3. Gut Epithelium Barrier and Immune System in IBD
4. Interactions between Diet, Gut Microbiota and Host Factors
4.1. Linking Diet with Gut Microbiota and Patient Factors
4.2. Gut Microbiome Can Affect the Patient Immune System
4.3. Patient Factors Affecting the Gut Microbial Function
4.4. Regulation of Mucosal Factors
4.5. Mucosal Biomarkers Reflecting Diet–Gut Microbes–Patient Interactions
5. Conclusions and Future Directions
6. Search Strategy and Selection Criteria
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Chang, J.T. Pathophysiology of Inflammatory Bowel Diseases. N. Engl. J. Med. 2020, 383, 2652–2664. [Google Scholar] [CrossRef]
- Kobayashi, T.; Siegmund, B.; Le Berre, C.; Wei, S.C.; Ferrante, M.; Shen, B.; Bernstein, C.N.; Danese, S.; Peyrin-Biroulet, L.; Hibi, T. Ulcerative colitis. Nat. Rev. Dis. Prim. 2020, 6, 74. [Google Scholar] [CrossRef] [PubMed]
- Glick, L.R.; Cifu, A.S.; Feld, L. Ulcerative Colitis in Adults. JAMA 2020, 324, 1205–1206. [Google Scholar] [CrossRef] [PubMed]
- Roda, G.; Chien Ng, S.; Kotze, P.G.; Argollo, M.; Panaccione, R.; Spinelli, A.; Kaser, A.; Peyrin-Biroulet, L.; Danese, S. Crohn’s disease. Nat. Rev. Dis. Prim. 2020, 6, 22. [Google Scholar] [CrossRef]
- Kaplan, G.G.; Windsor, J.W. The four epidemiological stages in the global evolution of inflammatory bowel disease. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 56–66. [Google Scholar] [CrossRef]
- Agrawal, M.; Jess, T. Implications of the changing epidemiology of inflammatory bowel disease in a changing world. United Eur. Gastroenterol. J. 2022, 10, 1113–1120. [Google Scholar] [CrossRef]
- Plevris, N.; Lees, C.W. Disease Monitoring in Inflammatory Bowel Disease: Evolving Principles and Possibilities. Gastroenterology 2022, 162, 1456–1475.e1451. [Google Scholar] [CrossRef] [PubMed]
- Verstockt, B.; Bressler, B.; Martinez-Lozano, H.; McGovern, D.; Silverberg, M.S. Time to Revisit Disease Classification in Inflammatory Bowel Disease: Is the Current Classification of Inflammatory Bowel Disease Good Enough for Optimal Clinical Management? Gastroenterology 2022, 162, 1370–1382. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, M.; Spencer, E.A.; Colombel, J.F.; Ungaro, R.C. Approach to the Management of Recently Diagnosed Inflammatory Bowel Disease Patients: A User’s Guide for Adult and Pediatric Gastroenterologists. Gastroenterology 2021, 161, 47–65. [Google Scholar] [CrossRef]
- Jayasooriya, N.; Baillie, S.; Blackwell, J.; Bottle, A.; Petersen, I.; Creese, H.; Saxena, S.; Pollok, R.C. Systematic review with meta-analysis: Time to diagnosis and the impact of delayed diagnosis on clinical outcomes in inflammatory bowel disease. Aliment. Pharmacol. Ther. 2023, 57, 635–652. [Google Scholar] [CrossRef]
- Kobayashi, T.; Hibi, T. Improving IBD outcomes in the era of many treatment options. Nat. Rev. Gastroenterol. Hepatol. 2023, 20, 79–80. [Google Scholar] [CrossRef] [PubMed]
- Baumgart, D.C.; Le Berre, C. Newer Biologic and Small-Molecule Therapies for Inflammatory Bowel Disease. N. Engl. J. Med. 2021, 385, 1302–1315. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Gao, Z.; Li, K. Controversy of Preoperative Exposure to Tumor Necrosis Factor Inhibitors in Surgical and Infectious Complications of Inflammatory Bowel Disease. Gastroenterology 2023, 164, 307–308. [Google Scholar] [CrossRef]
- Burisch, J.; Zhao, M.; Odes, S.; De Cruz, P.; Vermeire, S.; Bernstein, C.N.; Kaplan, G.G.; Duricova, D.; Greenberg, D.; Melberg, H.O.; et al. The cost of inflammatory bowel disease in high-income settings: A Lancet Gastroenterology & Hepatology Commission. Lancet. Gastroenterol. Hepatol. 2023, 8, 458–492. [Google Scholar] [CrossRef]
- van Linschoten, R.C.A.; Visser, E.; Niehot, C.D.; van der Woude, C.J.; Hazelzet, J.A.; van Noord, D.; West, R.L. Systematic review: Societal cost of illness of inflammatory bowel disease is increasing due to biologics and varies between continents. Aliment. Pharmacol. Ther. 2021, 54, 234–248. [Google Scholar] [CrossRef]
- Verstockt, B.; Verstockt, S.; Cremer, J.; Sabino, J.; Ferrante, M.; Vermeire, S.; Sudhakar, P. Distinct transcriptional signatures in purified circulating immune cells drive heterogeneity in disease location in IBD. BMJ Open Gastroenterol. 2023, 10, e001003. [Google Scholar] [CrossRef]
- Brooks-Warburton, J.; Modos, D.; Sudhakar, P.; Madgwick, M.; Thomas, J.P.; Bohar, B.; Fazekas, D.; Zoufir, A.; Kapuy, O.; Szalay-Beko, M.; et al. A systems genomics approach to uncover patient-specific pathogenic pathways and proteins in ulcerative colitis. Nat. Commun. 2022, 13, 2299. [Google Scholar] [CrossRef]
- Adams, A.; Gupta, V.; Mohsen, W.; Chapman, T.P.; Subhaharan, D.; Kakkadasam Ramaswamy, P.; Kumar, S.; Kedia, S.; McGregor, C.G.; Ambrose, T.; et al. Early management of acute severe UC in the biologics era: Development and international validation of a prognostic clinical index to predict steroid response. Gut 2023, 72, 433–442. [Google Scholar] [CrossRef]
- Laharie, D.; Riviere, P. Editorial: Selecting therapy for ulcerative colitis-think a step ahead. Aliment. Pharmacol. Ther. 2023, 57, 161–162. [Google Scholar] [CrossRef]
- Caron, B.; D’Amico, F.; Jairath, V.; Netter, P.; Danese, S.; Peyrin-Biroulet, L. Available Methods for Benefit-risk Assessment: Lessons for Inflammatory Bowel Disease Drugs. J. Crohn’s Colitis 2023, 17, 137–143. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, H.K.; Bording-Jorgensen, M.; Santer, D.M.; Zhang, Z.; Valcheva, R.; Rieger, A.M.; Sung-Ho Kim, J.; Dijk, S.I.; Mahmood, R.; Ogungbola, O.; et al. Unfermented β-fructan Fibers Fuel Inflammation in Select Inflammatory Bowel Disease Patients. Gastroenterology 2023, 164, 228–240. [Google Scholar] [CrossRef]
- Villablanca, E.J.; Selin, K.; Hedin, C.R.H. Mechanisms of mucosal healing: Treating inflammatory bowel disease without immunosuppression? Nat. Rev. Gastroenterol. Hepatol. 2022, 19, 493–507. [Google Scholar] [CrossRef] [PubMed]
- Lamb, C.A.; Saifuddin, A.; Powell, N.; Rieder, F. The Future of Precision Medicine to Predict Outcomes and Control Tissue Remodeling in Inflammatory Bowel Disease. Gastroenterology 2022, 162, 1525–1542. [Google Scholar] [CrossRef]
- Metwaly, A.; Reitmeier, S.; Haller, D. Microbiome risk profiles as biomarkers for inflammatory and metabolic disorders. Nat. Rev. Gastroenterol. Hepatol. 2022, 19, 383–397. [Google Scholar] [CrossRef]
- Bjerrum, J.T.; Wang, Y.L.; Seidelin, J.B.; Nielsen, O.H. IBD metabonomics predicts phenotype, disease course, and treatment response. eBioMedicine 2021, 71, 103551. [Google Scholar] [CrossRef] [PubMed]
- Akdis, C.A. Does the epithelial barrier hypothesis explain the increase in allergy, autoimmunity and other chronic conditions? Nat. Rev. Immunol. 2021, 21, 739–751. [Google Scholar] [CrossRef]
- Yu, Y.; Yang, W.; Li, Y.; Cong, Y. Enteroendocrine Cells: Sensing Gut Microbiota and Regulating Inflammatory Bowel Diseases. Inflamm. Bowel Dis. 2020, 26, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Narula, N.; Wong, E.C.L.; Dehghan, M.; Mente, A.; Rangarajan, S.; Lanas, F.; Lopez-Jaramillo, P.; Rohatgi, P.; Lakshmi, P.V.M.; Varma, R.P.; et al. Association of ultra-processed food intake with risk of inflammatory bowel disease: Prospective cohort study. BMJ 2021, 374, n1554. [Google Scholar] [CrossRef] [PubMed]
- Peters, V.; Spooren, C.; Pierik, M.J.; Weersma, R.K.; van Dullemen, H.M.; Festen, E.A.M.; Visschedijk, M.C.; Masclee, A.A.M.; Hendrix, E.M.B.; Almeida, R.J.; et al. Dietary Intake Pattern is Associated with Occurrence of Flares in IBD Patients. J. Crohn’s Colitis 2021, 15, 1305–1315. [Google Scholar] [CrossRef]
- Dong, C.; Chan, S.S.M.; Jantchou, P.; Racine, A.; Oldenburg, B.; Weiderpass, E.; Heath, A.K.; Tong, T.Y.N.; Tjønneland, A.; Kyrø, C.; et al. Meat Intake Is Associated with a Higher Risk of Ulcerative Colitis in a Large European Prospective Cohort Studyø. J. Crohn’s Colitis 2022, 16, 1187–1196. [Google Scholar] [CrossRef]
- Fritsch, J.; Garces, L.; Quintero, M.A.; Pignac-Kobinger, J.; Santander, A.M.; Fernandez, I.; Ban, Y.J.; Kwon, D.; Phillips, M.C.; Knight, K.; et al. Low-Fat, High-Fiber Diet Reduces Markers of Inflammation and Dysbiosis and Improves Quality of Life in Patients With Ulcerative Colitis. Clin. Gastroenterol. Hepatol. 2021, 19, 1189–1199.e30. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Taylor, L.; Shommu, N.; Ghosh, S.; Reimer, R.; Panaccione, R.; Kaur, S.; Hyun, J.E.; Cai, C.; Deehan, E.C.; et al. A Diversified Dietary Pattern Is Associated With a Balanced Gut Microbial Composition of Faecalibacterium and Escherichia/Shigella in Patients With Crohn’s Disease in Remission. J. Crohn’s Colitis 2020, 14, 1547–1557. [Google Scholar] [CrossRef] [PubMed]
- Ventin-Holmberg, R.; Eberl, A.; Saqib, S.; Korpela, K.; Virtanen, S.; Sipponen, T.; Salonen, A.; Saavalainen, P.; Nissila, E. Bacterial and Fungal Profiles as Markers of Infliximab Drug Response in Inflammatory Bowel Disease. J. Crohn’s Colitis 2021, 15, 1019–1031. [Google Scholar] [CrossRef] [PubMed]
- Caenepeel, C.; Sadat Seyed Tabib, N.; Vieira-Silva, S.; Vermeire, S. Review article: How the intestinal microbiota may reflect disease activity and influence therapeutic outcome in inflammatory bowel disease. Aliment. Pharmacol. Ther. 2020, 52, 1453–1468. [Google Scholar] [CrossRef]
- Ananthakrishnan, A.N. Microbiome-Based Biomarkers for IBD. Inflamm. Bowel Dis. 2020, 26, 1463–1469. [Google Scholar] [CrossRef]
- Rees, N.P.; Shaheen, W.; Quince, C.; Tselepis, C.; Horniblow, R.D.; Sharma, N.; Beggs, A.D.; Iqbal, T.H.; Quraishi, M.N. Systematic review of donor and recipient predictive biomarkers of response to faecal microbiota transplantation in patients with ulcerative colitis. eBioMedicine 2022, 81, 104088. [Google Scholar] [CrossRef]
- Sokol, H.; Brot, L.; Stefanescu, C.; Auzolle, C.; Barnich, N.; Buisson, A.; Fumery, M.; Pariente, B.; Le Bourhis, L.; Treton, X.; et al. Prominence of ileal mucosa-associated microbiota to predict postoperative endoscopic recurrence in Crohn’s disease. Gut 2020, 69, 462–472. [Google Scholar] [CrossRef]
- Wiredu Ocansey, D.K.; Hang, S.; Yuan, X.; Qian, H.; Zhou, M.; Valerie Olovo, C.; Zhang, X.; Mao, F. The diagnostic and prognostic potential of gut bacteria in inflammatory bowel disease. Gut Microbes 2023, 15, 2176118. [Google Scholar] [CrossRef]
- Lee, J.W.J.; Plichta, D.; Hogstrom, L.; Borren, N.Z.; Lau, H.; Gregory, S.M.; Tan, W.; Khalili, H.; Clish, C.; Vlamakis, H.; et al. Multi-omics reveal microbial determinants impacting responses to biologic therapies in inflammatory bowel disease. Cell Host Microbe 2021, 29, 1294–1304.e1294. [Google Scholar] [CrossRef]
- Federici, S.; Kredo-Russo, S.; Valdés-Mas, R.; Kviatcovsky, D.; Weinstock, E.; Matiuhin, Y.; Silberberg, Y.; Atarashi, K.; Furuichi, M.; Oka, A.; et al. Targeted suppression of human IBD-associated gut microbiota commensals by phage consortia for treatment of intestinal inflammation. Cell 2022, 185, 2879–2898.e2824. [Google Scholar] [CrossRef]
- Mansour, S.; Asrar, T.; Elhenawy, W. The multifaceted virulence of adherent-invasive Escherichia coli. Gut Microbes 2023, 15, 2172669. [Google Scholar] [CrossRef] [PubMed]
- Privitera, G.; Pugliese, D.; Rapaccini, G.L.; Gasbarrini, A.; Armuzzi, A.; Guidi, L. Predictors and Early Markers of Response to Biological Therapies in Inflammatory Bowel Diseases. J. Clin. Med. 2021, 10, 853–873. [Google Scholar] [CrossRef] [PubMed]
- Cui, G.; Fan, Q.; Li, Z.; Goll, R.; Florholmen, J. Evaluation of anti-TNF therapeutic response in patients with inflammatory bowel disease: Current and novel biomarkers. eBioMedicine 2021, 66, 103329. [Google Scholar] [CrossRef] [PubMed]
- Nanki, K.; Fujii, M.; Shimokawa, M.; Matano, M.; Nishikori, S.; Date, S.; Takano, A.; Toshimitsu, K.; Ohta, Y.; Takahashi, S.; et al. Somatic inflammatory gene mutations in human ulcerative colitis epithelium. Nature 2020, 577, 254–259. [Google Scholar] [CrossRef] [PubMed]
- Kakiuchi, N.; Yoshida, K.; Uchino, M.; Kihara, T.; Akaki, K.; Inoue, Y.; Kawada, K.; Nagayama, S.; Yokoyama, A.; Yamamoto, S.; et al. Frequent mutations that converge on the NFKBIZ pathway in ulcerative colitis. Nature 2020, 577, 260–265. [Google Scholar] [CrossRef]
- Olafsson, S.; McIntyre, R.E.; Coorens, T.; Butler, T.; Jung, H.; Robinson, P.S.; Lee-Six, H.; Sanders, M.A.; Arestang, K.; Dawson, C.; et al. Somatic Evolution in Non-neoplastic IBD-Affected Colon. Cell 2020, 182, 672–684. [Google Scholar] [CrossRef]
- Zhang, J.; Song, B.; Zeng, Y.; Xu, C.; Gao, L.; Guo, Y.; Liu, J. m6A modification in inflammatory bowel disease provides new insights into clinical applications. Biomed. Pharm. 2023, 159, 114298. [Google Scholar] [CrossRef]
- Mata-Garrido, J.; Xiang, Y.; Chang-Marchand, Y.; Reisacher, C.; Ageron, E.; Guerrera, I.C.; Casafont, I.; Bruneau, A.; Cherbuy, C.; Treton, X.; et al. The Heterochromatin protein 1 is a regulator in RNA splicing precision deficient in ulcerative colitis. Nat. Commun. 2022, 13, 6834. [Google Scholar] [CrossRef]
- Jukic, A.; Bakiri, L.; Wagner, E.F.; Tilg, H.; Adolph, T.E. Calprotectin: From biomarker to biological function. Gut 2021, 70, 1978–1988. [Google Scholar] [CrossRef]
- Torres, J.; Petralia, F.; Sato, T.; Wang, P.; Telesco, S.E.; Choung, R.S.; Strauss, R.; Li, X.J.; Laird, R.M.; Gutierrez, R.L.; et al. Serum Biomarkers Identify Patients Who Will Develop Inflammatory Bowel Diseases Up to 5 Years Before Diagnosis. Gastroenterology 2020, 159, 96–104. [Google Scholar] [CrossRef]
- Kawamura, T.; Yamamura, T.; Nakamura, M.; Maeda, K.; Sawada, T.; Ishikawa, E.; Iida, T.; Mizutani, Y.; Ishikawa, T.; Kakushima, N.; et al. Accuracy of Serum Leucine-Rich Alpha-2 Glycoprotein in Evaluating Endoscopic Disease Activity in Crohn’s Disease. Inflamm. Bowel Dis. 2023, 29, 245–253. [Google Scholar] [CrossRef]
- Bourgonje, A.R.; Kloska, D.; Grochot-Przęczek, A.; Feelisch, M.; Cuadrado, A.; van Goor, H. Personalized redox medicine in inflammatory bowel diseases: An emerging role for HIF-1α and NRF2 as therapeutic targets. Redox Biol. 2023, 60, 102603. [Google Scholar] [CrossRef]
- Rana, N.; Privitera, G.; Kondolf, H.C.; Bulek, K.; Lechuga, S.; De Salvo, C.; Corridoni, D.; Antanaviciute, A.; Maywald, R.L.; Hurtado, A.M.; et al. GSDMB is increased in IBD and regulates epithelial restitution/repair independent of pyroptosis. Cell 2022, 185, 283–298.e217. [Google Scholar] [CrossRef]
- Privitera, G.; Rana, N.; Armuzzi, A.; Pizarro, T.T. The gasdermin protein family: Emerging roles in gastrointestinal health and disease. Nat. Rev. Gastroenterol. Hepatol. 2023, 20, 366–387. [Google Scholar] [CrossRef]
- Dragoni, G.; De Hertogh, G.; Vermeire, S. The Role of Citrullination in Inflammatory Bowel Disease: A Neglected Player in Triggering Inflammation and Fibrosis? Inflamm. Bowel Dis. 2021, 27, 134–144. [Google Scholar] [CrossRef] [PubMed]
- Domínguez-Díaz, C.; Varela-Trinidad, G.U.; Muñoz-Sánchez, G.; Solórzano-Castanedo, K.; Avila-Arrezola, K.E.; Iñiguez-Gutiérrez, L.; Delgado-Rizo, V.; Fafutis-Morris, M. To Trap a Pathogen: Neutrophil Extracellular Traps and Their Role in Mucosal Epithelial and Skin Diseases. Cells 2021, 10, 1469. [Google Scholar] [CrossRef]
- Salas, A. What good can neutrophils do in UC? Gut 2022, 71, 2375–2376. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Saikam, V.; Skrada, K.A.; Merlin, D.; Iyer, S.S. Inflammatory bowel disease biomarkers. Med. Res. Rev. 2022, 42, 1856–1887. [Google Scholar] [CrossRef] [PubMed]
- Swaminathan, A.; Borichevsky, G.M.; Edwards, T.S.; Hirschfeld, E.; Mules, T.C.; Frampton, C.M.A.; Day, A.S.; Hampton, M.B.; Kettle, A.J.; Gearry, R.B. Faecal Myeloperoxidase as a Biomarker of Endoscopic Activity in Inflammatory Bowel Disease. J. Crohn’s Colitis 2022, 16, 1862–1873. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Zhang, C.; Wu, W.; Chen, H.; Lin, R.; Sun, R.; Gao, X.; Li, G.; He, Q.; Gao, H.; et al. MCPIP1 restrains mucosal inflammation by orchestrating the intestinal monocyte to macrophage maturation via an ATF3-AP1S2 axis. Gut 2023, 72, 882–895. [Google Scholar] [CrossRef]
- Turpin, W.; Lee, S.H.; Raygoza Garay, J.A.; Madsen, K.L.; Meddings, J.B.; Bedrani, L.; Power, N.; Espin-Garcia, O.; Xu, W.; Smith, M.I.; et al. Increased Intestinal Permeability Is Associated With Later Development of Crohn’s Disease. Gastroenterology 2020, 159, 2092–2100.e2095. [Google Scholar] [CrossRef]
- Fitzpatrick, J.A.; Melton, S.L.; Yao, C.K.; Gibson, P.R.; Halmos, E.P. Dietary management of adults with IBD—The emerging role of dietary therapy. Nat. Rev. Gastroenterol. Hepatol. 2022, 19, 652–669. [Google Scholar] [CrossRef]
- Sudhakar, P.; Wellens, J.; Verstockt, B.; Ferrante, M.; Sabino, J.; Vermeire, S. Holistic healthcare in inflammatory bowel disease: Time for patient-centric approaches? Gut 2023, 72, 192–204. [Google Scholar] [CrossRef] [PubMed]
- Adolph, T.E.; Meyer, M.; Schwarzler, J.; Mayr, L.; Grabherr, F.; Tilg, H. The metabolic nature of inflammatory bowel diseases. Nat. Rev. Gastroenterol. Hepatol. 2022, 19, 753–767. [Google Scholar] [CrossRef]
- Lopes, E.W.; Chan, S.S.M.; Song, M.; Ludvigsson, J.F.; Hakansson, N.; Lochhead, P.; Clark, A.; Burke, K.E.; Ananthakrishnan, A.N.; Cross, A.J.; et al. Lifestyle factors for the prevention of inflammatory bowel disease. Gut 2022. [Google Scholar] [CrossRef]
- Sun, Y.; Yuan, S.; Chen, X.; Sun, J.; Kalla, R.; Yu, L.; Wang, L.; Zhou, X.; Kong, X.; Hesketh, T.; et al. The Contribution of Genetic Risk and Lifestyle Factors in the Development of Adult-Onset Inflammatory Bowel Disease: A Prospective Cohort Study. Am. J. Gastroenterol. 2023, 118, 511–522. [Google Scholar] [CrossRef] [PubMed]
- Turpin, W.; Dong, M.; Sasson, G.; Raygoza Garay, J.A.; Espin-Garcia, O.; Lee, S.H.; Neustaeter, A.; Smith, M.I.; Leibovitzh, H.; Guttman, D.S.; et al. Mediterranean-Like Dietary Pattern Associations With Gut Microbiome Composition and Subclinical Gastrointestinal Inflammation. Gastroenterology 2022, 163, 685–698. [Google Scholar] [CrossRef] [PubMed]
- Peters, V.; Dijkstra, G.; Campmans-Kuijpers, M.J.E. Are all dietary fibers equal for patients with inflammatory bowel disease? A systematic review of randomized controlled trials. Nutr. Rev. 2022, 80, 1179–1193. [Google Scholar] [CrossRef] [PubMed]
- Narula, N.; Wong, E.C.L.; Dehghan, M.; Marshall, J.K.; Moayyedi, P.; Yusuf, S. Does a High-inflammatory Diet Increase the Risk of Inflammatory Bowel Disease? Results From the Prospective Urban Rural Epidemiology (PURE) Study: A Prospective Cohort Study. Gastroenterology 2021, 161, 1333–1335. [Google Scholar] [CrossRef]
- Gubatan, J.; Kulkarni, C.V.; Talamantes, S.M.; Temby, M.; Fardeen, T.; Sinha, S.R. Dietary Exposures and Interventions in Inflammatory Bowel Disease: Current Evidence and Emerging Concepts. Nutrients 2023, 15, 579. [Google Scholar] [CrossRef]
- Schirmer, M.; Garner, A.; Vlamakis, H.; Xavier, R.J. Microbial genes and pathways in inflammatory bowel disease. Nat. Rev. Microbiol. 2019, 17, 497–511. [Google Scholar] [CrossRef] [PubMed]
- Pittayanon, R.; Lau, J.T.; Leontiadis, G.I.; Tse, F.; Yuan, Y.; Surette, M.; Moayyedi, P. Differences in Gut Microbiota in Patients With vs Without Inflammatory Bowel Diseases: A Systematic Review. Gastroenterology 2020, 158, 930–946.e931. [Google Scholar] [CrossRef]
- Lloyd-Price, J.; Arze, C.; Ananthakrishnan, A.N.; Schirmer, M.; Avila-Pacheco, J.; Poon, T.W.; Andrews, E.; Ajami, N.J.; Bonham, K.S.; Brislawn, C.J.; et al. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature 2019, 569, 655–662. [Google Scholar] [CrossRef] [PubMed]
- Kurilshikov, A.; Medina-Gomez, C.; Bacigalupe, R.; Radjabzadeh, D.; Wang, J.; Demirkan, A.; Le Roy, C.I.; Raygoza Garay, J.A.; Finnicum, C.T.; Liu, X.; et al. Large-scale association analyses identify host factors influencing human gut microbiome composition. Nat. Genet. 2021, 53, 156–165. [Google Scholar] [CrossRef]
- Yu, S.; Balasubramanian, I.; Laubitz, D.; Tong, K.; Bandyopadhyay, S.; Lin, X.; Flores, J.; Singh, R.; Liu, Y.; Macazana, C.; et al. Paneth Cell-Derived Lysozyme Defines the Composition of Mucolytic Microbiota and the Inflammatory Tone of the Intestine. Immunity 2020, 53, 398–416. [Google Scholar] [CrossRef]
- Vujkovic-Cvijin, I.; Sklar, J.; Jiang, L.; Natarajan, L.; Knight, R.; Belkaid, Y. Host variables confound gut microbiota studies of human disease. Nature 2020, 587, 448–454. [Google Scholar] [CrossRef] [PubMed]
- Fenneman, A.C.; Weidner, M.; Chen, L.A.; Nieuwdorp, M.; Blaser, M.J. Antibiotics in the pathogenesis of diabetes and inflammatory diseases of the gastrointestinal tract. Nat. Rev. Gastroenterol. Hepatol. 2023, 20, 81–100. [Google Scholar] [CrossRef]
- Iliev, I.D. Mycobiota-host immune interactions in IBD: Coming out of the shadows. Nat. Rev. Gastroenterol. Hepatol. 2022, 19, 91–92. [Google Scholar] [CrossRef]
- Guzzo, G.L.; Andrews, J.M.; Weyrich, L.S. The Neglected Gut Microbiome: Fungi, Protozoa, and Bacteriophages in Inflammatory Bowel Disease. Inflamm. Bowel Dis. 2022, 28, 1112–1122. [Google Scholar] [CrossRef]
- Li, X.V.; Leonardi, I.; Putzel, G.G.; Semon, A.; Fiers, W.D.; Kusakabe, T.; Lin, W.Y.; Gao, I.H.; Doron, I.; Gutierrez-Guerrero, A.; et al. Immune regulation by fungal strain diversity in inflammatory bowel disease. Nature 2022, 603, 672–678. [Google Scholar] [CrossRef]
- Shih, T.; Yusung, S.; Gonsky, R.; Dutra-Clarke, R.; Ziring, D.; Rabizadeh, S.; Kugathasan, S.; Denson, L.A.; Li, D.; Braun, J. Environmental Interaction of Resolved Human Cytomegalovirus Infection With Crohn’s Disease Location. Inflamm. Bowel Dis. 2023, 29, 328–331. [Google Scholar] [CrossRef] [PubMed]
- Hansson, G.C. Mucins and the Microbiome. Annu. Rev. Biochem. 2020, 89, 769–793. [Google Scholar] [CrossRef] [PubMed]
- Yao, D.; Dai, W.; Dong, M.; Dai, C.; Wu, S. MUC2 and related bacterial factors: Therapeutic targets for ulcerative colitis. eBioMedicine 2021, 74, 103751. [Google Scholar] [CrossRef] [PubMed]
- Motta, J.P.; Wallace, J.L.; Buret, A.G.; Deraison, C.; Vergnolle, N. Gastrointestinal biofilms in health and disease. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 314–334. [Google Scholar] [CrossRef] [PubMed]
- Buisson, A.; Sokol, H.; Hammoudi, N.; Nancey, S.; Treton, X.; Nachury, M.; Fumery, M.; Hébuterne, X.; Rodrigues, M.; Hugot, J.P.; et al. Role of adherent and invasive Escherichia coli in Crohn’s disease: Lessons from the postoperative recurrence model. Gut 2023, 72, 39–48. [Google Scholar] [CrossRef]
- Iliev, I.D.; Cadwell, K. Effects of Intestinal Fungi and Viruses on Immune Responses and Inflammatory Bowel Diseases. Gastroenterology 2021, 160, 1050–1066. [Google Scholar] [CrossRef]
- Abraham, C.; Abreu, M.T.; Turner, J.R. Pattern Recognition Receptor Signaling and Cytokine Networks in Microbial Defenses and Regulation of Intestinal Barriers: Implications for Inflammatory Bowel Disease. Gastroenterology 2022, 162, 1602–1616. [Google Scholar] [CrossRef]
- Kimura, I.; Ichimura, A.; Ohue-Kitano, R.; Igarashi, M. Free Fatty Acid Receptors in Health and Disease. Physiol. Rev. 2020, 100, 171–210. [Google Scholar] [CrossRef]
- Melhem, H.; Kaya, B.; Ayata, C.K.; Hruz, P.; Niess, J.H. Metabolite-Sensing G Protein-Coupled Receptors Connect the Diet-Microbiota-Metabolites Axis to Inflammatory Bowel Disease. Cells 2019, 8, 450–469. [Google Scholar] [CrossRef]
- Nystrom, E.E.L.; Martinez-Abad, B.; Arike, L.; Birchenough, G.M.H.; Nonnecke, E.B.; Castillo, P.A.; Svensson, F.; Bevins, C.L.; Hansson, G.C.; Johansson, M.E.V. An intercrypt subpopulation of goblet cells is essential for colonic mucus barrier function. Science 2021, 372, eabb1590. [Google Scholar] [CrossRef]
- Smillie, C.S.; Biton, M.; Ordovas-Montanes, J.; Sullivan, K.M.; Burgin, G.; Graham, D.B.; Herbst, R.H.; Rogel, N.; Slyper, M.; Waldman, J.; et al. Intra- and Inter-cellular Rewiring of the Human Colon during Ulcerative Colitis. Cell 2019, 178, 714–730. [Google Scholar] [CrossRef]
- Parikh, K.; Antanaviciute, A.; Fawkner-Corbett, D.; Jagielowicz, M.; Aulicino, A.; Lagerholm, C.; Davis, S.; Kinchen, J.; Chen, H.H.; Alham, N.K.; et al. Colonic epithelial cell diversity in health and inflammatory bowel disease. Nature 2019, 567, 49–55. [Google Scholar] [CrossRef]
- Hendel, S.K.; Kellermann, L.; Hausmann, A.; Bindslev, N.; Jensen, K.B.; Nielsen, O.H. Tuft Cells and Their Role in Intestinal Diseases. Front. Immunol. 2022, 13, 822867. [Google Scholar] [CrossRef]
- Howitt, M.R.; Lavoie, S.; Michaud, M.; Blum, A.M.; Tran, S.V.; Weinstock, J.V.; Gallini, C.A.; Redding, K.; Margolskee, R.F.; Osborne, L.C.; et al. Tuft cells, taste-chemosensory cells, orchestrate parasite type 2 immunity in the gut. Science 2016, 351, 1329–1333. [Google Scholar] [CrossRef] [PubMed]
- Zundler, S.; Günther, C.; Kremer, A.E.; Zaiss, M.M.; Rothhammer, V.; Neurath, M.F. Gut immune cell trafficking: Inter-organ communication and immune-mediated inflammation. Nat. Rev. Gastroenterol. Hepatol. 2023, 20, 50–64. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, T.S.; Rampelli, S.; Jeffery, I.B.; Santoro, A.; Neto, M.; Capri, M.; Giampieri, E.; Jennings, A.; Candela, M.; Turroni, S.; et al. Mediterranean diet intervention alters the gut microbiome in older people reducing frailty and improving health status: The NU-AGE 1-year dietary intervention across five European countries. Gut 2020, 69, 1218–1228. [Google Scholar] [CrossRef] [PubMed]
- Meslier, V.; Laiola, M.; Roager, H.M.; De Filippis, F.; Roume, H.; Quinquis, B.; Giacco, R.; Mennella, I.; Ferracane, R.; Pons, N.; et al. Mediterranean diet intervention in overweight and obese subjects lowers plasma cholesterol and causes changes in the gut microbiome and metabolome independently of energy intake. Gut 2020, 69, 1258–1268. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.D.; Nguyen, L.H.; Li, Y.; Yan, Y.; Ma, W.; Rinott, E.; Ivey, K.L.; Shai, I.; Willett, W.C.; Hu, F.B.; et al. The gut microbiome modulates the protective association between a Mediterranean diet and cardiometabolic disease risk. Nat. Med. 2021, 27, 333–343. [Google Scholar] [CrossRef]
- Sarbagili Shabat, C.; Scaldaferri, F.; Zittan, E.; Hirsch, A.; Mentella, M.C.; Musca, T.; Cohen, N.A.; Ron, Y.; Fliss Isakov, N.; Pfeffer, J.; et al. Use of Faecal Transplantation with a Novel Diet for Mild to Moderate Active Ulcerative Colitis: The CRAFT UC Randomised Controlled Trial. J. Crohn’s Colitis 2022, 16, 369–378. [Google Scholar] [CrossRef]
- Kedia, S.; Virmani, S.; Vuyyuru, S.K.; Kumar, P.; Kante, B.; Sahu, P.; Kaushal, K.; Farooqui, M.; Singh, M.; Verma, M.; et al. Faecal microbiota transplantation with anti-inflammatory diet (FMT-AID) followed by anti-inflammatory diet alone is effective in inducing and maintaining remission over 1 year in mild to moderate ulcerative colitis: A randomised controlled trial. Gut 2022, 71, 2401–2413. [Google Scholar] [CrossRef] [PubMed]
- Overgaard, S.H.; Sørensen, S.B.; Munk, H.L.; Nexøe, A.B.; Glerup, H.; Henriksen, R.H.; Guldmann, T.; Pedersen, N.; Saboori, S.; Hvid, L.; et al. Impact of fibre and red/processed meat intake on treatment outcomes among patients with chronic inflammatory diseases initiating biological therapy: A prospective cohort study. Front. Nutr. 2022, 9, 985732. [Google Scholar] [CrossRef] [PubMed]
- Shin, A.; Kashyap, P.C. Promote or Prevent? Gut Microbial Function and Immune Status May Determine the Effect of Fiber in Inflammatory Bowel Disease. Gastroenterology 2023, 164, 182–184. [Google Scholar] [CrossRef] [PubMed]
- Procházková, N.; Venlet, N.; Hansen, M.L.; Lieberoth, C.B.; Dragsted, L.O.; Bahl, M.I.; Licht, T.R.; Kleerebezem, M.; Lauritzen, L.; Roager, H.M. Effects of a wholegrain-rich diet on markers of colonic fermentation and bowel function and their associations with the gut microbiome: A randomised controlled cross-over trial. Front. Nutr. 2023, 10, 1187165. [Google Scholar] [CrossRef] [PubMed]
- Gill, S.K.; Rossi, M.; Bajka, B.; Whelan, K. Dietary fibre in gastrointestinal health and disease. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 101–116. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Collij, V.; Jaeger, M.; van den Munckhof, I.C.L.; Vich Vila, A.; Kurilshikov, A.; Gacesa, R.; Sinha, T.; Oosting, M.; Joosten, L.A.B.; et al. Gut microbial co-abundance networks show specificity in inflammatory bowel disease and obesity. Nat. Commun. 2020, 11, 4018. [Google Scholar] [CrossRef]
- Deleu, S.; Machiels, K.; Raes, J.; Verbeke, K.; Vermeire, S. Short chain fatty acids and its producing organisms: An overlooked therapy for IBD? eBioMedicine 2021, 66, 103293. [Google Scholar] [CrossRef]
- Verburgt, C.M.; Dunn, K.A.; Ghiboub, M.; Lewis, J.D.; Wine, E.; Sigall Boneh, R.; Gerasimidis, K.; Shamir, R.; Penny, S.; Pinto, D.M.; et al. Successful Dietary Therapy in Paediatric Crohn’s Disease is Associated with Shifts in Bacterial Dysbiosis and Inflammatory Metabotype Towards Healthy Controls. J. Crohn’s Colitis 2023, 17, 61–72. [Google Scholar] [CrossRef]
- van der Hee, B.; Wells, J.M. Microbial Regulation of Host Physiology by Short-chain Fatty Acids. Trends Microbiol. 2021, 29, 700–712. [Google Scholar] [CrossRef]
- Graham, D.B.; Xavier, R.J. Pathway paradigms revealed from the genetics of inflammatory bowel disease. Nature 2020, 578, 527–539. [Google Scholar] [CrossRef]
- Somineni, H.K.; Nagpal, S.; Venkateswaran, S.; Cutler, D.J.; Okou, D.T.; Haritunians, T.; Simpson, C.L.; Begum, F.; Datta, L.W.; Quiros, A.J.; et al. Whole-genome sequencing of African Americans implicates differential genetic architecture in inflammatory bowel disease. Am. J. Hum. Genet. 2021, 108, 431–445. [Google Scholar] [CrossRef]
- Uhlig, H.H.; Charbit-Henrion, F.; Kotlarz, D.; Shouval, D.S.; Schwerd, T.; Strisciuglio, C.; de Ridder, L.; van Limbergen, J.; Macchi, M.; Snapper, S.B.; et al. Clinical Genomics for the Diagnosis of Monogenic Forms of Inflammatory Bowel Disease: A Position Paper From the Paediatric IBD Porto Group of European Society of Paediatric Gastroenterology, Hepatology and Nutrition. J. Pediatr. Gastroenterol. Nutr. 2021, 72, 456–473. [Google Scholar] [CrossRef] [PubMed]
- Bolton, C.; Smillie, C.S.; Pandey, S.; Elmentaite, R.; Wei, G.; Argmann, C.; Aschenbrenner, D.; James, K.R.; McGovern, D.P.B.; Macchi, M.; et al. An Integrated Taxonomy for Monogenic Inflammatory Bowel Disease. Gastroenterology 2022, 162, 859–876. [Google Scholar] [CrossRef]
- Ntunzwenimana, J.C.; Boucher, G.; Paquette, J.; Gosselin, H.; Alikashani, A.; Morin, N.; Beauchamp, C.; Thauvette, L.; Rivard, M.E.; Dupuis, F.; et al. Functional screen of inflammatory bowel disease genes reveals key epithelial functions. Genome Med. 2021, 13, 181. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Liu, R.; Gao, H.; Jung, S.; Gao, X.; Sun, R.; Liu, X.; Kim, Y.; Lee, H.S.; Kawai, Y.; et al. Genetic architecture of the inflammatory bowel diseases across East Asian and European ancestries. Nat. Genet. 2023, 55, 796–806. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.B.; Lin, H.C. Role of Intestinal Alkaline Phosphatase in Innate Immunity. Biomolecules 2021, 11, 1784–1795. [Google Scholar] [CrossRef]
- Mishra, V.; Bose, A.; Kiran, S.; Banerjee, S.; Shah, I.A.; Chaukimath, P.; Reshi, M.M.; Srinivas, S.; Barman, A.; Visweswariah, S.S. Gut-associated cGMP mediates colitis and dysbiosis in a mouse model of an activating mutation in GUCY2C. J. Exp. Med. 2021, 218, e20210479. [Google Scholar] [CrossRef]
- Sazonovs, A.; Stevens, C.R.; Venkataraman, G.R.; Yuan, K.; Avila, B.; Abreu, M.T.; Ahmad, T.; Allez, M.; Ananthakrishnan, A.N.; Atzmon, G.; et al. Large-scale sequencing identifies multiple genes and rare variants associated with Crohn’s disease susceptibility. Nat. Genet. 2022, 54, 1275–1283. [Google Scholar] [CrossRef]
- Brand, E.C.; Klaassen, M.A.Y.; Gacesa, R.; Vich Vila, A.; Ghosh, H.; de Zoete, M.R.; Boomsma, D.I.; Hoentjen, F.; Horjus Talabur Horje, C.S.; van de Meeberg, P.C.; et al. Healthy Cotwins Share Gut Microbiome Signatures With Their Inflammatory Bowel Disease Twins and Unrelated Patients. Gastroenterology 2021, 160, 1970–1985. [Google Scholar] [CrossRef]
- Sharma, A.; Szymczak, S.; Ruhlemann, M.; Freitag-Wolf, S.; Knecht, C.; Enderle, J.; Schreiber, S.; Franke, A.; Lieb, W.; Krawczak, M.; et al. Linkage analysis identifies novel genetic modifiers of microbiome traits in families with inflammatory bowel disease. Gut Microbes 2022, 14, 2024415. [Google Scholar] [CrossRef]
- Ruhlemann, M.C.; Hermes, B.M.; Bang, C.; Doms, S.; Moitinho-Silva, L.; Thingholm, L.B.; Frost, F.; Degenhardt, F.; Wittig, M.; Kassens, J.; et al. Genome-wide association study in 8,956 German individuals identifies influence of ABO histo-blood groups on gut microbiome. Nat. Genet. 2021, 53, 147–155. [Google Scholar] [CrossRef]
- Joustra, V.; Hageman, I.L.; Satsangi, J.; Adams, A.; Ventham, N.T.; de Jonge, W.J.; Henneman, P.; D’Haens, G.R.; Li Yim, A.Y.F. Systematic Review and Meta-analysis of Peripheral Blood DNA Methylation Studies in Inflammatory Bowel Disease. J. Crohn’s Colitis 2023, 17, 185–198. [Google Scholar] [CrossRef]
- Sendinc, E.; Shi, Y. RNA m6A methylation across the transcriptome. Mol. Cell 2023, 83, 428–441. [Google Scholar] [CrossRef]
- Deputy, M.; Devanaboina, R.; Al Bakir, I.; Burns, E.; Faiz, O. The role of faecal calprotectin in the diagnosis of inflammatory bowel disease. BMJ 2023, 380, e068947. [Google Scholar] [CrossRef]
- Fousert, E.; Toes, R.; Desai, J. Neutrophil Extracellular Traps (NETs) Take the Central Stage in Driving Autoimmune Responses. Cells 2020, 9, 915–934. [Google Scholar] [CrossRef]
- Kirov, S.; Sasson, A.; Zhang, C.; Chasalow, S.; Dongre, A.; Steen, H.; Stensballe, A.; Andersen, V.; Birkelund, S.; Bennike, T.B. Degradation of the extracellular matrix is part of the pathology of ulcerative colitis. Mol. Omics 2019, 15, 67–76. [Google Scholar] [CrossRef] [PubMed]
- Dinallo, V.; Marafini, I.; Di Fusco, D.; Laudisi, F.; Franzè, E.; Di Grazia, A.; Figliuzzi, M.M.; Caprioli, F.; Stolfi, C.; Monteleone, I.; et al. Neutrophil Extracellular Traps Sustain Inflammatory Signals in Ulcerative Colitis. J. Crohn’s Colitis 2019, 13, 772–784. [Google Scholar] [CrossRef] [PubMed]
- Hindson, J. Gasdermin B in IBD and epithelial barrier repair. Nat. Rev. Gastroenterol. Hepatol. 2022, 19, 216. [Google Scholar] [CrossRef]
- Manfredi, M.; Van Hoovels, L.; Benucci, M.; De Luca, R.; Coccia, C.; Bernardini, P.; Russo, E.; Amedei, A.; Guiducci, S.; Grossi, V.; et al. Circulating Calprotectin (cCLP) in autoimmune diseases. Autoimmun. Rev. 2023, 22, 103295. [Google Scholar] [CrossRef] [PubMed]
- Takenaka, K.; Kitazume, Y.; Kawamoto, A.; Fujii, T.; Udagawa, Y.; Wanatabe, R.; Shimizu, H.; Hibiya, S.; Nagahori, M.; Ohtsuka, K.; et al. Serum Leucine-Rich alpha2 Glycoprotein: A Novel Biomarker for Transmural Inflammation in Crohn’s Disease. Am. J. Gastroenterol. 2022, 118, 1028–1035. [Google Scholar] [CrossRef]
- Vitali, R.; Palone, F.; Armuzzi, A.; Fulci, V.; Negroni, A.; Carissimi, C.; Cucchiara, S.; Stronati, L. Proteomic Analysis Identifies Three Reliable Biomarkers of Intestinal Inflammation in the Stools of Patients With Inflammatory Bowel Disease. J. Crohn’s Colitis 2023, 17, 92–102. [Google Scholar] [CrossRef]
- Park, J.; Jeong, G.H.; Song, M.; Yon, D.K.; Lee, S.W.; Koyanagi, A.; Jacob, L.; Kostev, K.; Dragioti, E.; Radua, J.; et al. The global, regional, and national burden of inflammatory bowel diseases, 1990-2019: A systematic analysis for the global burden of disease study 2019. Dig. Liver Dis. 2023. [Google Scholar] [CrossRef]
- Joossens, M.; Huys, G.; Cnockaert, M.; De Preter, V.; Verbeke, K.; Rutgeerts, P.; Vandamme, P.; Vermeire, S. Dysbiosis of the faecal microbiota in patients with Crohn’s disease and their unaffected relatives. Gut 2011, 60, 631. [Google Scholar] [CrossRef]
- Sedwick, C. A Novel Regulatory T Cell Population in the Gut. PLOS Biology 2014, 12, e1001834. [Google Scholar] [CrossRef] [PubMed]
- Machiels, K.; Joossens, M.; Sabino, J.; De Preter, V.; Arijs, I.; Eeckhaut, V.; Ballet, V.; Claes, K.; Van Immerseel, F.; Verbeke, K.; et al. A decrease of the butyrate-producing species Roseburia hominis and Faecalibacterium prausnitzii defines dysbiosis in patients with ulcerative colitis. Gut 2014, 63, 1275–1283. [Google Scholar] [CrossRef] [PubMed]
- Png, C.W.; Linden, S.K.; Gilshenan, K.S.; Zoetendal, E.G.; McSweeney, C.S.; Sly, L.I.; McGuckin, M.A.; Florin, T.H. Mucolytic bacteria with increased prevalence in IBD mucosa augment in vitro utilization of mucin by other bacteria. Am. J. Gastroenterol. 2010, 105, 2420–2428. [Google Scholar] [CrossRef] [PubMed]
- Morrison, D.J.; Preston, T. Formation of short chain fatty acids by the gut microbiota and their impact on human metabolism. Gut microbes 2016, 7, 189–200. [Google Scholar] [CrossRef]
- Crost, E.H.; Tailford, L.E.; Monestier, M.; Swarbreck, D.; Henrissat, B.; Crossman, L.C.; Juge, N. The mucin-degradation strategy of Ruminococcus gnavus: The importance of intramolecular trans-sialidases. Gut Microbes 2016, 7, 302–312. [Google Scholar] [CrossRef]
- Round, J.L.; Mazmanian, S.K. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc. Natl. Acad. Sci. USA 2010, 107, 12204–12209. [Google Scholar] [CrossRef]
- Mazmanian, S.K.; Round, J.L.; Kasper, D.L. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature 2008, 453, 620–625. [Google Scholar] [CrossRef] [PubMed]
- Jeon, S.G.; Kayama, H.; Ueda, Y.; Takahashi, T.; Asahara, T.; Tsuji, H.; Tsuji, N.M.; Kiyono, H.; Ma, J.S.; Kusu, T.; et al. Probiotic Bifidobacterium breve induces IL-10-producing Tr1 cells in the colon. PLoS Pathog. 2012, 8, e1002714. [Google Scholar] [CrossRef]
- Yu, Y.; Zhu, S.; Li, P.; Min, L.; Zhang, S. Helicobacter pylori infection and inflammatory bowel disease: A crosstalk between upper and lower digestive tract. Cell Death Dis. 2018, 9, 961. [Google Scholar] [CrossRef] [PubMed]
- Iljazovic, A.; Roy, U.; Gálvez, E.J.C.; Lesker, T.R.; Zhao, B.; Gronow, A.; Amend, L.; Will, S.E.; Hofmann, J.D.; Pils, M.C.; et al. Perturbation of the gut microbiome by Prevotella spp. enhances host susceptibility to mucosal inflammation. Mucosal Immunol. 2021, 14, 113–124. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.C.-H. Microbiota dysbiosis and barrier dysfunction in inflammatory bowel disease and colorectal cancers: Exploring a common ground hypothesis. J. Biomed. Sci. 2018, 25, 79. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, S.; Campbell, B.J.; Rhodes, J.M. Bacteria in the pathogenesis of inflammatory bowel disease. Curr. Opin. Infect. Dis. 2006, 19, 475–484. [Google Scholar] [CrossRef]
- Strauss, J.; Kaplan, G.G.; Beck, P.L.; Rioux, K.; Panaccione, R.; Devinney, R.; Lynch, T.; Allen-Vercoe, E. Invasive potential of gut mucosa-derived Fusobacterium nucleatum positively correlates with IBD status of the host. Inflamm. Bowel Dis. 2011, 17, 1971–1978. [Google Scholar] [CrossRef]
- Parlato, M.; Charbit-Henrion, F.; Pan, J.; Romano, C.; Duclaux-Loras, R.; Le Du, M.H.; Warner, N.; Francalanci, P.; Bruneau, J.; Bras, M.; et al. Human ALPI deficiency causes inflammatory bowel disease and highlights a key mechanism of gut homeostasis. EMBO Mol. Med. 2018, 10, e8483. [Google Scholar] [CrossRef]
- Crowley, E.; Warner, N.; Pan, J.; Khalouei, S.; Elkadri, A.; Fiedler, K.; Foong, J.; Turinsky, A.L.; Bronte-Tinkew, D.; Zhang, S.; et al. Prevalence and Clinical Features of Inflammatory Bowel Diseases Associated With Monogenic Variants, Identified by Whole-Exome Sequencing in 1000 Children at a Single Center. Gastroenterology 2020, 158, 2208–2220. [Google Scholar] [CrossRef]
- Cadwell, K.; Liu, J.Y.; Brown, S.L.; Miyoshi, H.; Loh, J.; Lennerz, J.K.; Kishi, C.; Kc, W.; Carrero, J.A.; Hunt, S.; et al. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature 2008, 456, 259–263. [Google Scholar] [CrossRef]
- Deuring, J.J.; Fuhler, G.M.; Konstantinov, S.R.; Peppelenbosch, M.P.; Kuipers, E.J.; de Haar, C.; van der Woude, C.J. Genomic ATG16L1 risk allele-restricted Paneth cell ER stress in quiescent Crohn’s disease. Gut 2014, 63, 1081–1091. [Google Scholar] [CrossRef]
- Salem, M.; Ammitzboell, M.; Nys, K.; Seidelin, J.B.; Nielsen, O.H. ATG16L1: A multifunctional susceptibility factor in Crohn disease. Autophagy 2015, 11, 585–594. [Google Scholar] [CrossRef]
- Rioux, J.D.; Xavier, R.J.; Taylor, K.D.; Silverberg, M.S.; Goyette, P.; Huett, A.; Green, T.; Kuballa, P.; Barmada, M.M.; Datta, L.W.; et al. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat. Genet. 2007, 39, 596–604. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Fang, M.; Jostins, L.; Umicevic Mirkov, M.; Boucher, G.; Anderson, C.A.; Andersen, V.; Cleynen, I.; Cortes, A.; Crins, F.; et al. Fine-mapping inflammatory bowel disease loci to single-variant resolution. Nature 2017, 547, 173–178. [Google Scholar] [CrossRef] [PubMed]
- Broquet, A.H.; Hirata, Y.; McAllister, C.S.; Kagnoff, M.F. RIG-I/MDA5/MAVS are required to signal a protective IFN response in rotavirus-infected intestinal epithelium. J. Immunol. 2011, 186, 1618–1626. [Google Scholar] [CrossRef] [PubMed]
- Jostins, L.; Ripke, S.; Weersma, R.K.; Duerr, R.H.; McGovern, D.P.; Hui, K.Y.; Lee, J.C.; Schumm, L.P.; Sharma, Y.; Anderson, C.A.; et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 2012, 491, 119–124. [Google Scholar] [CrossRef] [PubMed]
- Yang, E.; Shen, J. The roles and functions of Paneth cells in Crohn‘s disease: A critical review. Cell Prolif. 2021, 54, e12958. [Google Scholar] [CrossRef] [PubMed]
- Negroni, A.; Colantoni, E.; Vitali, R.; Palone, F.; Pierdomenico, M.; Costanzo, M.; Cesi, V.; Cucchiara, S.; Stronati, L. NOD2 induces autophagy to control AIEC bacteria infectiveness in intestinal epithelial cells. Inflamm. Res. 2016, 65, 803–813. [Google Scholar] [CrossRef]
- Lee, C.; Choi, C.; Kang, H.S.; Shin, S.W.; Kim, S.Y.; Park, H.C.; Hong, S.N. NOD2 Supports Crypt Survival and Epithelial Regeneration after Radiation-Induced Injury. Int. J. Mol. Sci. 2019, 20, 4297. [Google Scholar] [CrossRef]
- Saxena, A.; Lopes, F.; Poon, K.K.H.; McKay, D.M. Absence of the NOD2 protein renders epithelia more susceptible to barrier dysfunction due to mitochondrial dysfunction. Am. J. Physiol. Gastrointest. Liver Physio.l 2017, 313, G26–G38. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Kim, J.J.; Denou, E.; Gallagher, A.; Thornton, D.J.; Shajib, M.S.; Xia, L.; Schertzer, J.D.; Grencis, R.K.; Philpott, D.J.; et al. New Role of Nod Proteins in Regulation of Intestinal Goblet Cell Response in the Context of Innate Host Defense in an Enteric Parasite Infection. Infect. Immun. 2016, 84, 275–285. [Google Scholar] [CrossRef]
- Siggers, R.H.; Hackam, D.J. The role of innate immune-stimulated epithelial apoptosis during gastrointestinal inflammatory diseases. Cell Mol Life Sci 2011, 68, 3623–3634. [Google Scholar] [CrossRef]
- Fritz, T.; Niederreiter, L.; Adolph, T.; Blumberg, R.S.; Kaser, A. Crohn’s disease: NOD2, autophagy and ER stress converge. Gut 2011, 60, 1580–1588. [Google Scholar] [CrossRef] [PubMed]
- Ogura, Y.; Bonen, D.K.; Inohara, N.; Nicolae, D.L.; Chen, F.F.; Ramos, R.; Britton, H.; Moran, T.; Karaliuskas, R.; Duerr, R.H.; et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn’s disease. Nature 2001, 411, 603–606. [Google Scholar] [CrossRef] [PubMed]
- Hugot, J.P.; Chamaillard, M.; Zouali, H.; Lesage, S.; Cezard, J.P.; Belaiche, J.; Almer, S.; Tysk, C.; O’Morain, C.A.; Gassull, M.; et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn’s disease. Nature 2001, 411, 599–603. [Google Scholar] [CrossRef]
- Feng, Y.; Tsai, Y.H.; Xiao, W.; Ralls, M.W.; Stoeck, A.; Wilson, C.L.; Raines, E.W.; Teitelbaum, D.H.; Dempsey, P.J. Loss of ADAM17-Mediated Tumor Necrosis Factor Alpha Signaling in Intestinal Cells Attenuates Mucosal Atrophy in a Mouse Model of Parenteral Nutrition. Mol. Cell Biol. 2015, 35, 3604–3621. [Google Scholar] [CrossRef]
- Fréour, T.; Jarry, A.; Bach-Ngohou, K.; Dejoie, T.; Bou-Hanna, C.; Denis, M.G.; Mosnier, J.F.; Laboisse, C.L.; Masson, D. TACE inhibition amplifies TNF-alpha-mediated colonic epithelial barrier disruption. Int. J. Mol. Med. 2009, 23, 41–48. [Google Scholar] [PubMed]
- Hilliard, V.C.; Frey, M.R.; Dempsey, P.J.; Peek, R.M., Jr.; Polk, D.B. TNF-α converting enzyme-mediated ErbB4 transactivation by TNF promotes colonic epithelial cell survival. Am. J. Physiol. Gastrointest. Liver Physiol. 2011, 301, G338–G346. [Google Scholar] [CrossRef]
- Shimoda, M.; Horiuchi, K.; Sasaki, A.; Tsukamoto, T.; Okabayashi, K.; Hasegawa, H.; Kitagawa, Y.; Okada, Y. Epithelial Cell-Derived a Disintegrin and Metalloproteinase-17 Confers Resistance to Colonic Inflammation Through EGFR Activation. EBioMedicine 2016, 5, 114–124. [Google Scholar] [CrossRef]
- Blaydon, D.C.; Biancheri, P.; Di, W.L.; Plagnol, V.; Cabral, R.M.; Brooke, M.A.; van Heel, D.A.; Ruschendorf, F.; Toynbee, M.; Walne, A.; et al. Inflammatory skin and bowel disease linked to ADAM17 deletion. N. Engl. J. Med. 2011, 365, 1502–1508. [Google Scholar] [CrossRef]
- Gettler, K.; Levantovsky, R.; Moscati, A.; Giri, M.; Wu, Y.; Hsu, N.Y.; Chuang, L.S.; Sazonovs, A.; Venkateswaran, S.; Korie, U.; et al. Common and Rare Variant Prediction and Penetrance of IBD in a Large, Multi-ethnic, Health System-based Biobank Cohort. Gastroenterology 2021, 160, 1546–1557. [Google Scholar] [CrossRef]
- Nakatsu, F.; Baskin, J.M.; Chung, J.; Tanner, L.B.; Shui, G.; Lee, S.Y.; Pirruccello, M.; Hao, M.; Ingolia, N.T.; Wenk, M.R.; et al. PtdIns4P synthesis by PI4KIIIα at the plasma membrane and its impact on plasma membrane identity. J. Cell Biol. 2012, 199, 1003–1016. [Google Scholar] [CrossRef]
- Avitzur, Y.; Guo, C.; Mastropaolo, L.A.; Bahrami, E.; Chen, H.; Zhao, Z.; Elkadri, A.; Dhillon, S.; Murchie, R.; Fattouh, R.; et al. Mutations in tetratricopeptide repeat domain 7A result in a severe form of very early onset inflammatory bowel disease. Gastroenterology 2014, 146, 1028–1039. [Google Scholar] [CrossRef]
- Jardine, S.; Dhingani, N.; Muise, A.M. TTC7A: Steward of Intestinal Health. Cell. Mol. Gastroenterol. Hepatol. 2019, 7, 555–570. [Google Scholar] [CrossRef]
- Mohanan, V.; Nakata, T.; Desch, A.N.; Levesque, C.; Boroughs, A.; Guzman, G.; Cao, Z.; Creasey, E.; Yao, J.; Boucher, G.; et al. C1orf106 is a colitis risk gene that regulates stability of epithelial adherens junctions. Science 2018, 359, 1161–1166. [Google Scholar] [CrossRef]
- Luong, P.; Hedl, M.; Yan, J.; Zuo, T.; Fu, T.M.; Jiang, X.; Thiagarajah, J.R.; Hansen, S.H.; Lesser, C.F.; Wu, H.; et al. INAVA-ARNO complexes bridge mucosal barrier function with inflammatory signaling. eLife 2018, 7, e38539. [Google Scholar] [CrossRef] [PubMed]
- Rivas, M.A.; Beaudoin, M.; Gardet, A.; Stevens, C.; Sharma, Y.; Zhang, C.K.; Boucher, G.; Ripke, S.; Ellinghaus, D.; Burtt, N.; et al. Deep resequencing of GWAS loci identifies independent rare variants associated with inflammatory bowel disease. Nat. Genet. 2011, 43, 1066–1073. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Jiang, Y.; Zhang, L.; Qian, W.; Hou, X.; Lin, R. Exogenous l-fucose protects the intestinal mucosal barrier depending on upregulation of FUT2-mediated fucosylation of intestinal epithelial cells. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2021, 35, e21699. [Google Scholar] [CrossRef] [PubMed]
- McGovern, D.P.; Jones, M.R.; Taylor, K.D.; Marciante, K.; Yan, X.; Dubinsky, M.; Ippoliti, A.; Vasiliauskas, E.; Berel, D.; Derkowski, C.; et al. Fucosyltransferase 2 (FUT2) non-secretor status is associated with Crohn’s disease. Hum. Mol. Genet. 2010, 19, 3468–3476. [Google Scholar] [CrossRef] [PubMed]
- Wacklin, P.; Makivuokko, H.; Alakulppi, N.; Nikkila, J.; Tenkanen, H.; Rabina, J.; Partanen, J.; Aranko, K.; Matto, J. Secretor genotype (FUT2 gene) is strongly associated with the composition of Bifidobacteria in the human intestine. PLoS ONE 2011, 6, e20113. [Google Scholar] [CrossRef]
- Rausch, P.; Rehman, A.; Kunzel, S.; Hasler, R.; Ott, S.J.; Schreiber, S.; Rosenstiel, P.; Franke, A.; Baines, J.F. Colonic mucosa-associated microbiota is influenced by an interaction of Crohn disease and FUT2 (Secretor) genotype. Proc. Natl. Acad. Sci. USA 2011, 108, 19030–19035. [Google Scholar] [CrossRef]
- Zhang, H.; Cui, Z.; Cheng, D.; Du, Y.; Guo, X.; Gao, R.; Chen, J.; Sun, W.; He, R.; Ma, X.; et al. RNF186 regulates EFNB1 (ephrin B1)-EPHB2-induced autophagy in the colonic epithelial cells for the maintenance of intestinal homeostasis. Autophagy 2021, 17, 3030–3047. [Google Scholar] [CrossRef]
- Ji, Y.; Tu, X.; Hu, X.; Wang, Z.; Gao, S.; Zhang, Q.; Zhang, W.; Zhang, H.; Chen, W. The role and mechanism of action of RNF186 in colorectal cancer through negative regulation of NF-κB. Cell Signal 2020, 75, 109764. [Google Scholar] [CrossRef]
- Fujimoto, K.; Kinoshita, M.; Tanaka, H.; Okuzaki, D.; Shimada, Y.; Kayama, H.; Okumura, R.; Furuta, Y.; Narazaki, M.; Tamura, A.; et al. Regulation of intestinal homeostasis by the ulcerative colitis-associated gene RNF186. Mucosal. Immunol. 2017, 10, 446–459. [Google Scholar] [CrossRef] [PubMed]
- Rivas, M.A.; Graham, D.; Sulem, P.; Stevens, C.; Desch, A.N.; Goyette, P.; Gudbjartsson, D.; Jonsdottir, I.; Thorsteinsdottir, U.; Degenhardt, F.; et al. A protein-truncating R179X variant in RNF186 confers protection against ulcerative colitis. Nat. Commun. 2016, 7, 12342. [Google Scholar] [CrossRef] [PubMed]
- Beaudoin, M.; Goyette, P.; Boucher, G.; Lo, K.S.; Rivas, M.A.; Stevens, C.; Alikashani, A.; Ladouceur, M.; Ellinghaus, D.; Törkvist, L.; et al. Deep resequencing of GWAS loci identifies rare variants in CARD9, IL23R and RNF186 that are associated with ulcerative colitis. PLoS Genet. 2013, 9, e1003723. [Google Scholar] [CrossRef]
- Amarachintha, S.; Harmel-Laws, E.; Steinbrecher, K.A. Guanylate cyclase C reduces invasion of intestinal epithelial cells by bacterial pathogens. Sci. Rep. 2018, 8, 1521. [Google Scholar] [CrossRef] [PubMed]
- Garin-Laflam, M.P.; Steinbrecher, K.A.; Rudolph, J.A.; Mao, J.; Cohen, M.B. Activation of guanylate cyclase C signaling pathway protects intestinal epithelial cells from acute radiation-induced apoptosis. Am. J. Physiol. Gastrointest. Liver Physiol. 2009, 296, G740–G749. [Google Scholar] [CrossRef] [PubMed]
- Tronstad, R.R.; Kummen, M.; Holm, K.; von Volkmann, H.L.; Anmarkrud, J.A.; Hoivik, M.L.; Moum, B.; Gilja, O.H.; Hausken, T.; Baines, J.; et al. Guanylate Cyclase C Activation Shapes the Intestinal Microbiota in Patients with Familial Diarrhea and Increased Susceptibility for Crohn’s Disease. Inflamm. Bowel Dis. 2017, 23, 1752–1761. [Google Scholar] [CrossRef]
| Function | Refs. | |
|---|---|---|
| Dietary | ||
| Ultra-processed foods, emulsifiers, | Associated with IBD | [28] |
| Western diet | Associated with IBD, flares | [29,30] |
| Mediterranean diet | Improve inflammation | [31] |
| Fibre intake | Feed anti-inflammatory bacteria | [31,32] |
| Function | Refs. | |
|---|---|---|
| Microbiome | ||
| SCFA producers, proinflammatory bacteria and fungi | Predict treatment response | [33,34] |
| Microbial composition and metabolites | Treatment response or progression | [35] |
| Faecal species richness, Candida or Caudovirales abundance, donor microbial profile similarity or biotin (vitamin B7) | FMT treatment response | [36] |
| Reduction in alpha diversity, abundance of Firmicutes | Predict postoperative recurrence in CD | [37] |
| Faecalibacterium and Bacteroides enrichment | Predict treatment response | [23] |
| Various specific bacteria | IBD diagnosis and prognosis | [38] |
| Microbial richness | Predict treatment response | [39] |
| Microbiome risk profiles | Predict treatment response | [24] |
| Klepsiella pneumonia | Associated to IBD | [40] |
| AIEC | Associated to IBD | [41] |
| Bacterial and Fungal Profiles | Predict treatment response | [33,39] |
| Metabolic profiles of bile acids, lipids and SCFAs | Predict treatment response | [25] |
| Patient factors | ||
| Genetic | ||
| Variants in TNFSF4/18, PLIN2, NOD2, ATG16L1, TLRs and IL23R | Predict treatment response | [42] |
| IL-1B, IL-6, IFN-gamma, TNFRSF1A, NLRP3, IL1RN, IL-18, JAK2, LR2, TLR4, NFKBI | Predict treatment response | [43] |
| NOD2, CARD9 and RIPK2 | Microbial sensing | [23] |
| C1orf106 and HNF4A | Intestinal barrier function | [23] |
| Variants in PIGR, NFKBIZ, IL17RA and TRAF3IP2 | Predict IBD-associated colon cancer | [44,45,46] |
| Epigenetics | ||
| m6A modification | Predict prognosis | [47] |
| RNA metabolism | ||
| HP1γ | Predict treatment response | [48] |
| Immunologic | ||
| Faecal and serum calprotectin | Discriminate between the inflammatory and noninflammatory gut; track disease activity; treatment response | [43,49] |
| Anti-microbial antibodies | Predict disease development | [50] |
| Blood calprotectin, S100A12 | Diagnosis and disease maintenance | [51] |
| Blood and faecal microRNAs | Predict treatment response | [43] |
| Redox biomarkers | Whole-body redox status | [52] |
| Mucosal | ||
| TNF-α, IL-17A, IL-17R, OSM, OSMR, TREM1 | Predict treatment response | [43] |
| Gasdermins | Intestinal barrier function | [53,54] |
| Mucosal FoxP3 | Predict treatment response | [43] |
| NETs | Track disease activity | [55,56,57] |
| MPO, lactoferrin | Track disease activity | [58,59] |
| Mucosal F. prausnitzii | Predict treatment response | [43] |
| MCPIP1 | Increase intestinal inflammation | [60] |
| Urine | ||
| LMR | Predict disease development | [61] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Andersen, V.; Bennike, T.B.; Bang, C.; Rioux, J.D.; Hébert-Milette, I.; Sato, T.; Hansen, A.K.; Nielsen, O.H. Investigating the Crime Scene—Molecular Signatures in Inflammatory Bowel Disease. Int. J. Mol. Sci. 2023, 24, 11217. https://doi.org/10.3390/ijms241311217
Andersen V, Bennike TB, Bang C, Rioux JD, Hébert-Milette I, Sato T, Hansen AK, Nielsen OH. Investigating the Crime Scene—Molecular Signatures in Inflammatory Bowel Disease. International Journal of Molecular Sciences. 2023; 24(13):11217. https://doi.org/10.3390/ijms241311217
Chicago/Turabian StyleAndersen, Vibeke, Tue B. Bennike, Corinna Bang, John D. Rioux, Isabelle Hébert-Milette, Toshiro Sato, Axel K. Hansen, and Ole H. Nielsen. 2023. "Investigating the Crime Scene—Molecular Signatures in Inflammatory Bowel Disease" International Journal of Molecular Sciences 24, no. 13: 11217. https://doi.org/10.3390/ijms241311217
APA StyleAndersen, V., Bennike, T. B., Bang, C., Rioux, J. D., Hébert-Milette, I., Sato, T., Hansen, A. K., & Nielsen, O. H. (2023). Investigating the Crime Scene—Molecular Signatures in Inflammatory Bowel Disease. International Journal of Molecular Sciences, 24(13), 11217. https://doi.org/10.3390/ijms241311217








