Genome-Wide Analysis of Exocyst Complex Subunit Exo70 Gene Family in Cucumber
Abstract
1. Introduction
2. Results
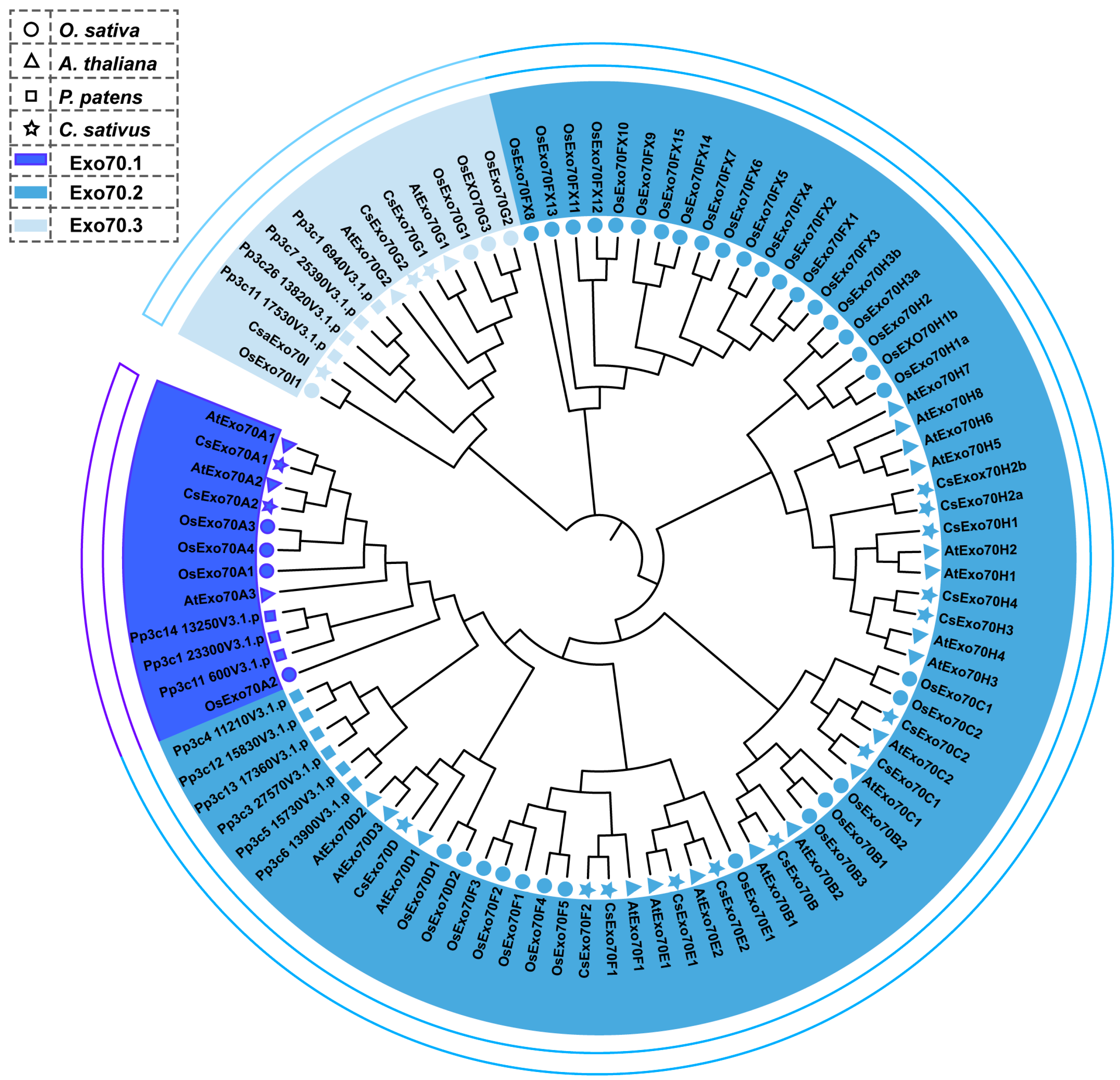
2.1. Identification and Characterization of CsExo70 Family in Cucumber
2.2. Chromosomal Localization and Collinearity Analysis of CsExo70 Genes in Cucumber
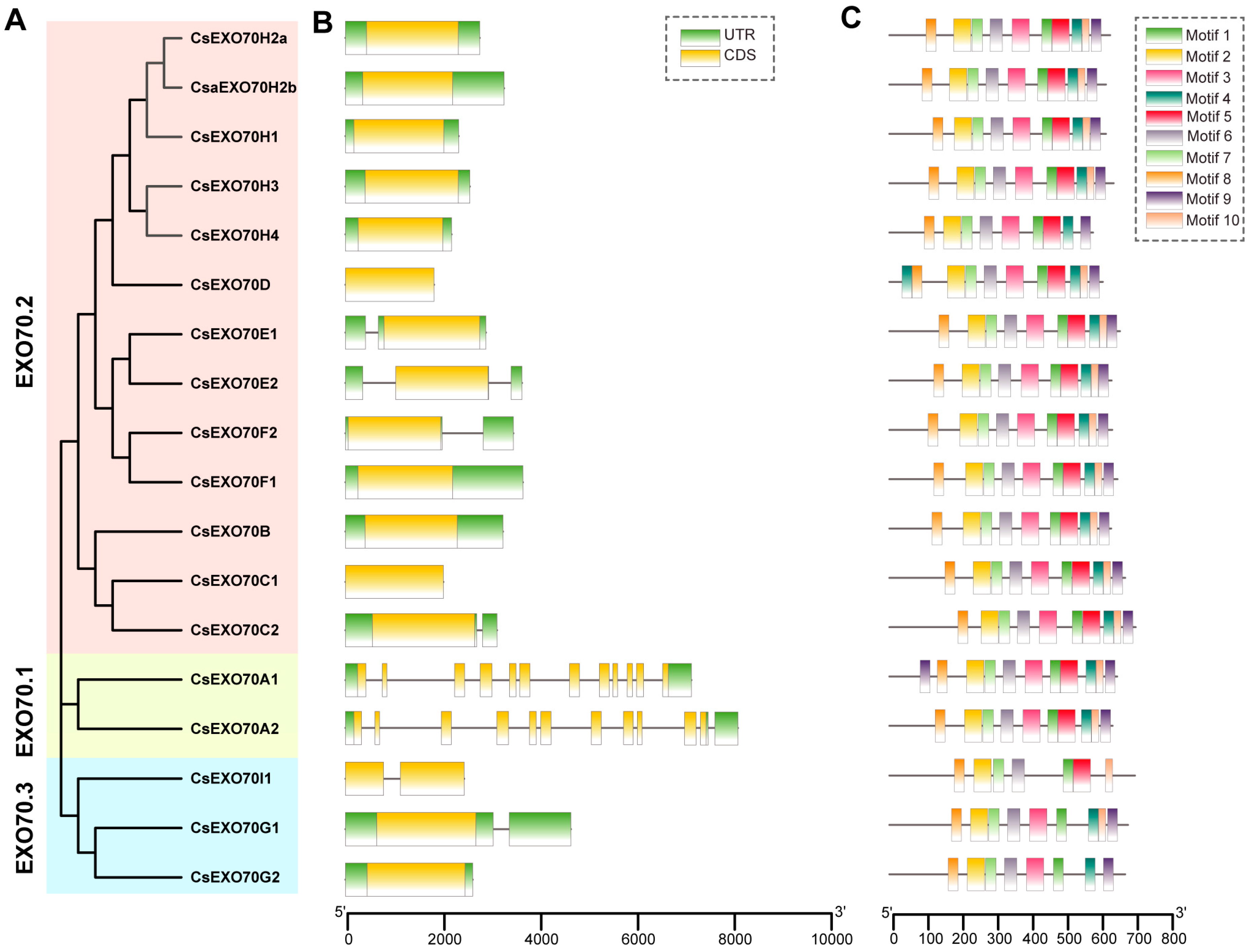
2.3. Gene Structure and Conserved Motif Analysis of CsExo70 Genes
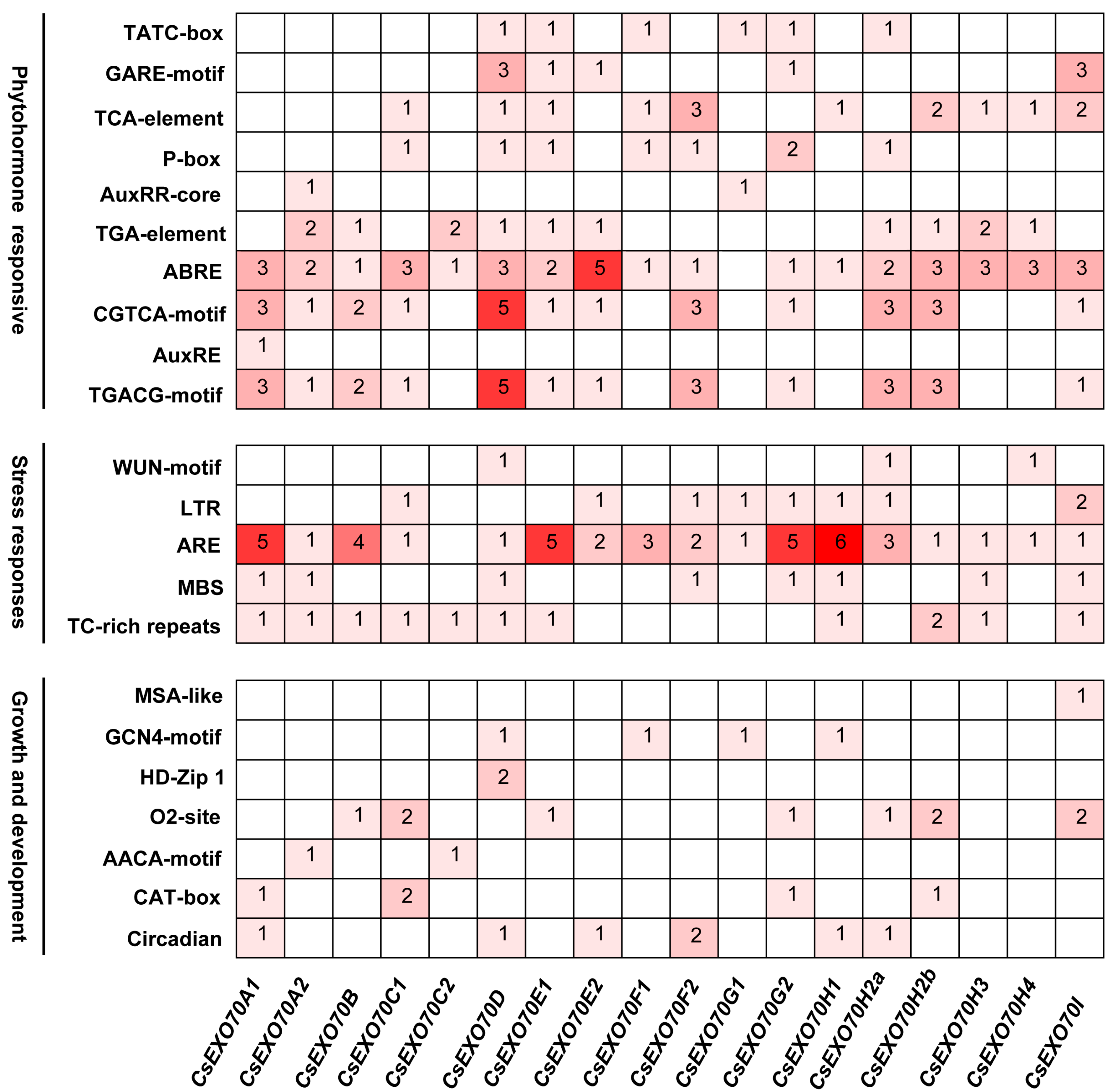
2.4. Analysis of Cis-Acting Elements on CsExo70 Promotors
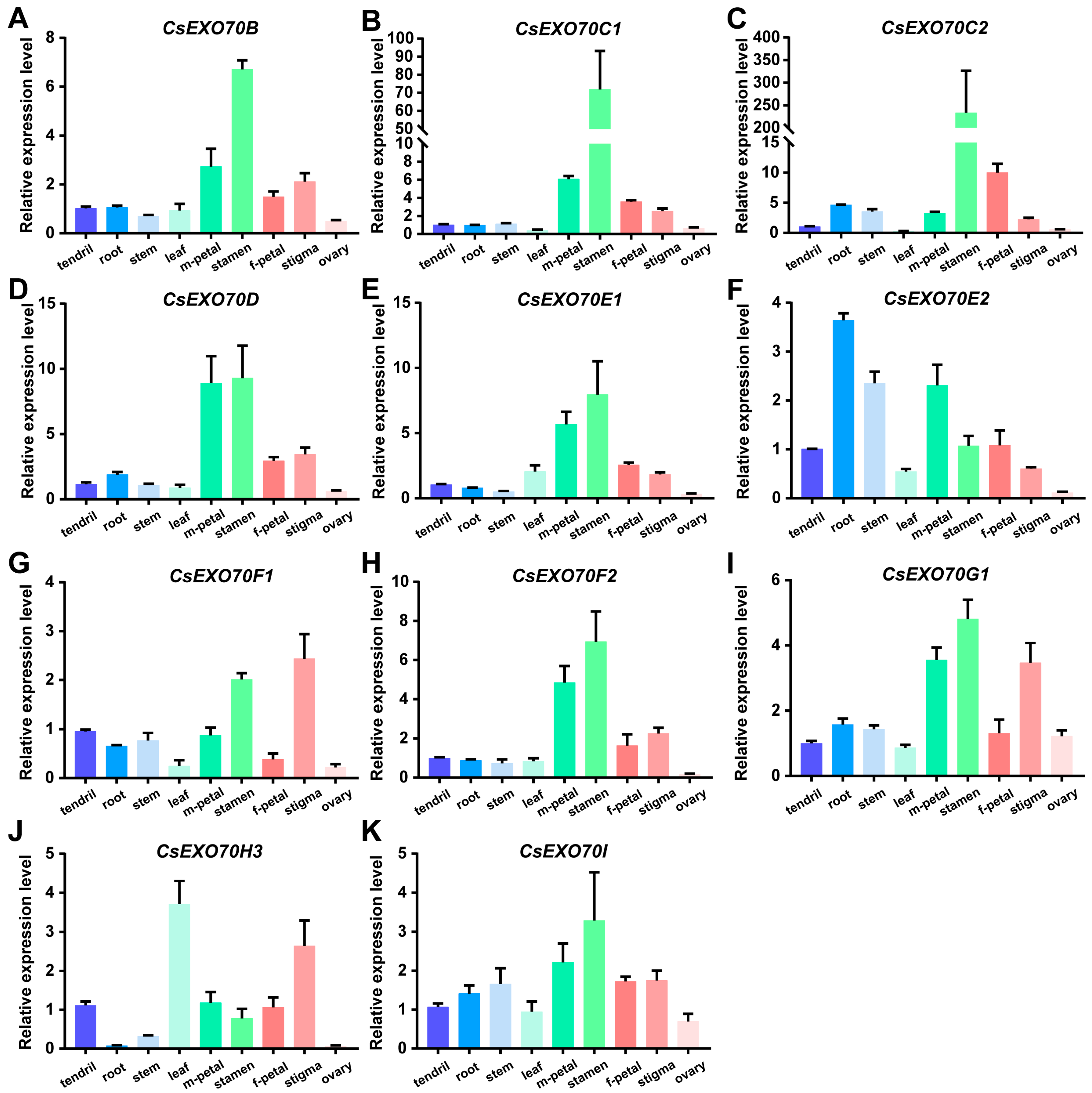
2.5. Expression Patterns of CsExo70 Genes in Cucumber
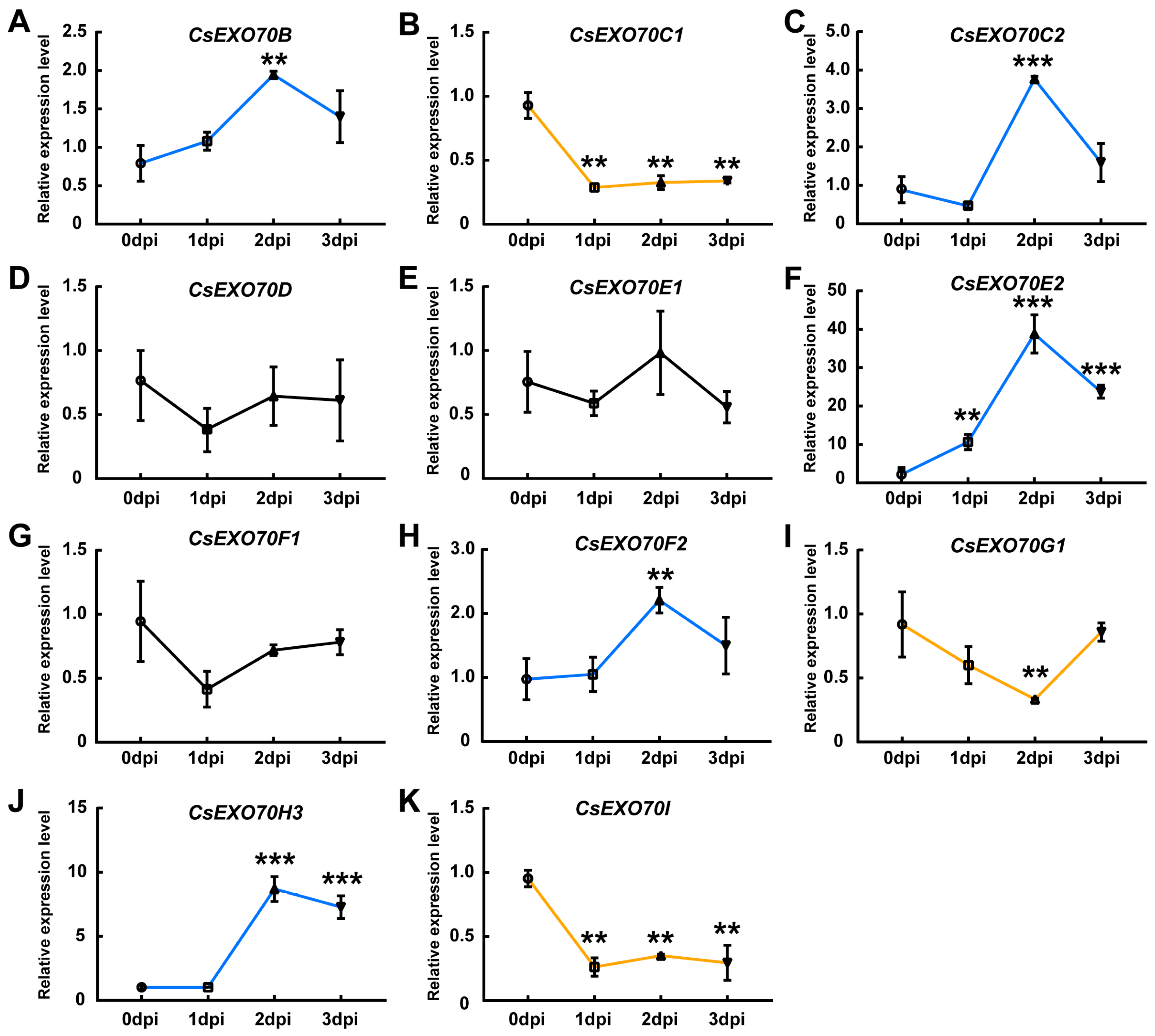
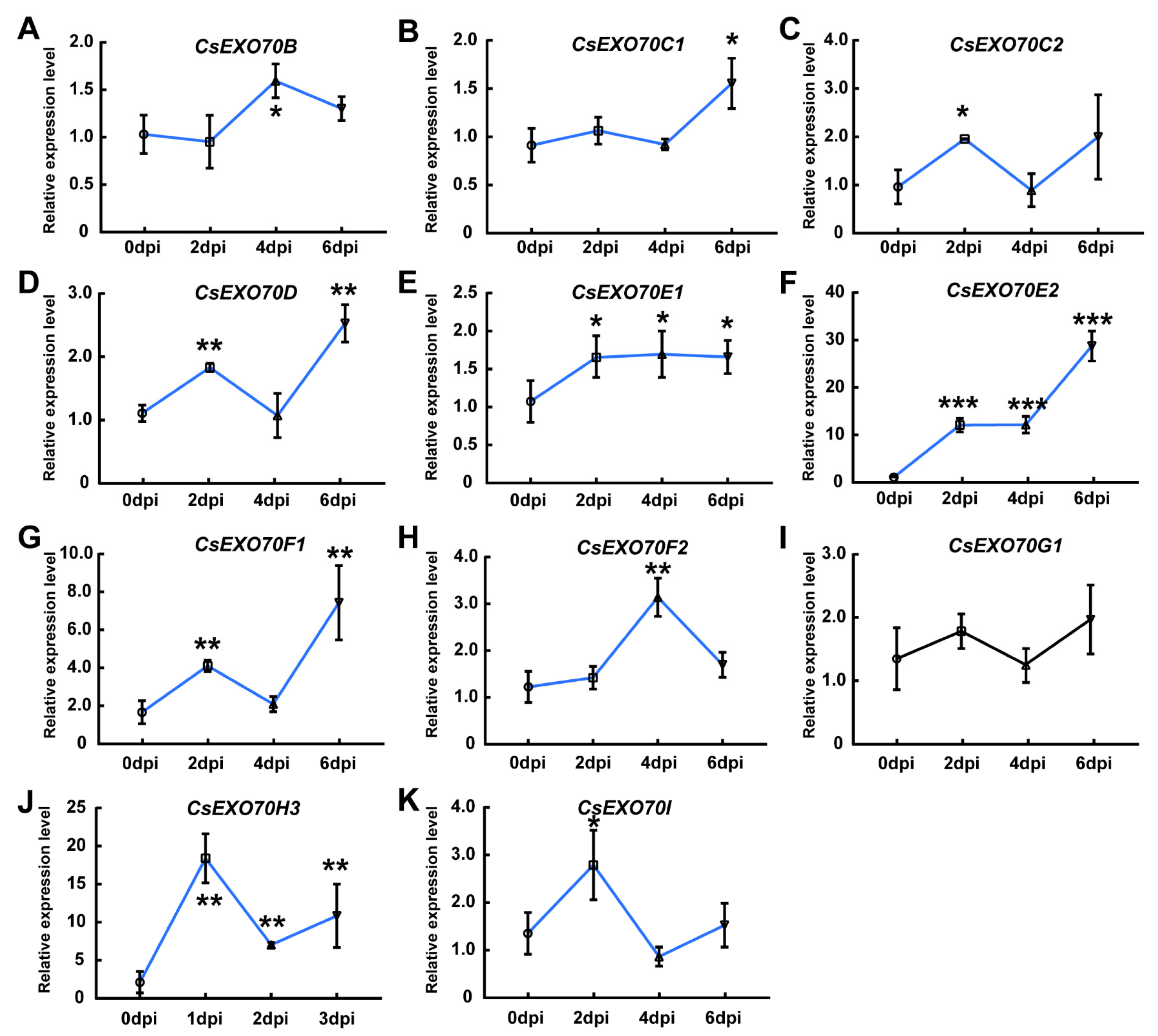
2.6. CsExo70 Gene Expression in Response to Different Pathogens
2.7. Subcellular Localization Analysis
3. Discussion
4. Material and Methods
4.1. Plant Materials
4.2. Identification and Phylogenetic Tree Construction of Exo70 Family
4.3. Chromosomal Location and Synteny Analysis
4.4. Gene Structures, Conserved Motifs, and Cis-Element Analysis
4.5. RNA Extraction and Gene Expression Analysis
4.6. Plant Pathogen Treatment
4.7. Subcellular Localization Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Saeed, B.; Brillada, C.; Trujillo, M. Dissecting the plant exocyst. Curr. Opin. Plant Biol. 2019, 52, 69–76. [Google Scholar] [CrossRef]
- Stegmann, M.; Monaghan, J.; Smakowska-Luzan, E.; Rovenich, H.; Lehner, A.; Holton, N.; Belkhadir, Y.; Zipfel, C. The receptor kinase FER is a RALF-regulated scaffold controlling plant immune signaling. Science 2017, 355, 287–289. [Google Scholar] [CrossRef] [PubMed]
- Cai, Q.; Qiao, L.; Wang, M.; He, B.; Lin, F.-M.; Palmquist, J.; Huang, S.-D.; Jin, H. Plants send small RNAs in extracellular vesicles to fungal pathogen to silence virulence genes. Science 2018, 360, 1126–1129. [Google Scholar] [CrossRef] [PubMed]
- Baldrich, P.; Rutter, B.D.; Karimi, H.Z.; Podicheti, R.; Meyers, B.C.; Innes, R.W. Plant extracellular vesicles contain diverse small RNA species and are enriched in 10- to 17-nucleotide “Tiny” RNAs. Plant Cell 2019, 31, 315–324. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.; Guo, W. The Exocyst at a Glance. J. Cell Sci. 2015, 128, 2957–2964. [Google Scholar] [CrossRef]
- TerBush, D.R.; Maurice, T.; Roth, D.; Novick, P. The Exocyst is a multiprotein complex required for exocytosis in Saccharomyces cerevisiae. EMBO J. 1996, 15, 6483–6494. [Google Scholar] [CrossRef]
- Pleskot, R.; Cwiklik, L.; Jungwirth, P.; Žárský, V.; Potocký, M. Membrane targeting of the yeast exocyst complex. Biochim. Biophys. Acta 2015, 1848, 1481–1489. [Google Scholar] [CrossRef]
- He, B.; Xi, F.; Zhang, X.; Zhang, J.; Guo, W. Exo70 interacts with phospholipids and mediates the targeting of the exocyst to the plasma membrane. EMBO J. 2007, 26, 4053–4065. [Google Scholar] [CrossRef]
- Boyd, C.; Hughes, T.; Pypaert, M.; Novick, P. Vesicles carry most exocyst subunits to exocytic sites marked by the remaining two subunits, Sec3p and Exo70p. J. Cell Biol. 2004, 167, 889–901. [Google Scholar] [CrossRef]
- Elias, M.; Drdova, E.; Ziak, D.; Bavlnka, B.; Hala, M.; Cvrckova, F.; Soukupova, H.; Zarsky, V. The exocyst complex in plants. Cell Biol. Int. 2003, 27, 199–201. [Google Scholar] [CrossRef]
- Žárský, V.; Sekereš, J.; Kubátová, Z.; Pečenková, T.; Cvrčková, F. Three subfamilies of exocyst EXO70 family subunits in land plants: Early divergence and ongoing functional specialization. J. Exp. Bot. 2020, 71, 49–62. [Google Scholar] [CrossRef]
- Cvrčková, F.; Grunt, M.; Bezvoda, R.; Hála, M.; Kulich, I.; Rawat, A.; Žárský, V. Evolution of the land plant exocyst complexes. Front. Plant Sci. 2012, 3, 159. [Google Scholar] [CrossRef]
- Chong, Y.T.; Gidda, S.K.; Sanford, C.; Parkinson, J.; Mullen, R.T.; Goring, D.R. Characterization of the Arabidopsis thaliana exocyst complex gene families by phylogenetic, expression profiling, and subcellular localization studies. New Phytol. 2010, 185, 401–419. [Google Scholar] [CrossRef]
- Synek, L.; Schlager, N.; Eliás, M.; Quentin, M.; Hauser, M.-T.; Žárský, V. AtEXO70A1, a member of a family of putative exocyst subunits specifically expanded in land plants, is important for polar growth and plant development. Plant J. 2006, 48, 54–72. [Google Scholar] [CrossRef] [PubMed]
- Drdová, E.J.; Synek, L.; Pečenková, T.; Hála, M.; Kulich, I.; Fowler, J.E.; Murphy, A.S.; Žárský, V. The exocyst complex contributes to PIN auxin efflux carrier recycling and polar auxin transport in Arabidopsis. Plant J. 2013, 73, 709–719. [Google Scholar] [CrossRef] [PubMed]
- Marković, V.; Cvrčková, F.; Potocký, M.; Kulich, I.; Pejchar, P.; Kollárová, E.; Synek, L.; Žárský, V. EXO70A2 is critical for exocyst complex function in pollen development. Plant Physiol. 2020, 184, 1823–1839. [Google Scholar] [CrossRef] [PubMed]
- Synek, L.; Vukašinović, N.; Kulich, I.; Hála, M.; Aldorfová, K.; Fendrych, M.; Žárský, V. EXO70C2 is a key regulatory factor for optimal tip growth of pollen. Plant Physiol. 2017, 174, 223–240. [Google Scholar] [CrossRef]
- Kulich, I.; Vojtíková, Z.; Glanc, M.; Ortmannová, J.; Rasmann, S.; Žárský, V. Cell Wall Maturation of Arabidopsis trichomes is dependent on exocyst subunit EXO70H4 and involves callose deposition. Plant Physiol. 2015, 168, 120–131. [Google Scholar] [CrossRef]
- Chen, C.; Liu, M.; Jiang, L.; Liu, X.; Zhao, J.; Yan, S.; Yang, S.; Ren, H.; Liu, R.; Zhang, X. Transcriptome profiling reveals roles of meristem regulators and polarity genes during fruit trichome development in cucumber (Cucumis sativus L.). J. Exp. Bot. 2014, 65, 4943–4958. [Google Scholar] [CrossRef]
- Pecenkova, T.; Hala, M.; Kulich, I.; Kocourkova, D.; Drdova, E.; Fendrych, M.; Toupalova, H.; Zarsky, V. The role for the exocyst complex subunits Exo70B2 and Exo70H1 in the plant-pathogen interaction. J. Exp. Bot. 2011, 62, 2107–2116. [Google Scholar] [CrossRef]
- Wang, W.; Liu, N.; Gao, C.; Rui, L.; Tang, D. The Pseudomonas syringae Effector AvrPtoB associates with and ubiquitinates Arabidopsis exocyst subunit EXO70B1. Front. Plant Sci. 2019, 10, 1027. [Google Scholar] [CrossRef]
- Stegmann, M.; Anderson, R.G.; Westphal, L.; Rosahl, S.; McDowell, J.M.; Trujillo, M. The exocyst subunit Exo70B1 is involved in the immune response of Arabidopsis thaliana to different pathogens and cell death. Plant Signal. Behav. 2013, 8, e27421. [Google Scholar] [CrossRef] [PubMed]
- Stegmann, M.; Anderson, R.G.; Ichimura, K.; Pecenkova, T.; Reuter, P.; Žársky, V.; McDowell, J.M.; Shirasu, K.; Trujillo, M. The ubiquitin ligase PUB22 targets a subunit of the exocyst complex required for PAMP-triggered responses in Arabidopsis. Plant Cell 2012, 24, 4703–4716. [Google Scholar] [CrossRef] [PubMed]
- Zhao, T.; Rui, L.; Li, J.; Nishimura, M.T.; Vogel, J.P.; Liu, N.; Liu, S.; Zhao, Y.; Dangl, J.L.; Tang, D. A truncated NLR protein, TIR-NBS2, is required for activated defense responses in the exo70B1 Mutant. PLoS Genet. 2015, 11, e1004945. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Liu, N.; Gao, C.; Cai, H.; Romeis, T.; Tang, D. The Arabidopsis exocyst subunits EXO70B1 and EXO70B2 regulate FLS2 homeostasis at the plasma membrane. New Phytol. 2020, 227, 529–544. [Google Scholar] [CrossRef]
- Hou, H.; Fang, J.; Liang, J.; Diao, Z.; Wang, W.; Yang, D.; Li, S.; Tang, D. OsExo70B1 positively regulates disease resistance to Magnaporthe oryzae in rice. Int. J. Mol. Sci. 2020, 21, 7049. [Google Scholar] [CrossRef]
- Sharif, R.; Su, L.; Chen, X.; Qi, X. Involvement of auxin in growth and stress response of cucumber. Veg. Res. 2022, 2, 13. [Google Scholar] [CrossRef]
- Robinson, R.W.; Decker-Walters, D.S. Cucurbits; CAB International: Oxford, UK; New York, NY, USA, 1997. [Google Scholar]
- Rodriguez-Granados, N.Y.; Lemhemdi, A.; Choucha, F.A.; Latrasse, D.; Benhamed, M.; Boualem, A.; Bendahmane, A. Sex determination in cucumis. In Genetics and Genomics of Cucurbitaceae; Springer International Publishing: Cham, Switzerland, 2017; pp. 307–319. [Google Scholar]
- Foucart, C.; Boualem, A.; Lasseur, B.; Eleblu, J.; Fahraj, I.; Bendahmane, A. Sex determination in cucurbits. Biol. Aujourd'hui 2012, 206, 57–62. [Google Scholar] [CrossRef]
- Yang, L.; Koo, D.-H.; Li, Y.; Zhang, X.; Luan, F.; Havey, M.J.; Jiang, J.; Weng, Y. Chromosome rearrangements during domestication of cucumber as revealed by high-density genetic mapping and draft genome assembly. Plant J. 2012, 71, 895–906. [Google Scholar] [CrossRef]
- Sebastian, P.; Schaefer, H.; Telford, I.R.H.; Renner, S.S. Cucumber (Cucumis sativus) and melon (C. melo) have numerous wild relatives in Asia and Australia, and the sister species of melon is from Australia. Proc. Natl. Acad. Sci. USA 2010, 107, 14269–14273. [Google Scholar] [CrossRef]
- Naegele, R.P.; Wehner, T.C. Genetic resources of cucumber. In Genetics and Genomics of Cucurbitaceae; Springer International Publishing: Cham, Switzerland, 2017; pp. 61–86. [Google Scholar]
- Cui, Y.; Li, S.; Dong, Y.; Wu, H.; Gao, Y.; Feng, Z.; Zhao, X.; Shan, L.; Zhang, Z.; Liu, Z.; et al. Genetic regulation and molecular mechanism of immature cucumber peel color: A review. Veg. Res. 2023, 3, 9. [Google Scholar] [CrossRef]
- Miao, H.; Zhang, S.; Wang, X.; Zhang, Z.; Li, M.; Mu, S.; Cheng, Z.; Zhang, R.; Huang, S.; Xie, B.; et al. A linkage map of cultivated cucumber (Cucumis sativus L.) with 248 microsatellite marker loci and seven genes for horticulturally important traits. Euphytica 2011, 182, 167–176. [Google Scholar] [CrossRef]
- Grumet, R.; Lin, Y.C.; Rett-Cadman, S.; Malik, A. Morphological and genetic diversity of Cucumber (Cucumis sativus L.) fruit development. Plants 2022, 12, 23. [Google Scholar] [CrossRef] [PubMed]
- Slomnicka, R.; Olczak-Woltman, H.; Sobczak, M.; Bartoszewski, G. Transcriptome profiling of Cucumber (Cucumis sativus L.) early response to Pseudomonas syringae pv. lachrymans. Int. J. Mol. Sci. 2021, 22, 4192. [Google Scholar] [CrossRef]
- Bhat, N.A.; Bhat, K.A.; Zargar, M.Y.; Teli, M.A.; Nazir, M.; Zargar, S.M. Current status of angular leaf spot (Pseudomonas syringae pv. lachrymans) of cucumber: A review. Int. J. Curr. Res. 2010, 8, 1–11. [Google Scholar]
- Raza, W.; Ling, N.; Zhang, R.; Huang, Q.; Xu, Y.; Shen, Q. Success evaluation of the biological control of Fusarium wilts of cucumber, banana, and tomato since 2000 and future research strategies. Crit. Rev. Biotechnol. 2017, 37, 202–212. [Google Scholar] [CrossRef] [PubMed]
- Ahammed, G.J.; Mao, Q.; Yan, Y.; Wu, M.; Wang, Y.; Ren, J.; Guo, P.; Liu, A.; Chen, S. Role of melatonin in arbuscular mycorrhizal fungi-induced resistance to Fusarium wilt in cucumber. Phytopathology 2020, 110, 999–1009. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.-Q.; Qiu, L.; Liu, L.-L.; Luo, L.; Han, X.-P.; Zhai, Y.-H.; Wang, W.-J.; Ren, M.-Z.; Xing, Y.-D. Identification and comprehensive structural and functional analyses of the EXO70 gene family in cotton. Genes 2021, 12, 1594. [Google Scholar] [CrossRef]
- Zhao, J.; Zhang, X.; Wan, W.; Zhang, H.; Liu, J.; Li, M.; Wang, H.; Xiao, J.; Wang, X. Identification and characterization of the EXO70 gene family in polyploid wheat and related species. Int. J. Mol. Sci. 2018, 20, 60. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Ma, Z.H.; Mao, J.; Chen, B.H. Genome-wide identification and expression analysis of the EXO70 gene family in grape (Vitis vinifera L). PeerJ 2021, 9, e11176. [Google Scholar] [CrossRef]
- Marković, V.; Kulich, I.; Žárský, V. Functional specialization within the EXO70 gene family in Arabidopsis. Int. J. Mol. Sci. 2021, 22, 7595. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Guo, J.; Zhang, Q.; Shi, S.; Guan, W.; Zhou, C.; Chen, R.; Du, B.; Zhu, L.; He, G. Necessity of rice resistance to planthoppers for OsEXO70H3 regulating SAMSL excretion and lignin deposition in cell walls. New Phytol. 2022, 234, 1031–1046. [Google Scholar] [CrossRef] [PubMed]
- Seo, D.H.; Ahn, M.Y.; Park, K.Y.; Kim, E.Y.; Kim, W.T. The N-Terminal UND motif of the Arabidopsis U-Box E3 Ligase PUB18 is critical for the negative regulation of ABA-mediated stomatal movement and determines its ubiquitination specificity for exocyst subunit Exo70B1. Plant Cell 2016, 28, 2952–2973. [Google Scholar] [CrossRef] [PubMed]
- Kulich, I.; Pecenkova, T.; Sekeres, J.; Smetana, O.; Fendrych, M.; Foissner, I.; Hoeftberger, M.; Zarsky, V. Arabidopsis exocyst subcomplex containing subunit EXO70B1 is involved in autophagy-related transport to the vacuole. Traffic 2013, 14, 1155–1165. [Google Scholar] [CrossRef]
- Cannon, S.B.; Mitra, A.; Baumgarten, A.; Young, N.D.; May, G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004, 4, 10. [Google Scholar] [CrossRef]
- Teixeira, F.K.; Menezes-Benavente, L.; Margis, R.; Margis-Pinheiro, M. Analysis of the molecular evolutionary history of the ascorbate peroxidase gene family: Inferences from the rice genome. J. Mol. Evol. 2004, 59, 761–770. [Google Scholar] [CrossRef]
- Shinozaki, K.; Yamaguchi-Shinozaki, K. Gene expression and signal transduction in water-stress response. Plant Physiol. 1997, 115, 327–334. [Google Scholar] [CrossRef]
- Hadiarto, T.; Tran, L.S. Progress studies of drought-responsive genes in rice. Plant Cell Rep. 2011, 30, 297–310. [Google Scholar] [CrossRef]
- Pan, W.; Zheng, P.; Zhang, C.; Wang, W.; Li, Y.; Fan, T.; Liu, Y.; Cao, S. The effect of ABRE BINDING FACTOR 4-mediated FYVE1 on salt stress tolerance in Arabidopsis. Plant Sci. 2020, 296, 110489. [Google Scholar] [CrossRef]
- Nakashima, K.; Yamaguchi-Shinozaki, K. ABA signaling in stress-response and seed development. Plant Cell Rep. 2013, 32, 959–970. [Google Scholar] [CrossRef]
- Grobei, M.A.; Qeli, E.; Brunner, E.; Rehrauer, H.; Zhang, R.; Roschitzki, B.; Basler, K.; Ahrens, C.H.; Grossniklaus, U. Deterministic protein inference for shotgun proteomics data provides new insights into Arabidopsis pollen development and function. Genome Res. 2009, 19, 1786–1800. [Google Scholar] [CrossRef] [PubMed]
- Weng, Y.; Wehner, T.C. Cucumber Gene Catalog 2017. In Cucurbit Genetic Cooperative; USDA, U.S. Vegetable Laboratory: Charleston, SC, USA, 2017; pp. 17–29. [Google Scholar]
- Bartholomew, E.S.; Xu, S.; Zhang, Y.; Yin, S.; Feng, Z.; Chen, S.; Sun, L.; Yang, S.; Wang, Y.; Liu, P.; et al. A chitinase CsChi23 promoter polymorphism underlies cucumber resistance against Fusarium oxysporum f. sp. cucumerinum. New Phytol. 2022, 236, 1471–1486. [Google Scholar] [CrossRef] [PubMed]
- Sperschneider, J.; Catanzariti, A.M.; DeBoer, K.; Petre, B.; Gardiner, D.M.; Singh, K.B.; Dodds, P.N.; Taylor, J.M. LOCALIZER: Subcellular localization prediction of both plant and effector proteins in the plant cell. Sci. Rep. 2017, 7, 44598. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-Time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Zhang, J.; Li, W.; Xiang, T.; Liu, Z.; Laluk, K.; Ding, X.; Zou, Y.; Gao, M.; Zhang, X.; Chen, S.; et al. Receptor-like cytoplasmic kinases integrate signaling from multiple plant immune receptors and are targeted by a Pseudomonas syringae effector. Cell Host Microbe 2010, 7, 290–301. [Google Scholar] [CrossRef]
- Kokkirala, V.R.; Yonggang, P.; Abbagani, S.; Zhu, Z.; Umate, P. Subcellular localization of proteins of Oryza sativa L. in the model tobacco and tomato plants. Plant Signal. Behav. 2010, 5, 1336–1341. [Google Scholar] [CrossRef]


| Gene Name | Gene ID | CDS 1 | AA 2 | MW (Da) 3 | PI 4 | Group |
|---|---|---|---|---|---|---|
| CsExo70A1 | CsaV3_5G035240.1 | 1956 | 651 | 73573.72 | 8.37 | Exo70.3 |
| CsExo70A2 | CsaV3_6G041070.1 | 1917 | 638 | 72857.09 | 6.95 | Exo70.3 |
| CsExo70B | CsaV3_1G036490.1 | 1905 | 634 | 72173.20 | 5.13 | Exo70.2 |
| CsExo70C1 | CsaV3_4G002930.1 | 2025 | 674 | 76816.14 | 5.97 | Exo70.2 |
| CsExo70C2 | CsaV3_5G024390.1 | 2115 | 704 | 81346.62 | 4.95 | Exo70.2 |
| CsExo70D | CsaV3_3G030800.1 | 1833 | 610 | 69471.55 | 5.38 | Exo70.2 |
| CsExo70E1 | CsaV3_4G009180.1 | 1980 | 659 | 75539.35 | 4.94 | Exo70.2 |
| CsExo70E2 | CsaV3_1G005990.1 | 1908 | 635 | 72952.51 | 6.17 | Exo70.2 |
| CsExo70F1 | CsaV3_1G045240.1 | 1959 | 652 | 74088.89 | 5.00 | Exo70.2 |
| CsExo70F2 | CsaV3_1G009630.1 | 1914 | 637 | 72342.77 | 5.16 | Exo70.2 |
| CsExo70G1 | CsaV3_2G031330.1 | 2049 | 682 | 77109.98 | 8.87 | Exo70.1 |
| CsExo70G2 | CsaV3_7G033810.1 | 2025 | 674 | 76901.53 | 6.67 | Exo70.1 |
| CsExo70H1 | CsaV3_5G006610.1 | 1860 | 619 | 69563.75 | 5.75 | Exo70.2 |
| CsExo70H2a | CsaV3_6G022100.1 | 1896 | 631 | 71359.32 | 6.71 | Exo70.2 |
| CsExo70H2b | CsaV3_1G033630.1 | 1860 | 619 | 70481.59 | 5.83 | Exo70.2 |
| CsExo70H3 | CsaV3_3G035190.1 | 1926 | 641 | 72726.64 | 5.95 | Exo70.2 |
| CsExo70H4 | CsaV3_4G024610.1 | 1749 | 582 | 66156.59 | 6.14 | Exo70.2 |
| CsExo70I1 | CsaV3_2G026060.1 | 2109 | 702 | 80192.32 | 5.69 | Exo70.1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, L.; Gu, C.; Zhang, J.; Guo, J.; Zhang, X.; Zhou, Z. Genome-Wide Analysis of Exocyst Complex Subunit Exo70 Gene Family in Cucumber. Int. J. Mol. Sci. 2023, 24, 10929. https://doi.org/10.3390/ijms241310929
Liu L, Gu C, Zhang J, Guo J, Zhang X, Zhou Z. Genome-Wide Analysis of Exocyst Complex Subunit Exo70 Gene Family in Cucumber. International Journal of Molecular Sciences. 2023; 24(13):10929. https://doi.org/10.3390/ijms241310929
Chicago/Turabian StyleLiu, Liu, Chaoheng Gu, Jiahao Zhang, Jingyu Guo, Xiaolan Zhang, and Zhaoyang Zhou. 2023. "Genome-Wide Analysis of Exocyst Complex Subunit Exo70 Gene Family in Cucumber" International Journal of Molecular Sciences 24, no. 13: 10929. https://doi.org/10.3390/ijms241310929
APA StyleLiu, L., Gu, C., Zhang, J., Guo, J., Zhang, X., & Zhou, Z. (2023). Genome-Wide Analysis of Exocyst Complex Subunit Exo70 Gene Family in Cucumber. International Journal of Molecular Sciences, 24(13), 10929. https://doi.org/10.3390/ijms241310929





