CRISPR/Cas9-Mediated Cytosine Base Editing Using an Improved Transformation Procedure in Melon (Cucumis melo L.)
Abstract
1. Introduction
2. Results
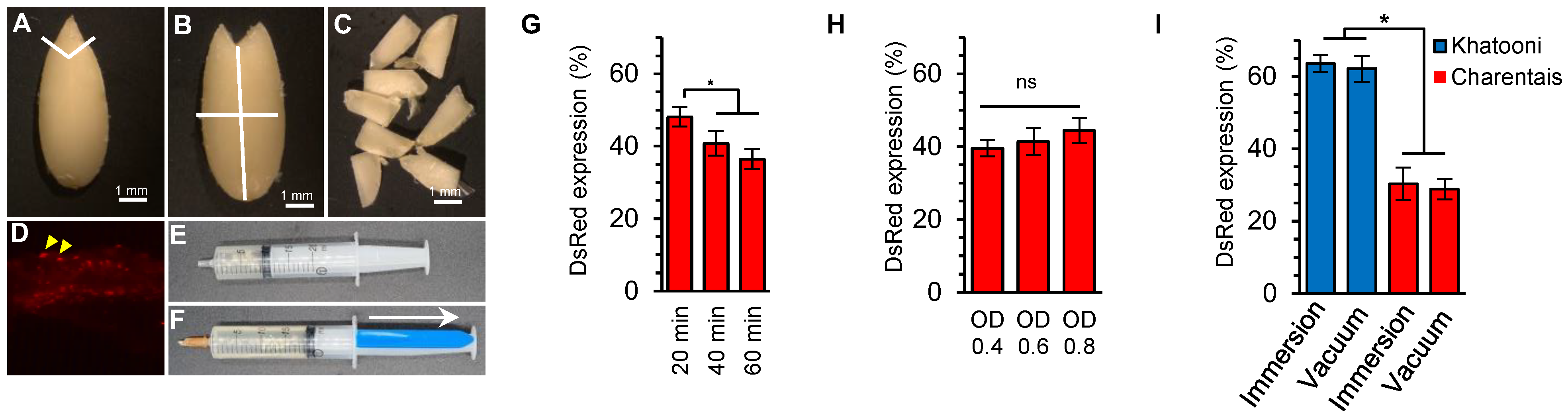
2.1. The Effects of Inoculation Time, Agrobacterium Concentration and Vacuum Infiltration on Explants Transformation Efficiency
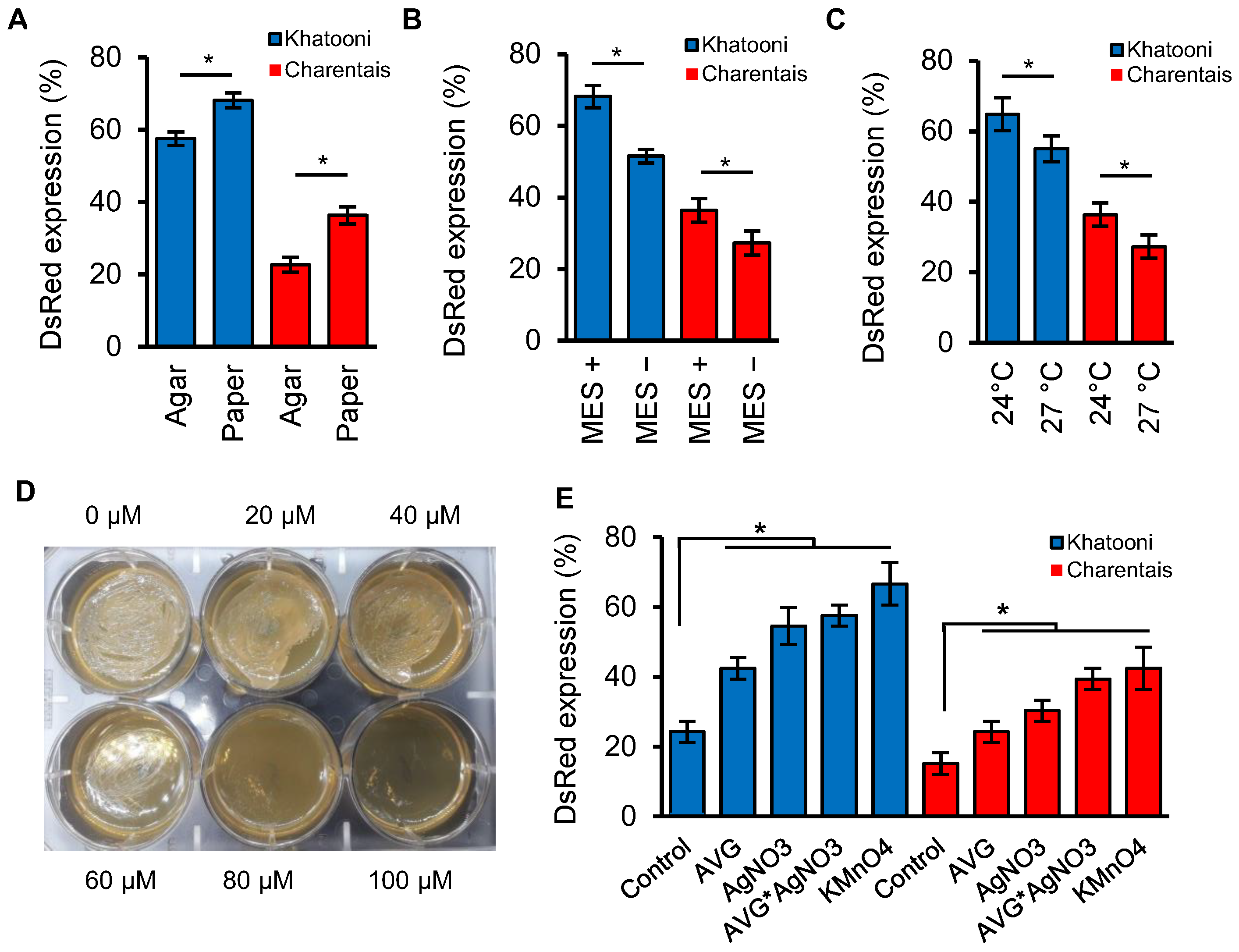
2.2. The Filter Paper, Reduce the Temperature and pH Stability in the Co-Culture Medium Increased Transformation Efficiency
2.3. Ethylene Inhibitors and Absorbers
2.3.1. Effect of AgNO3 on Agrobacterium Growth
2.3.2. Effects of Ethylene Inhibitors and Absorber on Transformation Efficiency
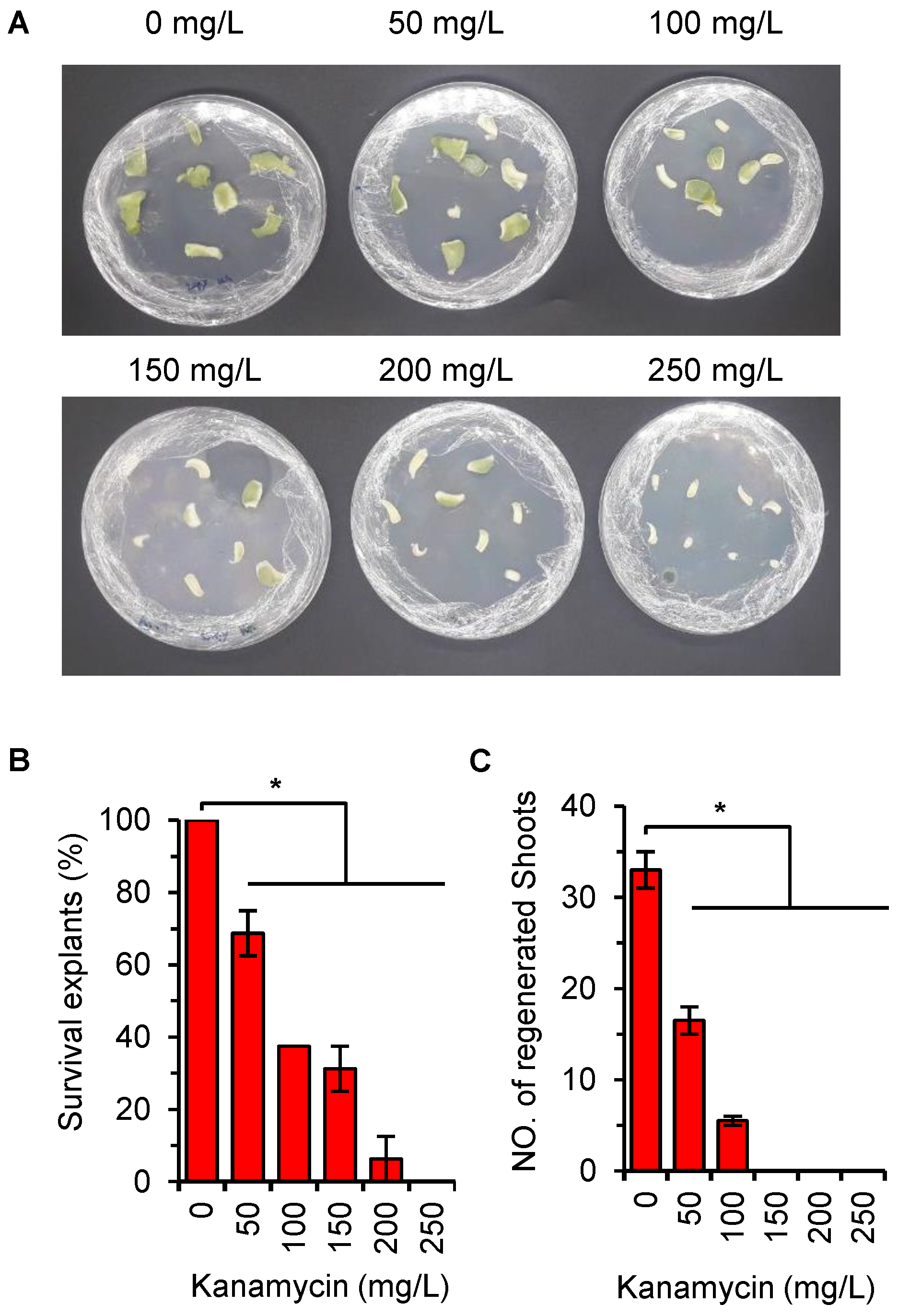
2.4. Optimization of Kanamycin Concentration for Efficient Selection of Transgenic Plant
2.5. Stable Transformation Optimization
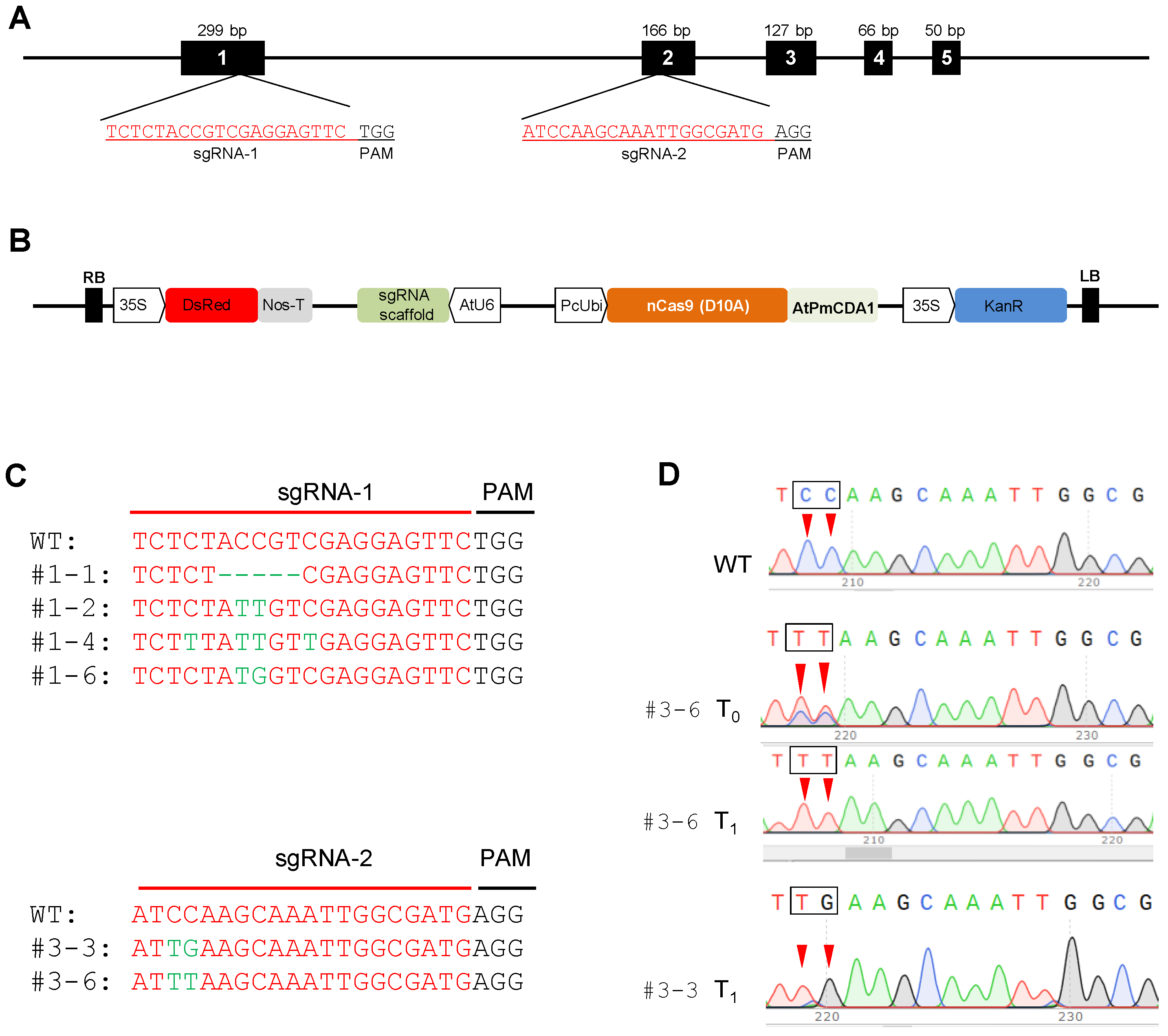
2.6. Genome Editing Using CRISPR-Cas9 Cytidine Deaminase Fusion
3. Discussion
4. Materials and Methods
4.1. Preparation of Explants
4.2. Agrobacterium Strain, Binary Vector and Inoculation Medium
4.3. Levels, Durations, and Methods of Explants Inoculation with Agrobacterium
4.4. Co-Culture Medium Experiments
4.5. Effect of Kanamycin on Explant Growth
4.6. Detection of DsRed Expression by Fluorescence Stereomicroscopy
4.7. Stable Transformation
4.8. Flow Cytometry
4.9. Statistical Analysis
4.10. Amplifying the DsRed Reporter Gene and Cloning to Target-AID Vector
4.11. The sgRNA Designing and Cloning in the Target-AID-DsRed Vector and Agrobacterium Transformation
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pitrat, M.M. Handbook of Plant Breeding: Vegetable; Prohens, J., Nuez, F., Eds.; Springer Science: New York, NY, USA, 2008; Volume I, pp. 283–316. [Google Scholar]
- Chomicki, G.; Schaefer, H.; Renner, S.S. Origin and domestication of Cucurbitaceae crops: Insights from phylogenies, genomics and archaeology. New Phytol. 2020, 226, 1240–1255. [Google Scholar] [CrossRef] [PubMed]
- Karimzadeh, G.; Mousavi, S.H.; Jafarkhani-Kermani, M.; Jalali-Javaran, M. Karyological and Nuclear DNA Variation in Iranian Endemic Muskmelon (Cucumis melo var. inodorus). Cytologia 2010, 75, 451–461. [Google Scholar] [CrossRef]
- Garcia-Mas, J.; Benjak, A.; Sanseverino, W.; Bourgeois, M.; Mir, G.; González, V.M.; Hénaff, E.; Câmara, F.; Cozzuto, L.; Lowy, E.; et al. The genome of melon (Cucumis melo L.). Proc. Natl. Acad. Sci. USA 2012, 109, 11872–11877. [Google Scholar] [CrossRef] [PubMed]
- Faostat, F. FAOSTAT Statistical Database; FAO (Food and Agriculture Organization of the United Nations): Rome, Italy, 2010; Available online: http://faostat.fao.org (accessed on 1 November 2022).
- Faostat, F. FAOSTAT Statistical Database; FAO (Food and Agriculture Organization of the United Nations): Rome, Italy, 2019; Available online: http://faostat.fao.org (accessed on 1 November 2022).
- Ray, D.K.; Mueller, N.D.; West, P.C.; Foley, J.A. Yield trends are insufficient to double global crop production by 2050. PLoS ONE 2013, 8, e66428. [Google Scholar] [CrossRef]
- Fernández-Trujillo, J.P.; Picó, B.; Garcia-Mas, J.; Álvarez, J.M.; Monforte, A.J. Breeding for Fruit Quality in Melon. In Breeding for Fruit Quality; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2011; pp. 261–278. [Google Scholar] [CrossRef]
- Kesh, H.; Kaushik, P. Advances in melon (Cucumis melo L.) breeding: An update. Sci. Hortic. 2021, 282, 110045. [Google Scholar] [CrossRef]
- Xu, L.; He, Y.; Tang, L.; Xu, Y.; Zhao, G. Genetics, Genomics, and Breeding in Melon. Agronomy 2022, 12, 2891. [Google Scholar] [CrossRef]
- Rodríguez-Hernández, A.M.; Gosalvez, B.; Sempere, R.N.; Burgos, L.; Aranda, M.A.; Truniger, V. Melon RNA interference (RNAi) lines silenced for Cm-eIF4E show broad virus resistance. Mol. Plant Pathol. 2012, 13, 755–763. [Google Scholar] [CrossRef]
- Nuñez-Palenius, H.G.; Cantliffe, D.J.; Huber, D.J.; Ciardi, J.; Klee, H.J. Transformation of a muskmelon ‘Galia’ hybrid parental line (Cucumis melo L. var. reticulatus Ser.) with an antisense ACC oxidase gene. Plant Cell Rep. 2006, 25, 198–205. [Google Scholar] [CrossRef]
- Ren, Y.; Bang, H.; Curtis, I.S.; Gould, J.; Patil, B.S.; Crosby, K.M. Agrobacterium-mediated transformation and shoot regeneration in elite breeding lines of western shipper cantaloupe and honeydew melons (Cucumis melo L.). Plant Cell Tissue Organ Cult. 2011, 108, 147–158. [Google Scholar] [CrossRef]
- Guis, M.; Ben Amor, M.; Latché, A.; Pech, J.-C.; Roustan, J.-P. A reliable system for the transformation of cantaloupe charentais melon (Cucumis melo L. var. cantalupensis) leading to a majority of diploid regenerants. Sci. Hortic. 2000, 84, 91–99. [Google Scholar] [CrossRef]
- Choi, J.Y.; Shin, J.S.; Chung, Y.S.; Hyung, N.-I. An efficient selection and regeneration protocol for Agrobacterium-mediated transformation of oriental melon (Cucumis melo L. var. makuwa). Plant Cell Tissue Organ Cult. 2012, 110, 133–140. [Google Scholar] [CrossRef]
- Marone, D.; Mastrangelo, A.M.; Borrelli, G.M. From Transgenesis to Genome Editing in Crop Improvement: Applications, Marketing, and Legal Issues. Int. J. Mol. Sci. 2023, 24, 7122. [Google Scholar] [CrossRef] [PubMed]
- Altpeter, F.; Springer, N.M.; Bartley, L.E.; Blechl, A.E.; Brutnell, T.P.; Citovsky, V.; Conrad, L.J.; Gelvin, S.B.; Jackson, D.P.; Kausch, A.P.; et al. Advancing Crop Transformation in the Era of Genome Editing. Plant Cell 2016, 28, 1510–1520. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.; Zhang, X.; Liang, S.; Nie, Y.; Guo, X.; Huang, C. Factors affecting transformation efficiency of embryogenic callus of Upland cotton (Gossypium hirsutum) with Agrobacterium tumefaciens. Plant Cell Tissue Organ Cult. 2005, 81, 229–237. [Google Scholar] [CrossRef]
- Concepción-hernández, M.; Reyes, M.; Rodríguez, M.; Gómez, R. Effect of inoculation time on Agrobacterium-mediated transformation efficiency of Musa cv. ‘Grande naine’ (AAA ). Plant Biotechnol. 2017, 17, 171–180. [Google Scholar]
- Chovelon, V.; Restier, V.; Giovinazzo, N.; Dogimont, C.; Aarrouf, J. Histological study of organogenesis in Cucumis melo L. after genetic transformation: Why is it difficult to obtain transgenic plants? Plant Cell Rep. 2011, 30, 2001–2011. [Google Scholar] [CrossRef]
- Nanasato, Y.; Konagaya, K.-I.; Okuzaki, A.; Tsuda, M.; Tabei, Y. Improvement of Agrobacterium-mediated transformation of cucumber (Cucumis sativus L.) by combination of vacuum infiltration and co-cultivation on filter paper wicks. Plant Biotechnol. Rep. 2012, 7, 267–276. [Google Scholar] [CrossRef]
- Švábová, L.; Griga, M. The effect of cocultivation treatments on transformation efficiency in pea (Pisum sativum L.). Plant Cell Tissue Organ Cult. 2008, 95, 293–304. [Google Scholar] [CrossRef]
- Yang, L.; Hu, W.; Xie, Y.; Li, Y.; Deng, Z. Factors affecting Agrobacterium -mediated transformation efficiency of kumquat seedling internodal stem segments. Sci. Hortic. 2016, 209, 105–112. [Google Scholar] [CrossRef]
- García-Almodóvar, R.C.; Gosalvez, B.; Aranda, M.A.; Burgos, L. Production of transgenic diploid Cucumis melo plants. Plant Cell Tissue Organ Cult. 2017, 130, 323–333. [Google Scholar] [CrossRef]
- Uranbey, S.; Sevimay, C.S.; Kaya, M.D.; İpek, A.; Sancak, C.; Başalma, D.; Er, C.; Özcan, S. Influence of different co-cultivation temperatures, periods and media on Agrobacterium tumefaciens-mediated gene transfer. Biol. Plant. 2005, 49, 53–57. [Google Scholar] [CrossRef]
- Zhang, Z.; Finer, J.J. Low Agrobacterium tumefaciens inoculum levels and a long co-culture period lead to reduced plant defense responses and increase transgenic shoot production of sunflower (Helianthus annuus L.). Vitr. Cell. Dev. Biol.-Plant 2016, 52, 354–366. [Google Scholar] [CrossRef] [PubMed]
- Ezura, H.; Yuhashi, K.-I.; Yasuta, T.; Minamisawa, K. Effect of ethylene on Agrobacterium tumefaciens-mediated gene transfer to melon. Plant Breed. 2000, 119, 75–79. [Google Scholar] [CrossRef]
- Kumar, V.; Parvatam, G.; Ravishankar, G.A. AgNO3—A potential regulator of ethylene activity and plant growth modulator. Electron. J. Biotechnol. 2009, 12, 8–9. [Google Scholar] [CrossRef]
- Ptak, A.; Tahchy, A.E.; Wyżgolik, G.; Henry, M.; Laurain-Mattar, D. Effects of ethylene on somatic embryogenesis and galanthamine content in Leucojum aestivum L. cultures. Plant Cell Tissue Organ Cult. 2010, 102, 61–67. [Google Scholar] [CrossRef]
- Sgamma, T.; Thomas, B.; Muleo, R. Ethylene inhibitor silver nitrate enhances regeneration and genetic transformation of Prunus avium (L.) cv Stella. Plant Cell Tissue Organ Cult. 2015, 120, 79–88. [Google Scholar] [CrossRef]
- Li, S.; Cong, Y.; Liu, Y.; Wang, T.; Shuai, Q.; Chen, N.; Gai, J.; Li, Y. Optimization of Agrobacterium-Mediated Transformation in Soybean. Front. Plant Sci. 2017, 8, 246. [Google Scholar] [CrossRef] [PubMed]
- Malambane, G.; Nonaka, S.; Shiba, H.; Ezura, H.; Tsujimoto, H.; Akashi, K. Comparative effects of ethylene inhibitors on Agrobacterium-mediated transformation of drought-tolerant wild watermelon. Biosci. Biotechnol. Biochem. 2018, 82, 433–441. [Google Scholar] [CrossRef] [PubMed]
- Colijn-Hooymans, C.M.; Hakkert, J.C.; Jansen, J.; Custers, J.B.M. Competence for regeneration of cucumber cotyledons is restricted to specific developmental stages. Plant Cell Tissue Organ Cult. 1994, 39, 211–217. [Google Scholar] [CrossRef]
- Sebastiani, M.S.; Ficcadenti, N. In vitro plant regeneration from cotyledonary explants of Cucumis melo L. var. cantalupensis and genetic stability evaluation using RAPD analysis. Plant Cell Tissue Organ Cult. 2016, 124, 69–79. [Google Scholar] [CrossRef]
- Wang, H.; La Russa, M.; Qi, L.S. CRISPR/Cas9 in Genome Editing and Beyond. Annu. Rev. Biochem. 2016, 85, 227–264. [Google Scholar] [CrossRef]
- Osakabe, Y.; Osakabe, K. Genome Editing with Engineered Nucleases in Plants. Plant Cell Physiol. 2015, 56, 389–400. [Google Scholar] [CrossRef]
- Jaganathan, D.; Ramasamy, K.; Sellamuthu, G.; Jayabalan, S.; Venkataraman, G. CRISPR for Crop Improvement: An Update Review. Front. Plant Sci. 2018, 9, 985. [Google Scholar] [CrossRef]
- Chen, H.; Neubauer, M.; Wang, J.P. Enhancing HR Frequency for Precise Genome Editing in Plants. Front. Plant Sci. 2022, 13, 883421. [Google Scholar] [CrossRef] [PubMed]
- Gaudelli, N.M.; Komor, A.C.; Rees, H.A.; Packer, M.S.; Badran, A.H.; Bryson, D.I.; Liu, D.R. Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature 2017, 551, 464–471. [Google Scholar] [CrossRef]
- Zong, Y.; Wang, Y.; Li, C.; Zhang, R.; Chen, K.; Ran, Y.; Qiu, J.-L.; Wang, D.; Gao, C. Precise base editing in rice, wheat and maize with a Cas9-cytidine deaminase fusion. Nat. Biotechnol. 2017, 35, 438–440. [Google Scholar] [CrossRef]
- Bastet, A.; Zafirov, D.; Giovinazzo, N.; Guyon-Debast, A.; Nogué, F.; Robaglia, C.; Gallois, J.-L. Mimicking natural polymorphism in eIF4E by CRISPR-Cas9 base editing is associated with resistance to potyviruses. Plant Biotechnol. J. 2019, 17, 1736–1750. [Google Scholar] [CrossRef]
- Hunziker, J.; Nishida, K.; Kondo, A.; Kishimoto, S.; Ariizumi, T.; Ezura, H. Multiple gene substitution by Target-AID base-editing technology in tomato. Sci. Rep. 2020, 10, 20471. [Google Scholar] [CrossRef]
- Shimatani, Z.; Kashojiya, S.; Takayama, M.; Terada, R.; Arazoe, T.; Ishii, H.; Teramura, H.; Yamamoto, T.; Komatsu, H.; Miura, K.; et al. Targeted base editing in rice and tomato using a CRISPR-Cas9 cytidine deaminase fusion. Nat. Biotechnol. 2017, 35, 441–443. [Google Scholar] [CrossRef] [PubMed]
- Nieto, C.; Morales, M.; Orjeda, G.; Clepet, C.; Monfort, A.; Sturbois, B.; Puigdomènech, P.; Pitrat, M.; Caboche, M.; Dogimont, C.; et al. An eIF4E allele confers resistance to an uncapped and non-polyadenylated RNA virus in melon. Plant J. 2006, 48, 452–462. [Google Scholar] [CrossRef] [PubMed]
- Nieto, C.; Piron, F.; Dalmais, M.; Marco, C.F.; Moriones, E.; Gómez-Guillamón, M.L.; Truniger, V.; Gómez, P.; Garcia-Mas, J.; Aranda, A.M.; et al. EcoTILLING for the identification of allelic variants of melon eIF4E, a factor that controls virus susceptibility. BMC Plant Biol. 2007, 7, 34. [Google Scholar] [CrossRef]
- Ala-Poikela, M.; Goytia, E.; Haikonen, T.; Rajamäki, M.-L.; Valkonen, J.P.T. Helper Component Proteinase of the Genus Potyvirus Is an Interaction Partner of Translation Initiation Factors eIF(iso)4E and eIF4E and Contains a 4E Binding Motif. J. Virol. 2011, 85, 6784–6794. [Google Scholar] [CrossRef]
- Rajagopalan, P.A.; Perl-Treves, R. Improved Cucumber Transformation by a Modified Explant Dissection and Selection Protocol. Hortscience 2005, 40, 431–435. [Google Scholar] [CrossRef]
- Bakshi, S.; Sadhukhan, A.; Mishra, S.; Sahoo, L. Improved Agrobacterium-mediated transformation of cowpea via sonication and vacuum infiltration. Plant Cell Rep. 2011, 30, 2281–2292. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Zhou, B.; Yang, Y.; Mei, J.; Zhao, X.; Guo, X.; Huang, X.; Tang, D.; Liu, X. Piercing and vacuum infiltration of the mature embryo: A simplified method for Agrobacterium-mediated transformation of indica rice. Plant Cell Rep. 2009, 28, 1065–1074. [Google Scholar] [CrossRef] [PubMed]
- De Oliveira, M.L.P.; Febres, V.J.; Costa, M.G.C.; Moore, G.A.; Otoni, W.C. High-efficiency Agrobacterium-mediated transformation of citrus via sonication and vacuum infiltration. Plant Cell Rep. 2009, 28, 387–395. [Google Scholar] [CrossRef]
- Hooghvorst, I.; López-Cristoffanini, C.; Nogués, S. Efficient knockout of phytoene desaturase gene using CRISPR/Cas9 in melon. Sci. Rep. 2019, 9, 17077. [Google Scholar] [CrossRef]
- Subramanyam, K.; Rajesh, M.; Jaganath, B.; Vasuki, A.; Theboral, J.; Elayaraja, D.; Karthik, S.; Manickavasagam, M.; Ganapathi, A. Assessment of Factors Influencing the Agrobacterium-mediated in planta Seed Transformation of Brinjal (Solanum melongena L.). Appl. Biochem. Biotechnol. 2013, 171, 450–468. [Google Scholar] [CrossRef]
- Abeles, F.B.; Morgan, P.W.; Saltveit, M.E., Jr. Ethylene in Plant Biology, 2nd ed.; Academic Press: Cambridge, MA, USA, 2012. [Google Scholar]
- Spanu, P.; Boller, T. Ethylene Biosynthesis in Tomato Plants Infected by Phytophthora infestans. J. Plant Physiol. 1989, 134, 533–537. [Google Scholar] [CrossRef]
- Bezirganoglu, I.; Hwang, S.; Shaw, J.; Fang, T. Efficient production of transgenic melon via Agrobacterium-mediated transformation. Genet. Mol. Res. 2014, 13, 3218–3227. [Google Scholar] [CrossRef] [PubMed]
- Akasaka-Kennedy, Y.; Tomita, K.-O.; Ezura, H. Efficient plant regeneration and Agrobacterium-mediated transformation via somatic embryogenesis in melon (Cucumis melo L.). Plant Sci. 2004, 166, 763–769. [Google Scholar] [CrossRef]
- McHughen, A.; Jordan, M.; Feist, G. A Preculture Period Prior to Agrobacterium Inoculation Increases Production of Transgenic Plants. J. Plant Physiol. 1989, 135, 245–248. [Google Scholar] [CrossRef]
- Gaba, V.; Zelcer, A.; Gal-On, A. Cucurbit biotechnology-the importance of virus resistance. Vitr. Cell. Dev. Biol.-Plant 2004, 40, 346–358. [Google Scholar] [CrossRef]
- Chee, P.P. Plant Regeneration from Cotyledons of Cucumis melo ‘Topmark’. Hortscience 1991, 26, 908–910. [Google Scholar] [CrossRef]
- Gray, D.; McColley, D.; Compton, M.E. High-frequency Somatic Embryogenesis from Quiescent Seed Cotyledons of Cucumis melo Cultivars. J. Am. Soc. Hortic. Sci. 1993, 118, 425–432. [Google Scholar] [CrossRef]
- Kathal, R.; Bhatnagar, S.; Bhojwani, S.S. Plant regeneration from the callus derived from root explants of Cucumis melo L. cv. Pusa sharbati. Plant Sci. 1994, 96, 137–142. [Google Scholar] [CrossRef]
- Curuk, S.; Elman, C.; Schlarman, E.; Sagee, O.; Shomer, I.; Cetiner, S.; Gray, D.J.; Gaba, V. A novel pathway for rapid shoot regeneration from the proximal zone of the inpocotyl of melon (Cucumis melo L.). Vitr. Cell. Dev. Biol.-Plant 2002, 38, 260–267. [Google Scholar] [CrossRef]
- Wu, H.-W.; Yu, T.-A.; Raja, J.A.J.; Wang, H.-C.; Yeh, S.-D. Generation of transgenic oriental melon resistant to Zucchini yellow mosaic virus by an improved cotyledon-cutting method. Plant Cell Rep. 2009, 28, 1053–1064. [Google Scholar] [CrossRef] [PubMed]
- Raji, M.R.; Lotfi, M.; Tohidfar, M.; Zahedi, B.; Carra, A.; Abbate, L.; Carimi, F. Somatic embryogenesis of muskmelon (Cucumis melo L.) and genetic stability assessment of regenerants using flow cytometry and ISSR markers. Protoplasma 2018, 255, 873–883. [Google Scholar] [CrossRef]
- Souza, F.V.D.; Garcia-Sogo, B.; Souza, A.d.S.; San-Juán, A.P.; Moreno, V. Morphogenetic response of cotyledon and leaf explants of melon (Cucumis melo L.) cv. Amarillo Oro. Braz. Arch. Biol. Technol. 2006, 49, 21–27. [Google Scholar] [CrossRef]
- Nugent, P.E.; Ray, D.T. Spontaneous Tetraploid Melons. Hortscience 1992, 27, 47–50. [Google Scholar] [CrossRef]
- Sikora, P.; Chawade, A.; Larsson, M.; Olsson, J.; Olsson, O. Mutagenesis as a Tool in Plant Genetics, Functional Genomics, and Breeding. Int. J. Plant Genom. 2011, 2011, 314829. [Google Scholar] [CrossRef] [PubMed]
- Duprat, A.; Caranta, C.; Revers, F.; Menand, B.; Browning, K.S.; Robaglia, C. The Arabidopsis eukaryotic initiation factor (iso)4E is dispensable for plant growth but required for susceptibility to potyviruses. Plant J. 2002, 32, 927–934. [Google Scholar] [CrossRef]
- Lellis, A.D.; Kasschau, K.D.; Whitham, A.S.; Carrington, J.C. Loss-of-Susceptibility Mutants of Arabidopsis thaliana Reveal an Essential Role for eIF(iso)4E during Potyvirus Infection. Curr. Biol. 2002, 12, 1046–1051. [Google Scholar] [CrossRef]
- Sato, M.; Nakahara, K.; Yoshii, M.; Ishikawa, M.; Uyeda, I. Selective involvement of members of the eukaryotic initiation factor 4E family in the infection of Arabidopsis thaliana by potyviruses. FEBS Lett. 2005, 579, 1167–1171. [Google Scholar] [CrossRef] [PubMed]
- Ruffel, S.; Gallois, J.-L.; Moury, B.; Robaglia, C.; Palloix, A.; Caranta, C. Simultaneous mutations in translation initiation factors eIF4E and eIF(iso)4E are required to prevent pepper veinal mottle virus infection of pepper. J. Gen. Virol. 2006, 87, 2089–2098. [Google Scholar] [CrossRef] [PubMed]
- Pavan, S.; Jacobsen, E.; Visser, R.G.F.; Bai, Y. Loss of susceptibility as a novel breeding strategy for durable and broad-spectrum resistance. Mol. Breed. 2010, 25, 1–12. [Google Scholar] [CrossRef]
- Hiei, Y.; Komari, T.; Kubo, T. Transformation of rice mediated by Agrobacterium tumefaciens. Plant Mol. Biol. 1997, 35, 205–218. [Google Scholar] [CrossRef]
- Hu, B.; Li, D.; Liu, X.; Qi, J.; Gao, D.; Zhao, S.; Huang, S.; Sun, J.; Yang, L. Engineering Non-transgenic Gynoecious Cucumber Using an Improved Transformation Protocol and Optimized CRISPR/Cas9 System. Mol. Plant 2017, 10, 1575–1578. [Google Scholar] [CrossRef]
- Gyves, E.M.; Enea, M.; Fabrizio, P. Inhibition of Agrobacterium tumefaciens growth by silver nitrate. Int. J. Plant Dev. Biol. 2010, 4, 64–67. [Google Scholar]
- Park, J.; Bae, S.; Kim, J.-S. Cas-Designer: A web-based tool for choice of CRISPR-Cas9 target sites. Bioinformatics 2015, 31, 4014–4016. [Google Scholar] [CrossRef] [PubMed]


| Cultivar | Explants | Regeneration (%) 1 | Escapes 2 | Rooted Lines | Diploid | T.E 3 |
|---|---|---|---|---|---|---|
| Charentais mono | 134 | 62 (46.3%) | 55 | 5 | 5 | 3.73 |
| Khatooni | 145 | 79 (54.5%) | 70 | 8 | 7 | 4.83 |
| Nb of Progenies | Transformed | Non-Transformed | Segregation Ratio | χ2 | p-Value |
|---|---|---|---|---|---|
| 235 | 182 | 53 | 3:1 | 0.75 | 0.39 |
| 186 | 147 | 39 | 3:1 | 1.61 | 0.20 |
| Primer Name | Prime Sequence 5′ to 3′ | Amplified Region |
|---|---|---|
| For3melon | GGACGGATTGGTTTTAGGGTTC | eIF4E target |
| eIF4EIIR | CTGTTCTCCGATCATAGCAAGC | eIF4E target |
| sgRNA-R | CTTTGTACAAAAAAGCAGGCG | sgRNA |
| eIF-Target-1-R | CTAGAGTCGAAGTAGTGATTGTCTCTACCGTCGAGGAGTTCGTTTTAGAGCTAGAAATAGCAAG | sgRNA |
| eIF-Target-2-R | CTAGAGTCGAAGTAGTGATTGATCCAAGCAAATTGGCGATGGTTTTAGAGCTAGAAATAGCAAG | sgRNA |
| AtU6_R | GCCATAGAAAAGTTGGGTG | AtU6 promoter |
| AtU6-F | CAATCACTACTTCGACTCTAG | AtU6 promoter |
| PacI-For-DsRed | AACAGGGTAATttaattaaAGAAGGTAATTATCCAAGATGTAGCATC | DsRed |
| BamHI-Rev-DsRed | TTGATCACTAGTggatccTTTATCCTAGTTTGCGCGCTATAT | DsRed |
| SgRNA Target | Melon Variety | Total 2 | ||
|---|---|---|---|---|
| Samsuri | Charentais Mono | Vedrantais | ||
| sgRNA1-272 | 7 (7%) | 5 (5%) | 3 (3%) | 15 (5%) |
| sgRNA2-320 | 8 (8%) | 5 (5%) | 5 (5%) | 18 (6%) |
| Total 1 | 15 (7.5%) | 10 (5%) | 8 (4%) | 33 (5.5%) |
| SgRNA Target | Melon Variety | Total 2 | ||
|---|---|---|---|---|
| Samsuri | Charentais Mono | Vedrantais | ||
| sgRNA1-272 | 2 (28.57%) | 1 (20%) | 1 (33.33%) | 4 (26.67%) |
| sgRNA2-320 | 2 (25%) | 2 (40%) | 1 (20%) | 5 (27.78%) |
| Total 1 | 4 (26.67%) | 3 (30%) | 2 (25%) | 9 (27.3%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shirazi Parsa, H.; Sabet, M.S.; Moieni, A.; Shojaeiyan, A.; Dogimont, C.; Boualem, A.; Bendahmane, A. CRISPR/Cas9-Mediated Cytosine Base Editing Using an Improved Transformation Procedure in Melon (Cucumis melo L.). Int. J. Mol. Sci. 2023, 24, 11189. https://doi.org/10.3390/ijms241311189
Shirazi Parsa H, Sabet MS, Moieni A, Shojaeiyan A, Dogimont C, Boualem A, Bendahmane A. CRISPR/Cas9-Mediated Cytosine Base Editing Using an Improved Transformation Procedure in Melon (Cucumis melo L.). International Journal of Molecular Sciences. 2023; 24(13):11189. https://doi.org/10.3390/ijms241311189
Chicago/Turabian StyleShirazi Parsa, Hadi, Mohammad Sadegh Sabet, Ahmad Moieni, Abdolali Shojaeiyan, Catherine Dogimont, Adnane Boualem, and Abdelhafid Bendahmane. 2023. "CRISPR/Cas9-Mediated Cytosine Base Editing Using an Improved Transformation Procedure in Melon (Cucumis melo L.)" International Journal of Molecular Sciences 24, no. 13: 11189. https://doi.org/10.3390/ijms241311189
APA StyleShirazi Parsa, H., Sabet, M. S., Moieni, A., Shojaeiyan, A., Dogimont, C., Boualem, A., & Bendahmane, A. (2023). CRISPR/Cas9-Mediated Cytosine Base Editing Using an Improved Transformation Procedure in Melon (Cucumis melo L.). International Journal of Molecular Sciences, 24(13), 11189. https://doi.org/10.3390/ijms241311189







