Resveratrol Protects Rat Ovarian Luteinized Granulosa Cells from H2O2-Induced Dysfunction by Activating Autophagy
Abstract
1. Introduction
2. Results
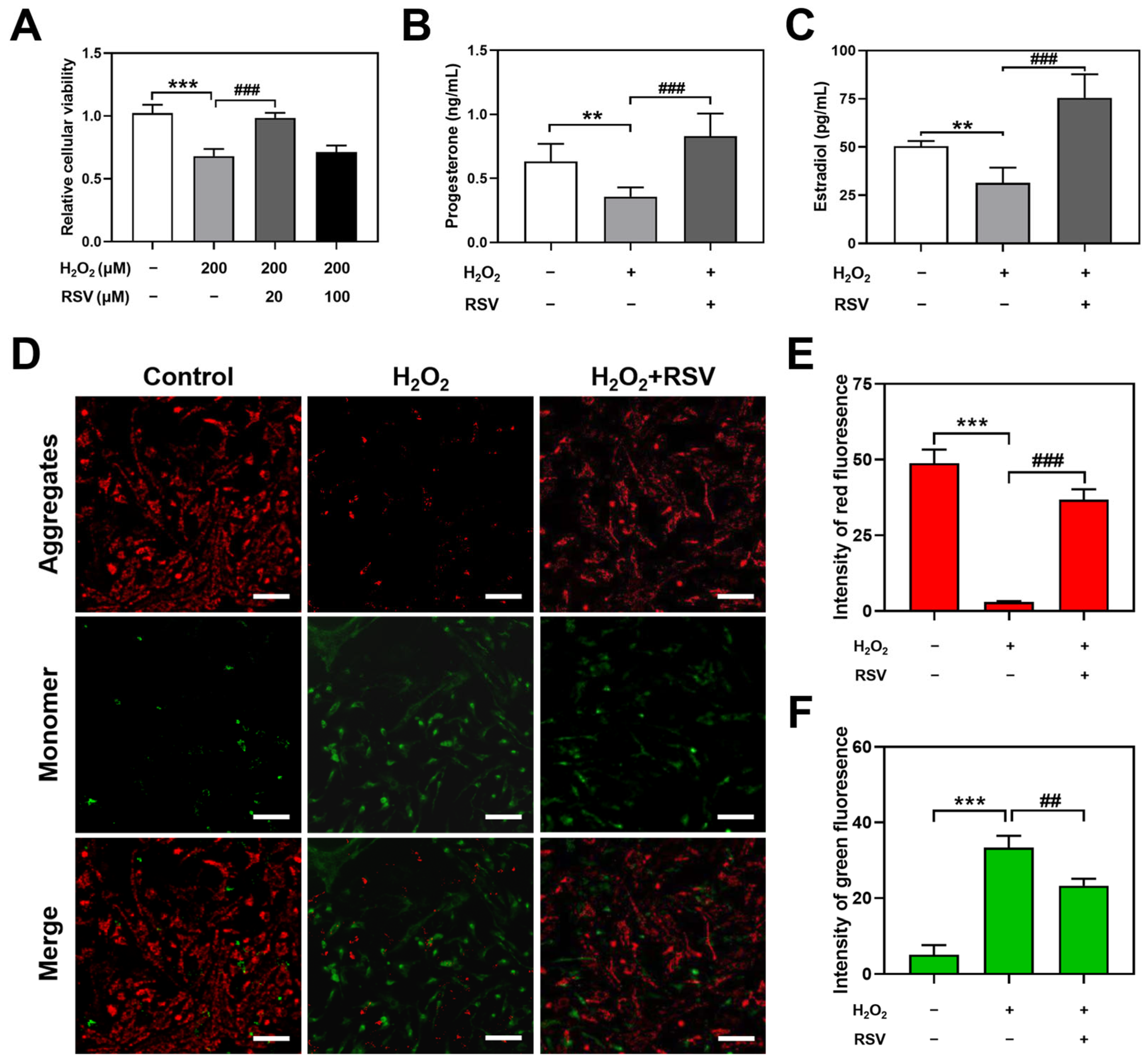
2.1. Resveratrol Protected H2O2-Induced Luteinized Granulosa Cell Dysfunction
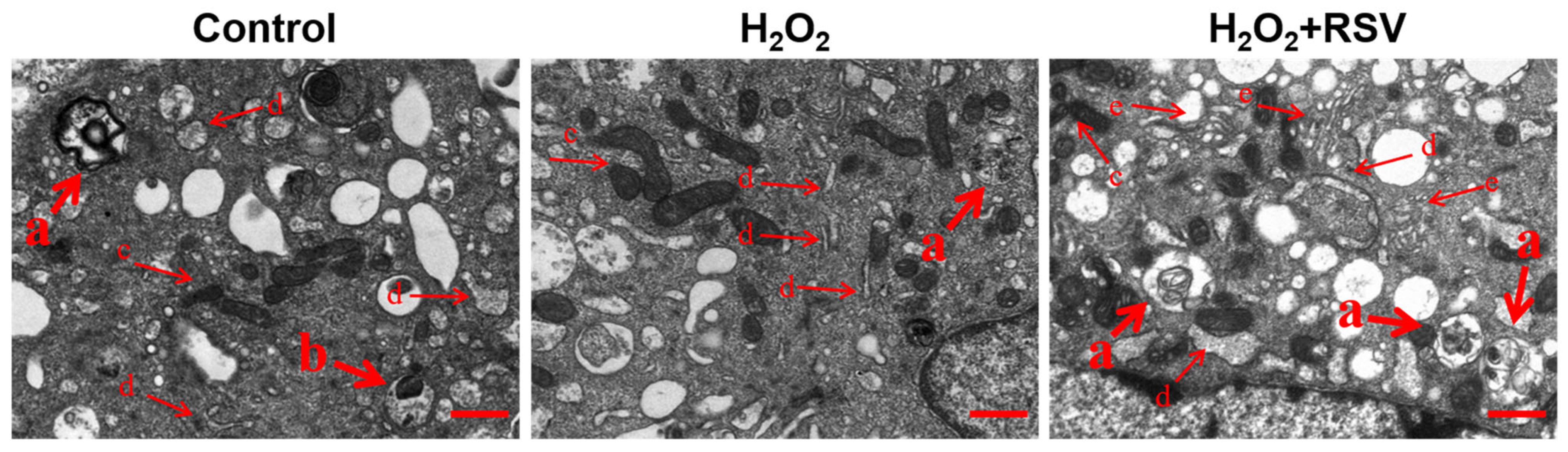
2.2. Resveratrol Enhanced Autophagy in H2O2-Induced Luteinized Granulosa Cells
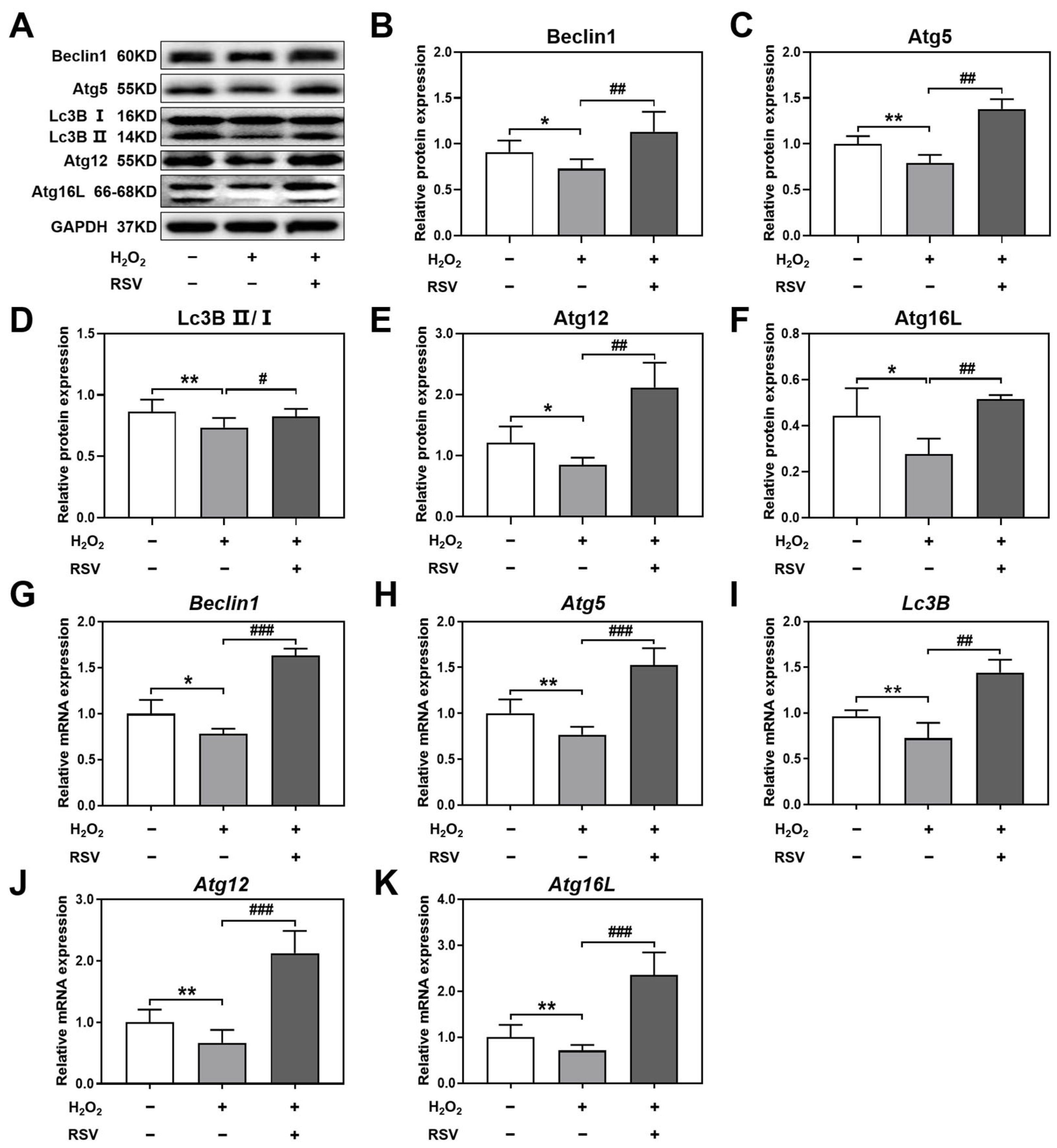
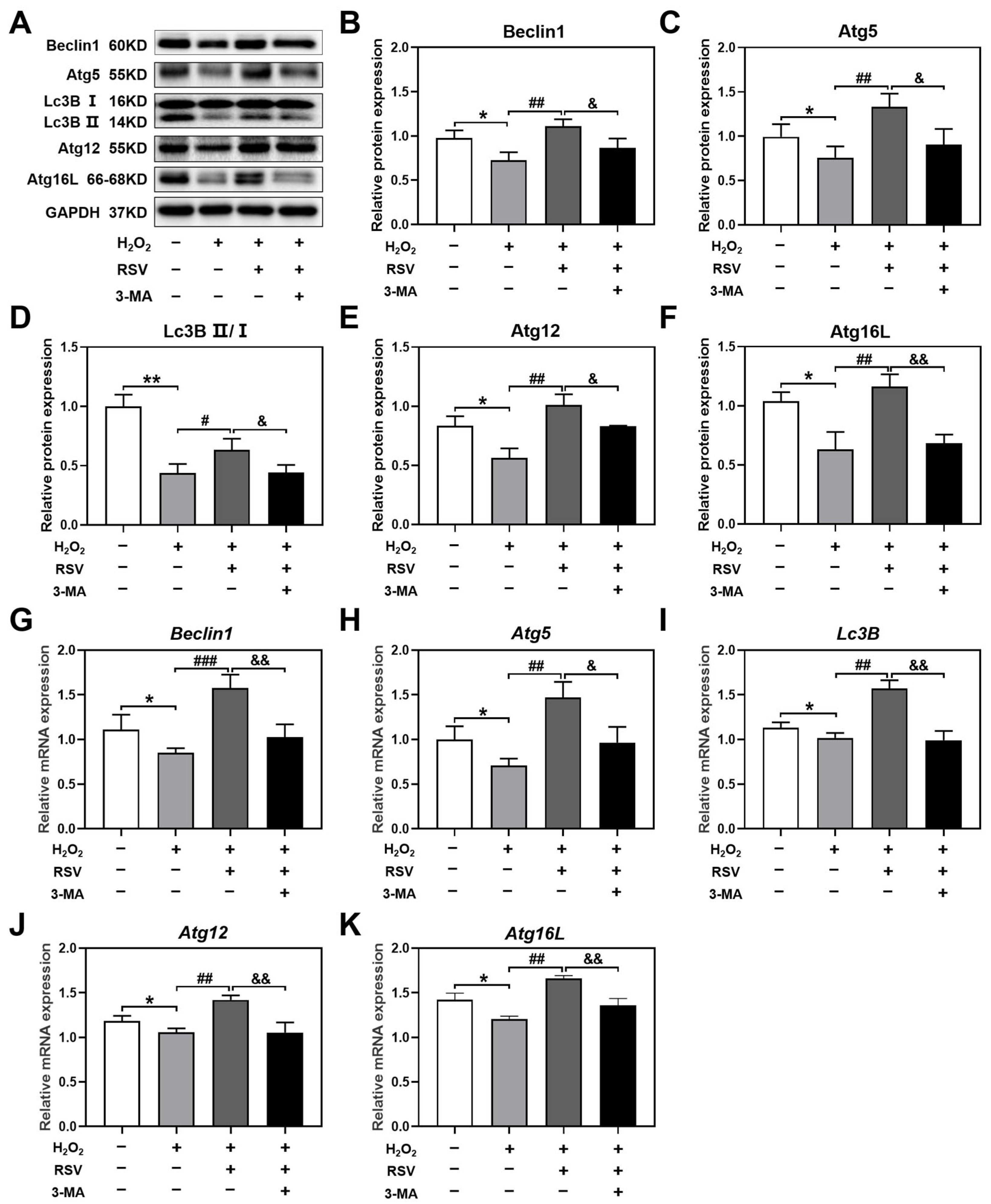
2.3. Resveratrol Activated Autophagy by Upregulating the Levels of Autophagy-Related Genes
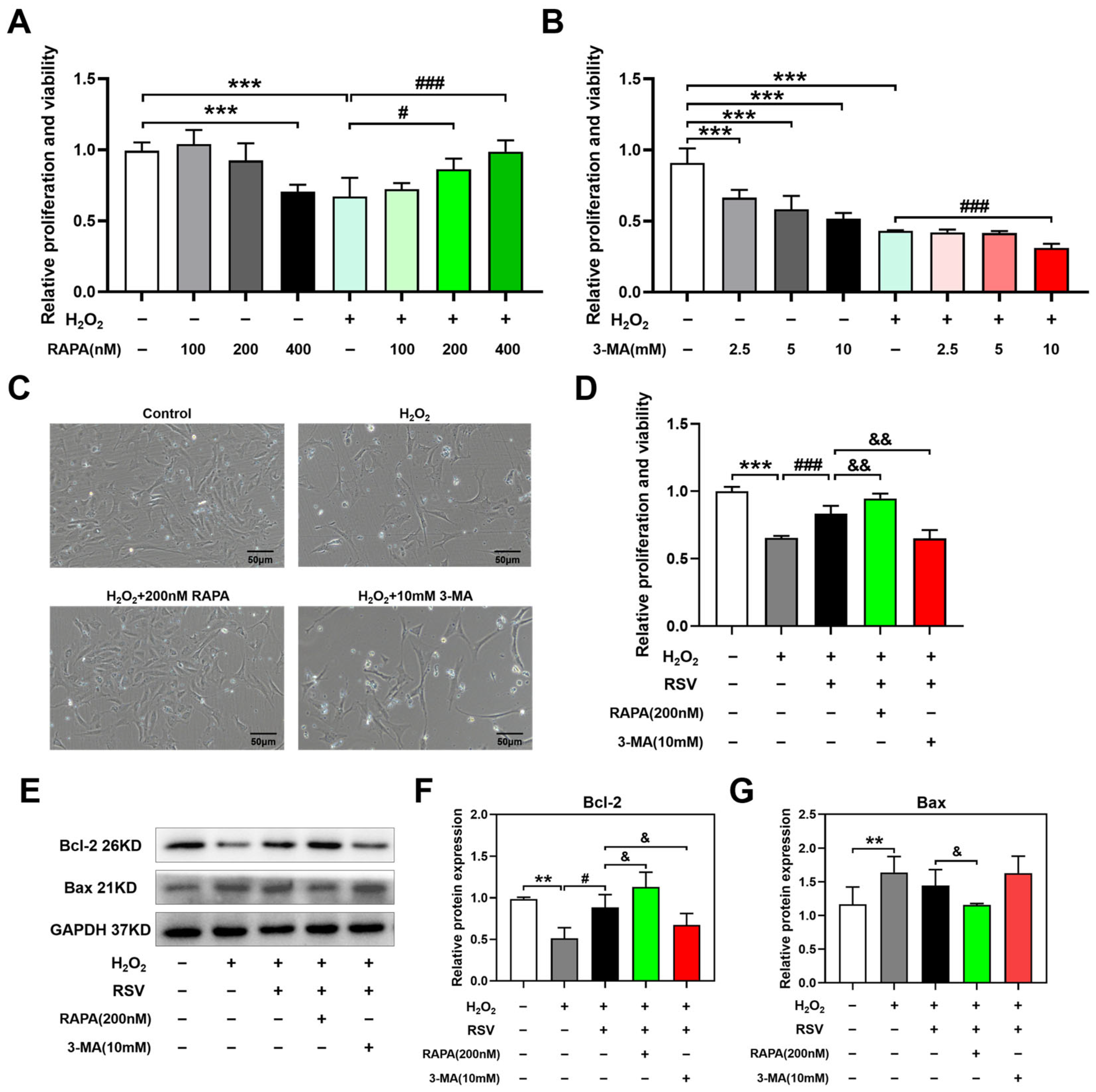
2.4. Autophagy Exerted Protective Effects on H2O2-Induced Luteinized Granulosa Cell Dysfunction
2.5. Rapamycin and 3-MA Regulated Autophagy in H2O2-Induced Luteinized Granulosa Cell Dysfunction
2.6. 3-MA Inhibited the Activation of Autophagy Induced by Resveratrol
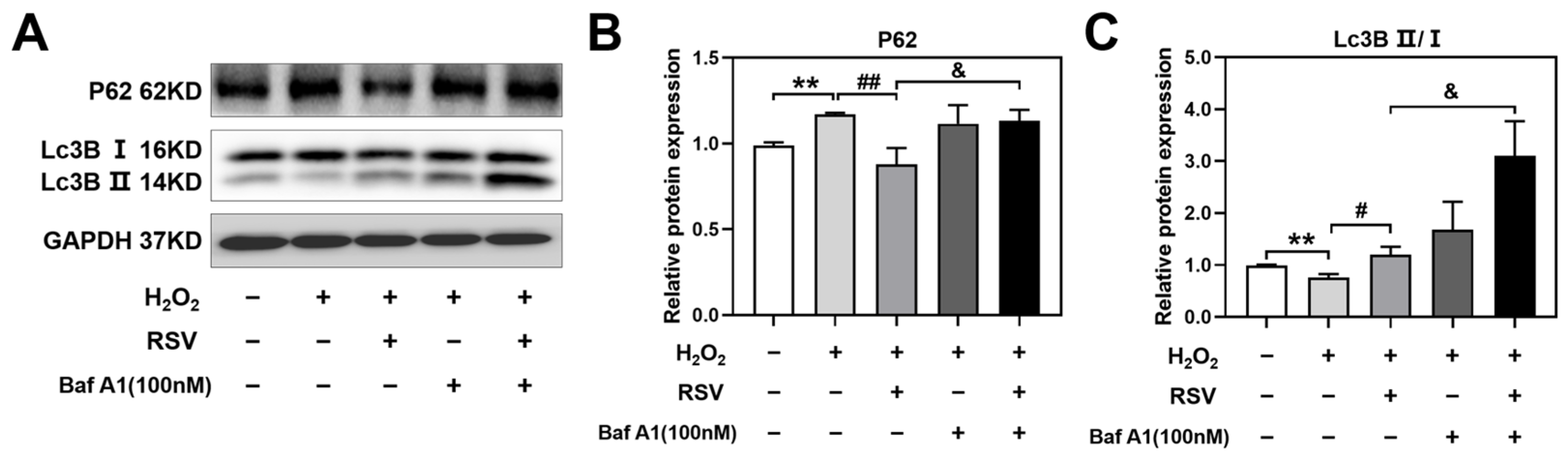
2.7. Resveratrol Controlled Autophagosome-Lysosome Fusion
3. Discussion
4. Materials and Methods
4.1. Animals
4.2. Rat Ovarian Luteinized Granulosa Cell Isolation and Culture
4.3. CCK-8 Assay
4.4. Hormone Detection
4.5. JC-1 Staining
4.6. Observation of Autophagy Structure
4.7. Western Blot
4.8. Real-Time PCR
4.9. Statistical Analysis
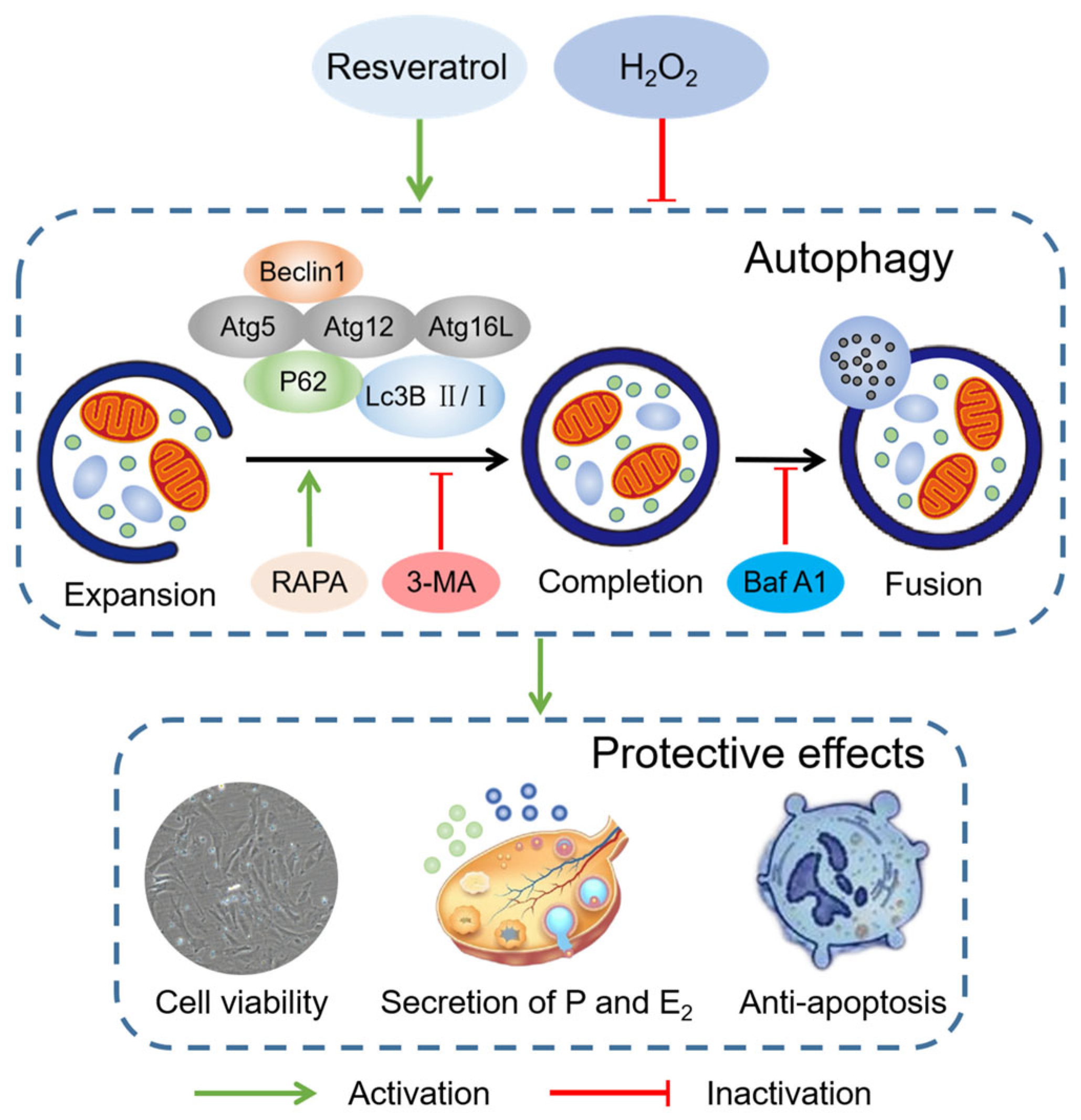
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chou, C.; Chen, M. The effect of steroid hormones on ovarian follicle development. Vitam. Horm. 2018, 107, 155–175. [Google Scholar]
- Toner, J.P. The corpus luteum is more than progesterone. Fertil. Steril. 2021, 115, 1432. [Google Scholar] [CrossRef]
- Przygrodzka, E.; Plewes, M.R.; Davis, J.S. Luteinizing hormone regulation of inter-organelle communication and fate of the corpus luteum. Int. J. Mol. Sci. 2021, 22, 9972. [Google Scholar] [CrossRef]
- Yao, S.; Lopez-Tello, J.; Sferruzzi-Perri, A.N. Developmental programming of the female reproductive system—A review. Biol. Reprod. 2021, 104, 745–770. [Google Scholar] [CrossRef]
- Medvediev, M.V.; Malvasi, A.; Gustapane, S.; Tinelli, A. Hemorrhagic corpus luteum: Clinical management update. Turk. J. Obstet. Gynecol. 2020, 17, 300–309. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Han, X.; Meng, Q.; Guo, Y.; Liu, B.; Song, T.; Jia, Y.; Fang, L.; Sun, Y. HB-EGF upregulates StAR expression and stimulates progesterone production through ERK1/2 signaling in human granulosa-lutein cells. Cell. Commun. Signal. 2022, 20, 166. [Google Scholar] [CrossRef] [PubMed]
- Yu, D.; Jiang, X.; Ge, W.; Qiao, B.; Zhang, D.; Liu, H.; Kuang, H. Gestational exposure to acrylamide suppresses luteal endocrine function through dysregulation of ovarian angiogenesis, oxidative stress and apoptosis in mice. Food Chem. Toxicol. 2022, 159, 112766. [Google Scholar] [CrossRef]
- Wang, L.; Tang, J.; Wang, L.; Tan, F.; Song, H.; Zhou, J.; Li, F. Oxidative stress in oocyte aging and female reproduction. J. Cell. Physiol. 2021, 236, 7966–7983. [Google Scholar] [CrossRef]
- Yan, F.; Zhao, Q.; Li, Y.; Zheng, Z.; Kong, X.; Shu, C.; Liu, Y.; Shi, Y. The role of oxidative stress in ovarian aging: A review. J. Ovarian Res. 2022, 15, 100. [Google Scholar] [CrossRef]
- Zheng, Z.; Wang, M.; Cheng, C.; Liu, D.; Wu, L.; Zhu, J.; Qian, X. Ginsenoside Rb1 reduces H2O2-induced HUVEC dysfunction by stimulating the sirtuin-1/AMP-activated protein kinase pathway. Mol. Med. Rep. 2020, 22, 247–256. [Google Scholar] [CrossRef] [PubMed]
- Jiang, F.; Xu, X.; Li, W.; Xia, K.; Wang, L.; Yang, X. Monotropein alleviates H2O2-induced inflammation, oxidative stress and apoptosis via NF-κB/AP-1 signaling. Mol. Med. Rep. 2020, 22, 4828–4836. [Google Scholar] [CrossRef] [PubMed]
- Sies, H.; Jones, D.P. Reactive oxygen species (ROS) as pleiotropic physiological signalling agents. Nat. Rev. Mol. Cell. Biol. 2020, 21, 363–383. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; He, S.; Ma, B. Autophagy and autophagy-related proteins in cancer. Mol. Cancer 2020, 19, 12. [Google Scholar] [CrossRef] [PubMed]
- Wong, S.Q.; Kumar, A.V.; Mills, J.; Lapierre, L.R. Autophagy in aging and longevity. Hum. Genet. 2020, 139, 277–290. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Xu, L.; Wang, X.; Zhang, D.; Sun, G.; Wang, M.; Wang, M.; Han, Y.; Chai, R.; Wang, H. PRDX1 activates autophagy via the PTEN-AKT signaling pathway to protect against cisplatin-induced spiral ganglion neuron damage. Autophagy 2021, 17, 4159–4181. [Google Scholar] [CrossRef] [PubMed]
- Bernard, M.; Yang, B.; Migneault, F.; Turgeon, J.; Dieudé, M.; Olivier, M.A.; Cardin, G.B.; El-Diwany, M.; Underwood, K.; Rodier, F.; et al. Autophagy drives fibroblast senescence through MTORC2 regulation. Autophagy 2020, 16, 2004–2016. [Google Scholar] [CrossRef]
- Zhang, J.; Ren, Q.; Chen, J.; Gao, B.; Wang, X.; Zhang, Z.; Wang, J.; Xu, Z.; Xing, B. Autophagy Contributes to Oxidative Stress-Induced Apoptosis in Porcine Granulosa Cells. Reprod. Sci. 2021, 28, 2147–2160. [Google Scholar] [CrossRef]
- Galiniak, S.; Aebisher, D.; Bartusik-Aebisher, D. Health benefits of resveratrol administration. Acta Biochim. Pol. 2019, 66, 13–21. [Google Scholar] [CrossRef]
- Zhou, D.D.; Luo, M.; Huang, S.Y.; Saimaiti, A.; Shang, A.; Gan, R.Y.; Li, H.B. Effects and Mechanisms of Resveratrol on Aging and Age-Related Diseases. Oxid. Med. Cell. Longev. 2021, 2021, 9932218. [Google Scholar] [CrossRef]
- Yong, W.; Jiao, J.; Kou, Z.; Wang, C.; Pang, W. Resveratrol ameliorates malathion-induced estrus cycle disorder through attenuating the ovarian tissue oxidative stress, autophagy and apoptosis. Reprod. Toxicol. 2021, 104, 8–15. [Google Scholar] [CrossRef]
- Jiao, J.; Gao, L.; Yong, W.; Kou, Z.; Ren, Z.; Cai, R.; Chu, G.; Pang, W. Resveratrol improves estrus disorder induced by bisphenol A through attenuating oxidative stress, autophagy, and apoptosis. J. Biochem. Mol. Toxicol. 2022, 36, e23120. [Google Scholar] [CrossRef] [PubMed]
- Sugiyama, M.; Kawahara-Miki, R.; Kawana, H.; Shirasuna, K.; Kuwayama, T.; Iwata, H. Resveratrol-induced mitochondrial synthesis and autophagy in oocytes derived from early antral follicles of aged cows. J. Reprod. Dev. 2015, 61, 251–259. [Google Scholar] [CrossRef] [PubMed]
- Cai, M.; Wang, J.; Sun, H.; Guo, Q.; Zhang, C.; Yao, H.; Zhao, C.; Jia, Y.; Zhu, H. Resveratrol Attenuates Hydrogen Peroxide-induced Dysfunction of Rat Ovarian Granulosa-lutein Cells by Resisting Oxidative Stress via the SIRT1/Nrf2/ARE Signaling Pathway. Curr. Pharm. Des. 2023, 29, 947–956. [Google Scholar]
- Mauthe, M.; Orhon, I.; Rocchi, C.; Zhou, X.; Luhr, M.; Hijlkema, K.J.; Coppes, R.P.; Engedal, N.; Mari, M.; Reggiori, F. Chloroquine inhibits autophagic flux by decreasing autophagosome-lysosome fusion. Autophagy 2018, 14, 1435–1455. [Google Scholar] [CrossRef]
- Ávila, J.; González-Fernández, R.; Rotoli, D.; Hernández, J.; Palumbo, A. Oxidative Stress in Granulosa-Lutein Cells from In Vitro Fertilization Patients. Reprod. Sci. 2016, 23, 1656–1661. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Pang, X.; Tang, Z.; Yin, D.; Wang, Z. Overexpression of hypoxia-inducible factor prolyl hydoxylase-2 attenuates hypoxia-induced vascular endothelial growth factor expression in luteal cells. Mol. Med. Rep. 2015, 12, 3809–3814. [Google Scholar] [CrossRef]
- Tungmahasuk, D.; Fungbun, N.; Laoharatchatathanin, T.; Terashima, R.; Kurusu, S.; Kawaminami, M. Effects of gonadotropin-releasing hormone agonist on human chorionic gonadotropin activity in granulosa cells of immature female rats. J. Reprod. Dev. 2018, 64, 129–134. [Google Scholar] [CrossRef]
- Wang, J.; Qian, X.; Gao, Q.; Lv, C.; Xu, J.; Jin, H.; Zhu, H. Quercetin increases the antioxidant capacity of the ovary in menopausal rats and in ovarian granulosa cell culture in vitro. J. Ovarian Res. 2018, 11, 51. [Google Scholar] [CrossRef]
- Gao, Q.; Guo, X.; Cao, Y.; Jia, X.; Xu, S.; Lu, C.; Zhu, H. Melatonin Protects HT22 Hippocampal Cells from H2O2-induced Injury by Increasing Beclin1 and Atg Protein Levels to Activate Autophagy. Curr. Pharm. Des. 2021, 27, 446–454. [Google Scholar] [CrossRef]
- Wang, M.; Li, Y.; Molenaar, A.; Li, Q.; Cao, Y.; Shen, Y.; Chen, P.; Yan, J.; Gao, Y.; Li, J. Vitamin E and selenium supplementation synergistically alleviate the injury induced by hydrogen peroxide in bovine granulosa cells. Theriogenology 2021, 170, 91–106. [Google Scholar] [CrossRef]
- Chen, Z.; Wang, C.; Yu, N.; Si, L.; Zhu, L.; Zeng, A.; Liu, Z.; Wang, X. INF2 regulates oxidative stress-induced apoptosis in epidermal HaCaT cells by modulating the HIF1 signaling pathway. Biomed. Pharmacother. 2019, 111, 151–161. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; Lee, Y.J. Synergistic anticancer activity of resveratrol in combination with docetaxel in prostate carcinoma cells. Nutr. Res. Pract. 2021, 15, 12–25. [Google Scholar] [CrossRef] [PubMed]
- Hsu, Y.; Chen, C.; Wang, Y.; Lin, E.; Chang, C.; Chen, J.; Wu, M.; Lin, H.; Wan, L. Anti-Inflammatory Effects of Resveratrol on Human Retinal Pigment Cells and a Myopia Animal Model. Curr. Issues Mol. Biol. 2021, 43, 716–727. [Google Scholar] [CrossRef]
- Deckmann, I.; Santos-Terra, J.; Fontes-Dutra, M.; Körbes-Rockenbach, M.; Bauer-Negrini, G.; Schwingel, G.B.; Riesgo, R.; Bambini-Junior, V.; Gottfried, C. Resveratrol prevents brain edema, blood-brain barrier permeability, and altered aquaporin profile in autism animal model. Int. J. Dev. Neurosci. 2021, 81, 579–604. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Zhang, J.; Tao, Y.; Ji, C.; Aniagu, S.; Jiang, Y.; Chen, T. AHR/ROS-mediated mitochondria apoptosis contributes to benzo[a]pyrene-induced heart defects and the protective effects of resveratrol. Toxicology 2021, 462, 152965. [Google Scholar] [CrossRef]
- Zhou, X.; Ruan, Q.; Ye, Z.; Chu, Z.; Xi, M.; Li, M.; Hu, W.; Guo, X.; Yao, P.; Xie, W. Resveratrol accelerates wound healing by attenuating oxidative stress-induced impairment of cell proliferation and migration. Burns 2021, 47, 133–139. [Google Scholar] [CrossRef]
- Moreira-Pinto, B.; Costa, L.; Felgueira, E.; Fonseca, B.M.; Rebelo, I. Low Doses of Resveratrol Protect Human Granulosa Cells from Induced-Oxidative Stress. Antioxidants 2021, 10, 561. [Google Scholar] [CrossRef]
- Qin, H.; Zhang, H.; Zhang, X.; Zhang, S.; Zhu, S.; Wang, H. Resveratrol attenuates radiation enteritis through the SIRT1/FOXO3a and PI3K/AKT signaling pathways. Biochem. Biophys. Res. Commun. 2021, 554, 199–205. [Google Scholar] [CrossRef]
- Levine, B.; Kroemer, G. Biological Functions of Autophagy Genes: A Disease Perspective. Cell 2019, 176, 11–42. [Google Scholar] [CrossRef]
- Yang, X.; Jiang, T.; Wang, Y.; Guo, L. The Role and Mechanism of SIRT1 in Resveratrol-regulated Osteoblast Autophagy in Osteoporosis Rats. Sci. Rep. 2019, 9, 18424. [Google Scholar] [CrossRef]
- Chen, M.; Yi, L.; Jin, X.; Liang, X.; Zhou, Y.; Zhang, T.; Xie, Q.; Zhou, X.; Chang, H.; Fu, Y.; et al. Resveratrol attenuates vascular endothelial inflammation by inducing autophagy through the cAMP signaling pathway. Autophagy 2013, 9, 2033–2045. [Google Scholar] [CrossRef] [PubMed]
- Hollenstein, D.M.; Kraft, C. Autophagosomes are formed at a distinct cellular structure. Curr. Opin. Cell Biol. 2020, 65, 50–57. [Google Scholar] [CrossRef] [PubMed]
- Leidal, A.M.; Levine, B.; Debnath, J. Autophagy and the cell biology of age-related disease. Nat. Cell Biol. 2018, 20, 1338–1348. [Google Scholar] [CrossRef] [PubMed]
- 4Romanov, J.; Walczak, M.; Ibiricu, I.; Schuchner, S.; Ogris, E.; Kraft, C.; Martens, S. Mechanism and functions of membrane binding by the Atg5-Atg12/Atg16 complex during autophagosome formation. EMBO J. 2012, 31, 4304–4317. [Google Scholar] [CrossRef]
- Walczak, M.; Martens, S. Dissecting the role of the Atg12-Atg5-Atg16 complex during autophagosome formation. Autophagy 2013, 9, 424–425. [Google Scholar] [CrossRef] [PubMed]
- Schaaf, M.B.; Keulers, T.G.; Vooijs, M.A.; Rouschop, K.M. LC3/GABARAP family proteins: Autophagy-(un)related functions. FASEB J. 2016, 30, 3961–3978. [Google Scholar] [CrossRef]
- Shao, T.; Ke, H.; Liu, R.; Xu, L.; Han, S.; Zhang, X.; Dang, Y.; Jiao, X.; Li, W.; Chen, Z.J.; et al. Autophagy regulates differentiation of ovarian granulosa cells through degradation of WT1. Autophagy 2022, 18, 1864–1878. [Google Scholar] [CrossRef]
- Lin, M.; Hua, R.; Ma, J.; Zhou, Y.; Li, P.; Xu, X.; Yu, Z.; Quan, S. Bisphenol A promotes autophagy in ovarian granulosa cells by inducing AMPK/mTOR/ULK1 signalling pathway. Environ. Int. 2021, 147, 106298. [Google Scholar] [CrossRef]
- Lu, H.; Yang, H.; Zhou, W.; Lai, Z.; Qiu, X.; Fu, Q.; Zhao, J.; Wang, J.; Li, D.; Li, M. Rapamycin prevents spontaneous abortion by triggering decidual stromal cell autophagy-mediated NK cell residence. Autophagy 2021, 17, 2511–2527. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, H. Regulation of Autophagy by mTOR Signaling Pathway. Adv. Exp. Med. Biol. 2019, 1206, 67–83. [Google Scholar]
- Bao, J.; Chen, Z.; Xu, L.; Wu, L.; Xiong, Y. Rapamycin protects chondrocytes against IL-18-induced apoptosis and ameliorates rat osteoarthritis. Aging 2020, 12, 5152–5167. [Google Scholar] [CrossRef] [PubMed]
- Sotthibundhu, A.; McDonagh, K.; von Kriegsheim, A.; Garcia-Munoz, A.; Klawiter, A.; Thompson, K.; Chauhan, K.D.; Krawczyk, J.; McInerney, V.; Dockery, P.; et al. Rapamycin regulates autophagy and cell adhesion in induced pluripotent stem cells. Stem Cell Res. Ther. 2016, 7, 166. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Wu, Y.; Zhao, G.; Ye, Z.; Xing, C.; Yang, X. Inhibition of autophagy by 3-MA promotes hypoxia-induced apoptosis in human colorectal cancer cells. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 1047–1054. [Google Scholar]
- Chen, W.; Xian, G.; Gu, M.; Pan, B.; Wu, X.; Ye, Y.; Zheng, L.; Zhang, Z.; Sheng, P. Autophagy inhibitors 3-MA and LY294002 repress osteoclastogenesis and titanium particle-stimulated osteolysis. Biomater. Sci. 2021, 9, 4922–4935. [Google Scholar] [CrossRef]
- Zong, D.D.; Liu, X.M.; Li, J.H.; Ouyang, R.Y.; Long, Y.J.; Chen, P.; Chen, Y. Resveratrol attenuates cigarette smoke induced endothelial apoptosis by activating Notch1 signaling mediated autophagy. Respir. Res. 2021, 22, 22. [Google Scholar] [CrossRef]
- Huang, S.S.; Ding, D.F.; Chen, S.; Dong, C.L.; Ye, X.L.; Yuan, Y.G.; Feng, Y.M.; You, N.; Xu, J.R.; Miao, H.; et al. Resveratrol protects podocytes against apoptosis via stimulation of autophagy in a mouse model of diabetic nephropathy. Sci. Rep. 2017, 7, 45692. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Li, Y.; Huang, S.; Xu, H.; Li, H.; Liu, B. Resveratrol-primed exosomes strongly promote the recovery of motor function in SCI rats by activating autophagy and inhibiting apoptosis via the PI3K signaling pathway. Neurosci. Lett. 2020, 736, 135262. [Google Scholar] [CrossRef]
- Gao, J.; Zhang, Q.; Song, L. Resveratrol enhances matrix biosynthesis of nucleus pulposus cells through activating autophagy via the PI3K/Akt pathway under oxidative damage. Biosci. Rep. 2018, 38, BSR20180544. [Google Scholar] [CrossRef]
- Duan, L.; Li, S.; Wang, L.; Jing, Y.; Li, G.; Sun, Y.; Sun, W.; Li, Y.; Zhao, L.; Xin, S. Melatonin Plays a Critical Protective Role in Nicotine-Related Abdominal Aortic Aneurysm. Front. Physiol. 2020, 11, 866. [Google Scholar] [CrossRef]
- Wu, B.; Zeng, W.; Ouyang, W.; Xu, Q.; Chen, J.; Wang, B.; Zhang, X. Quercetin induced NUPR1-dependent autophagic cell death by disturbing reactive oxygen species homeostasis in osteosarcoma cells. J. Clin. Biochem. Nutr. 2020, 67, 137–145. [Google Scholar] [CrossRef]
- Wang, Q.; Han, W.; Ma, C.; Wang, T.; Zhong, J. Western blot normalization: Time to choose a proper loading control seriously. Electrophoresis 2023, 44, 854–863. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–428. [Google Scholar] [CrossRef] [PubMed]

| Gene | Type | Sequences (5′ → 3′) |
|---|---|---|
| Beclin1 | Forward Primer | GCCTCTGAAACTGGACACG |
| Beclin1 | Reverse Primer | CCTCTTCCTCCTGGCTCTCT |
| Atg5 | Forward Primer | CACTGGGACTTCTGCTCCTG |
| Atg5 | Reverse Primer | TTCTTCAACCAAGCCAAACC |
| Lc3B | Forward Primer | GGTGTTTTTCTCCTGGTTTGG |
| Lc3B | Reverse Primer | GCACTTGGACTTCAGCCTTC |
| Atg12 | Forward Primer | AAACGAAGAAATGGGCTGTG |
| Atg12 | Reverse Primer | GAAGGGGCAAAGGACTGATT |
| Atg16L | Forward Primer | CTGTGCTTTTCCCGTCTTTC |
| Atg16L | Reverse Primer | GCCCTGATTTGGTTTCCAC |
| β-actin | Forward Primer | AGCCATGTACGTAGCCATCC |
| β-actin | Reverse Primer | GCTGTGGTGGTGAAGCTGTA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cai, M.; Sun, H.; Huang, Y.; Yao, H.; Zhao, C.; Wang, J.; Zhu, H. Resveratrol Protects Rat Ovarian Luteinized Granulosa Cells from H2O2-Induced Dysfunction by Activating Autophagy. Int. J. Mol. Sci. 2023, 24, 10914. https://doi.org/10.3390/ijms241310914
Cai M, Sun H, Huang Y, Yao H, Zhao C, Wang J, Zhu H. Resveratrol Protects Rat Ovarian Luteinized Granulosa Cells from H2O2-Induced Dysfunction by Activating Autophagy. International Journal of Molecular Sciences. 2023; 24(13):10914. https://doi.org/10.3390/ijms241310914
Chicago/Turabian StyleCai, Minghui, Haijuan Sun, Yujia Huang, Haixu Yao, Chen Zhao, Jiao Wang, and Hui Zhu. 2023. "Resveratrol Protects Rat Ovarian Luteinized Granulosa Cells from H2O2-Induced Dysfunction by Activating Autophagy" International Journal of Molecular Sciences 24, no. 13: 10914. https://doi.org/10.3390/ijms241310914
APA StyleCai, M., Sun, H., Huang, Y., Yao, H., Zhao, C., Wang, J., & Zhu, H. (2023). Resveratrol Protects Rat Ovarian Luteinized Granulosa Cells from H2O2-Induced Dysfunction by Activating Autophagy. International Journal of Molecular Sciences, 24(13), 10914. https://doi.org/10.3390/ijms241310914





