Ester Bonds for Modulation of the Mechanical Properties of Protein Hydrogels
Abstract
1. Introduction
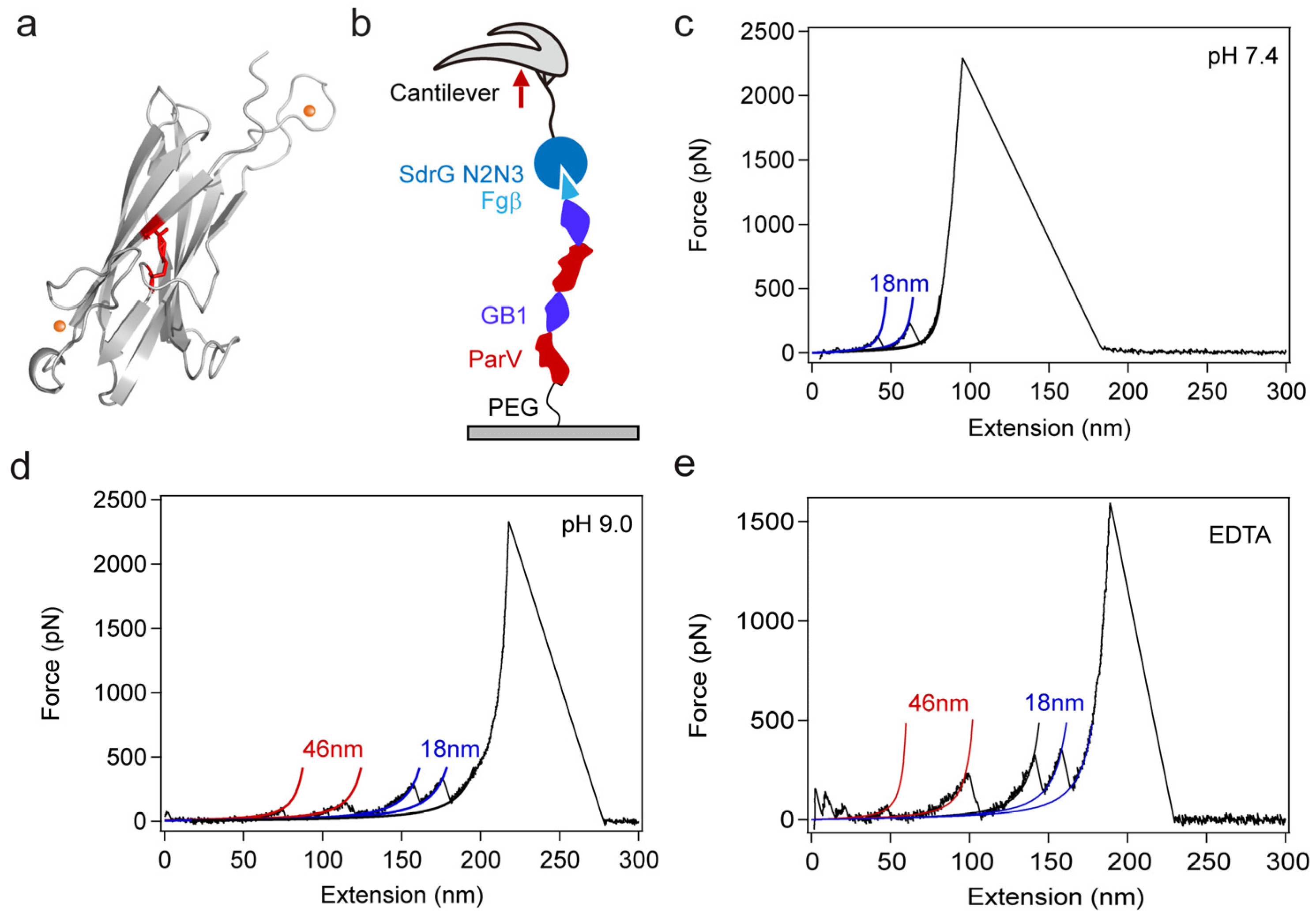
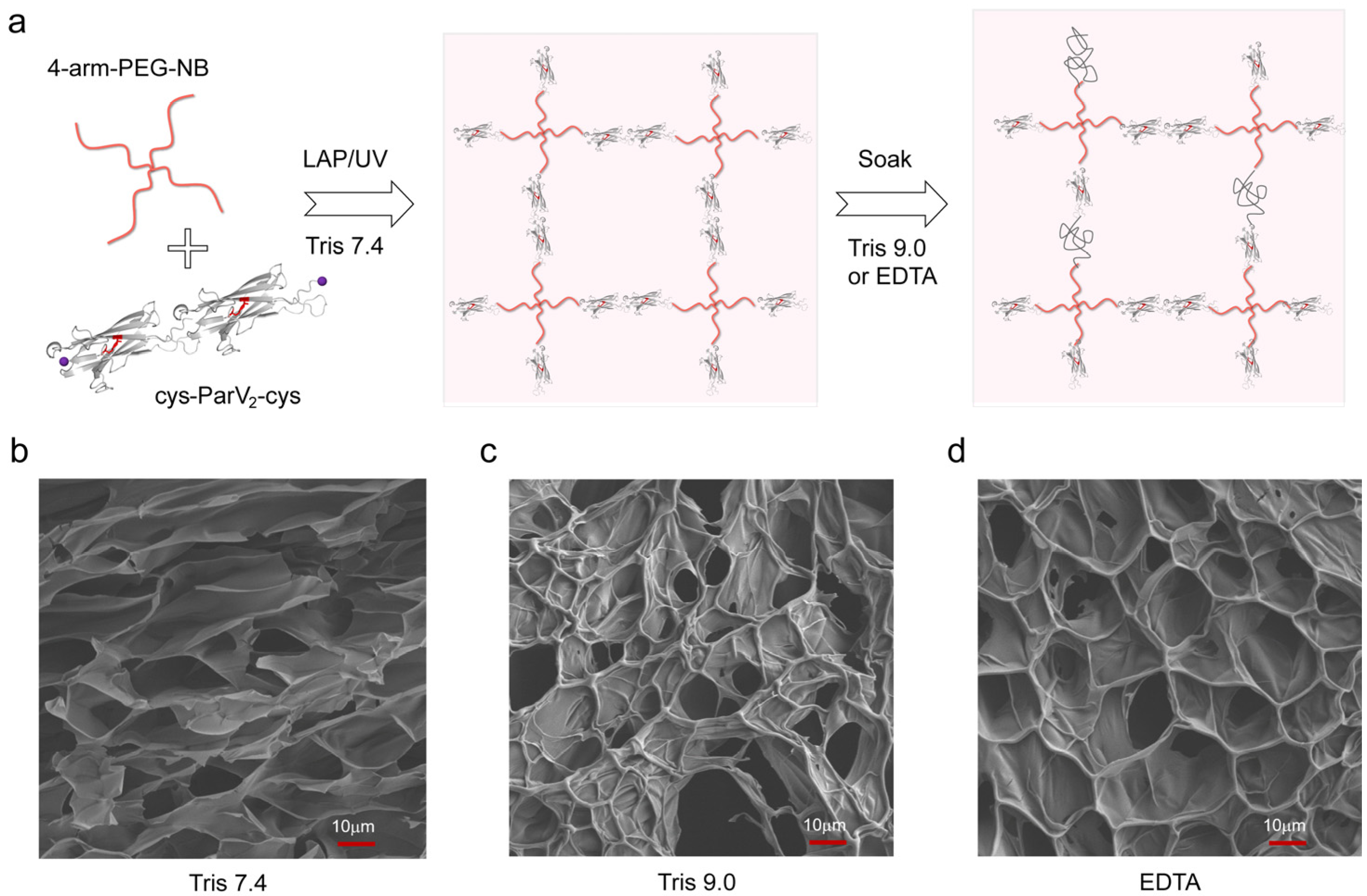
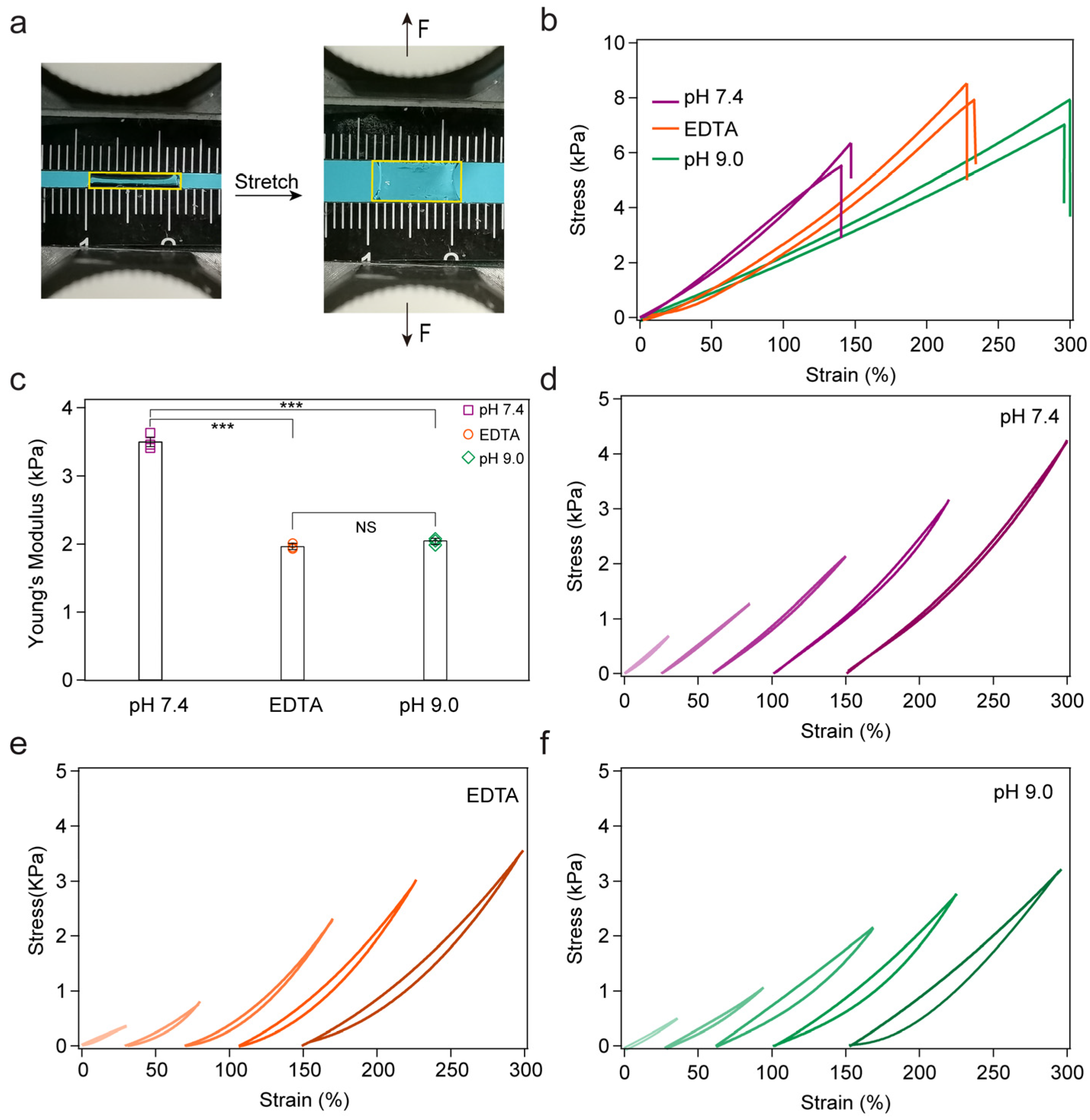
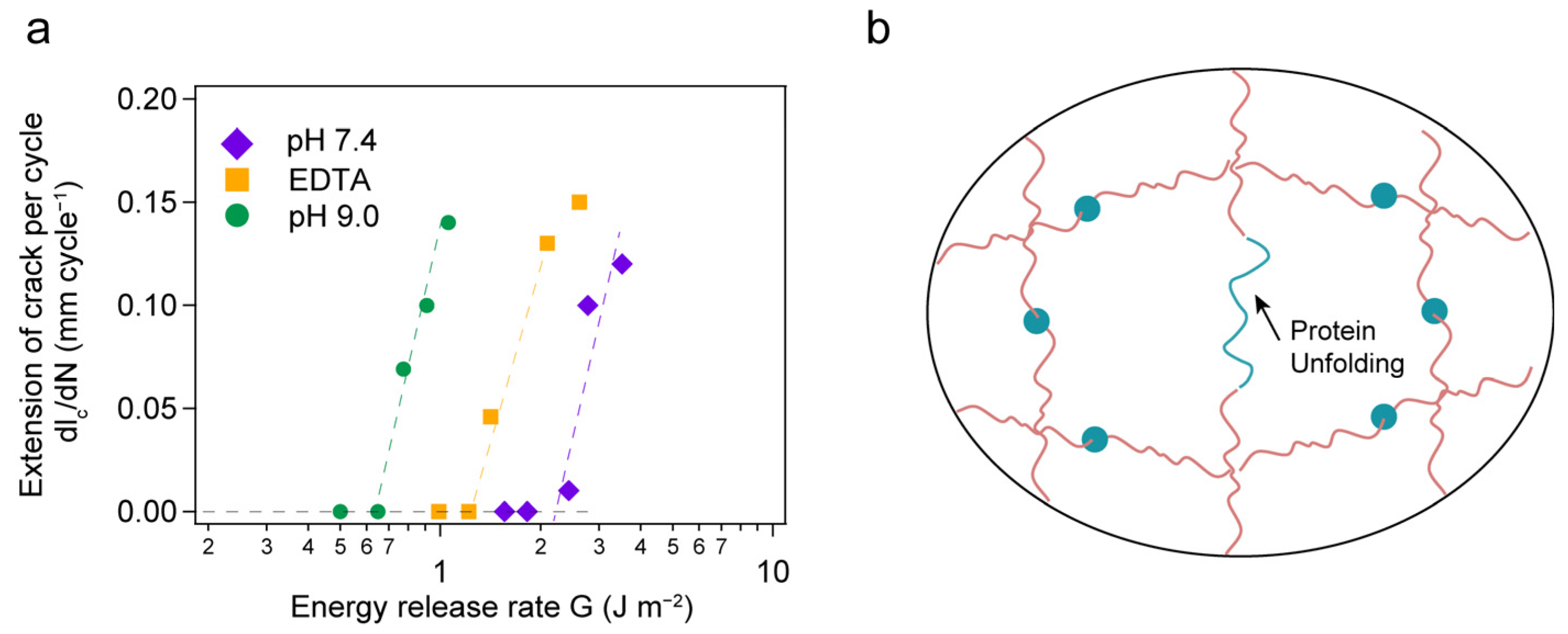
2. Results and Discussion
3. Materials and Methods
3.1. Materials
3.2. Protein Expression and Purification
3.3. Single-Molecule Force Spectroscopy (SMFS) Experiments
3.4. Preparation of the Hydrogels
3.5. Tensile Test
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wu, B.Y.; Xue, Y.T.; Ali, I.; Lu, H.H.; Yang, Y.M.; Yang, X.X.; Lu, W.; Zheng, Y.F.; Chen, T. The Dynamic Mortise-and-Tenon Interlock Assists Hydrated Soft Robots Toward Off-Road Locomotion. Research 2022, 2022, 0015. [Google Scholar] [CrossRef]
- Zhu, Q.L.; Du, C.; Dai, Y.H.; Daab, M.; Matejdes, M.; Breu, J.; Hong, W.; Zheng, Q.; Wu, Z.L. Light-steered locomotion of muscle-like hydrogel by self-coordinated shape change and friction modulation. Nat. Commun. 2020, 11, 5166. [Google Scholar] [CrossRef] [PubMed]
- Salih, A.E.; Elsherif, M.; Alam, F.; Yetisen, A.K.; Butt, H. Gold Nanocomposite Contact Lenses for Color Blindness Management. ACS Nano 2021, 15, 4870–4880. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Yao, H.Y.; Zhao, G.N.; Ameer, G.A.; Sun, W.; Yang, J.; Mi, S.L. Flexible, wearable microfluidic contact lens with capillary networks for tear diagnostics. J. Mater. Sci. 2020, 55, 9551–9561. [Google Scholar] [CrossRef]
- Fischer, A.; Lilienthal, S.; Vazquez-Gonzalez, M.; Fadeev, M.; Sohn, Y.S.; Nechushtai, R.; Willner, I. Triggered Release of Loads from Microcapsule-in-Microcapsule Hydrogel Microcarriers: En-Route to an “Artificial Pancreas”. J. Am. Chem. Soc. 2020, 142, 4223–4234. [Google Scholar] [CrossRef] [PubMed]
- Zong, S.Y.; Wen, H.K.; Lv, H.; Li, T.; Tang, R.L.; Liu, L.J.; Jiang, J.X.; Wang, S.P.; Duan, J.F. Intelligent hydrogel with both redox and thermo-response based on cellulose nanofiber for controlled drug delivery. Carbohydr. Polym. 2020, 278, 118943. [Google Scholar] [CrossRef] [PubMed]
- Sun Han Chang, R.A.; Shanley, J.F.; Kersh, M.E.; Harley, B.A.C. Tough and tunable scaffold-hydrogel composite biomaterial for soft-to-hard musculoskeletal tissue interfaces. Sci. Adv. 2020, 6, abb6763. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.B.; Dai, J.J.; Huang, Y.F.; Li, X.M.; Yang, J.M.; Zheng, Y.Q.; Shi, X.A. Extracellular matrix mimicking dynamic interpenetrating network hydrogel for skin tissue engineering. Chem. Eng. J. 2023, 457, 141362. [Google Scholar] [CrossRef]
- Chen, Q.; Zhu, L.; Chen, H.; Yan, H.L.; Huang, L.; Yang, J.; Zheng, J. A Novel Design Strategy for Fully Physically Linked Double Network Hydrogels with Tough, Fatigue Resistant, and Self-Healing Properties. Adv. Funct. Mater. 2015, 25, 1598–1607. [Google Scholar] [CrossRef]
- Liu, X.Y.; He, X.; Yang, B.; Lai, L.; Chen, N.X.; Hu, J.G.; Lu, Q.Y. Dual Physically Cross-Linked Hydrogels Incorporating Hydrophobic Interactions with Promising Repairability and Ultrahigh Elongation. Adv. Funct. Mater. 2021, 31, 2008187. [Google Scholar] [CrossRef]
- Luo, Y.C.; Yu, M.L.; Zhang, Y.T.; Wang, Y.Y.; Long, L.; Tan, H.H.; Li, N.; Xu, L.J.; Xu, J.X. Highly sensitive strain sensor and self-powered triboelectric nanogenerator using a fully physical crosslinked double-network conductive hydrogel. Nano Energy 2022, 104, 107955. [Google Scholar] [CrossRef]
- Tang, L.; Wu, S.J.; Xu, Y.; Cui, T.; Li, Y.H.; Wang, W.; Gong, L.; Tang, J.X. High toughness fully physical cross-linked double network organohydrogels for strain sensors with anti-freezing and anti-fatigue properties. Mater. Adv. 2021, 2, 6655–6664. [Google Scholar] [CrossRef]
- Huynh, N.T.; Jeon, Y.S.; Kim, D.; Kim, J.H. Preparation and swelling properties of “click” hydrogel from polyaspartamide derivatives using tri-arm PEG and PEG-co-poly(amino urethane) azides as crosslinking agents. Polymer 2013, 54, 1341–1349. [Google Scholar] [CrossRef]
- Lee, S.; Park, Y.H.; Ki, C.S. Fabrication of PEG-carboxymethylcellulose hydrogel by thiol-norbornene photo-click chemistry. Int. J. Biol. Macromol. 2016, 83, 1–8. [Google Scholar] [CrossRef]
- Wang, L.Y.; Shan, G.Y.; Pan, P.J. A strong and tough interpenetrating network hydrogel with ultrahigh compression resistance. Soft Matter 2014, 10, 3850–3856. [Google Scholar] [CrossRef]
- Cao, Y.; Yao, Y.Q.; Li, Y.; Yang, X.X.; Cao, Z.J.; Yang, G. Tunable keratin hydrogel based on disulfide shuffling strategy for drug delivery and tissue engineering. J. Colloid Interface Sci. 2019, 544, 121–129. [Google Scholar] [CrossRef]
- Meng, H.; Zheng, J.; Wen, X.F.; Cai, Z.Q.; Zhang, J.W.; Chen, T. pH- and Sugar-Induced Shape Memory Hydrogel Based on Reversible Phenylboronic Acid-Diol Ester Bonds. Macromol. Rapid Commun. 2015, 36, 533–537. [Google Scholar] [CrossRef]
- Gupta, B.; Anjum, S.; Ikram, S. Physicochemical studies of crosslinked thiolated polyvinyl alcohol hydrogels. Polym. Bull. 2013, 70, 2437–2450. [Google Scholar] [CrossRef]
- Lei, H.; Dong, L.; Li, Y.; Zhang, J.S.; Chen, H.Y.; Wu, J.H.; Zhang, Y.; Fan, Q.Y.; Xue, B.; Qin, M.; et al. Stretchable hydrogels with low hysteresis and anti-fatigue fracture based on polyprotein cross-linkers. Nat. Commun. 2020, 11, 4032. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Cui, L.; Wang, H.Z.; Chen, Q.B.; Guan, Y.; Zhang, Y.J. Tough, Resilient, Adhesive, and Anti-Freezing Hydrogels Cross-Linked with a Macromolecular Cross-Linker for Wearable Strain Sensors. ACS Appl. Mater. Interfaces 2021, 13, 42052–42062. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.D.; Xiang, L.; Diaz-Dussan, D.; Zhang, J.W.; Yang, W.S.; Gong, L.; Chen, J.S.; Narain, R.; Zeng, H.B. Dynamic Flexible Hydrogel Network with Biological Tissue-like Self-Protective Functions. Chem. Mater. 2020, 32, 10545–10555. [Google Scholar] [CrossRef]
- Li, X.Y.; Cui, K.P.; Kurokawa, T.; Ye, Y.N.; Sun, T.L.; Yu, C.T.; Creton, C.; Gong, J.P. Effect of mesoscale phase contrast on fatigue-delaying behavior of self-healing hydrogels. Sci. Adv. 2021, 7, abe8210. [Google Scholar] [CrossRef] [PubMed]
- Chavda, H.; Patel, C. Effect of crosslinker concentration on characteristics of superporous hydrogel. Int. J. Pharm. Investig. 2011, 1, 17–21. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Beech, H.K.; Bowser, B.H.; Kouznetsova, T.B.; Olsen, B.D.; Rubinstein, M.; Craig, S.L. Mechanism Dictates Mechanics: A Molecular Substituent Effect in the Macroscopic Fracture of a Covalent Polymer Network. J. Am. Chem. Soc. 2021, 143, 3714–3718. [Google Scholar] [CrossRef]
- Creusen, G.; Schmidt, R.S.; Walther, A. One-Component DNA Mechanoprobes for Facile Mechanosensing in Photopolymerized Hydrogels and Elastomers. ACS Macro Lett. 2021, 10, 671–678. [Google Scholar] [CrossRef]
- Cui, J.H.; Wu, D.; Sun, Q.; Yang, X.Z.; Wang, D.D.; Zhuang, M.; Zhang, Y.H.; Gan, M.Z.; Luo, D. A PEGDA/DNA Hybrid Hydrogel for Cell-Free Protein Synthesis. Front. Chem. 2020, 8, 28. [Google Scholar] [CrossRef]
- Gačanin, J.; Synatschke, C.V.; Weil, T. Biomedical Applications of DNA-Based Hydrogels. Adv. Funct. Mater. 2019, 30, 1906253. [Google Scholar] [CrossRef]
- Kandatsu, D.; Cervantes-Salguero, K.; Kawamata, I.; Hamada, S.; Nomura, S.M.; Fujimoto, K.; Murata, S. Reversible Gel-Sol Transition of a Photo-Responsive DNA Gel. ChemBioChem 2016, 17, 1118–1121. [Google Scholar] [CrossRef]
- Li, H.B.; Kong, N.; Laver, B.; Liu, J.Q. Hydrogels Constructed from Engineered Proteins. Small 2016, 12, 973–987. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Liu, Y.; Yang, G.Z.; Kong, H.; Guo, L.; Wei, G. Recent advances in protein hydrogels: From design, structural and functional regulations to healthcare applications. Chem. Eng. J. 2023, 451, 138494. [Google Scholar] [CrossRef]
- Li, Y.J.; Ding, Y.Q.; Yang, B.; Cao, T.Y.; Xu, J.F.; Dong, Y.C.; Chen, Q.; Xu, L.J.; Liu, D.S. Reinforcing DNA Supramolecular Hydrogel with Polymeric Multiple-Unit Linker. CCS Chem. 2023, 5, 434–444. [Google Scholar] [CrossRef]
- Ikebe, M. Regulation of the function of mammalian myosin and its conformational change. Biochem. Biophys. Res. Commun. 2008, 369, 157–164. [Google Scholar] [CrossRef]
- Lv, S.S.; Dudek, D.M.; Cao, Y.; Balamurali, M.M.; Gosline, J.; Li, H.B. Designed biomaterials to mimic the mechanical properties of muscles. Nature 2010, 465, 69–73. [Google Scholar] [CrossRef]
- Martinez, M.G.; Bullock, A.J.; MacNeil, S.; Rehman, I.U. Characterisation of structural changes in collagen with Raman spectroscopy. Appl. Spectrosc. Rev. 2019, 54, 509–542. [Google Scholar] [CrossRef]
- Wang, R.; Yang, Z.G.; Luo, J.R.; Hsing, I.M.; Sun, F. B12-dependent photoresponsive protein hydrogels for controlled stem cell/protein release. Proc. Natl. Acad. Sci. USA 2017, 114, 5912–5917. [Google Scholar] [CrossRef] [PubMed]
- Bian, Q.Y.; Fu, L.L.; Li, H.B. Engineering shape memory and morphing protein hydrogels based on protein unfolding and folding. Nat. Commun. 2022, 13, 137. [Google Scholar] [CrossRef]
- Kim, S.H.; Hong, H.; Ajiteru, O.; Sultan, M.T.; Lee, Y.J.; Lee, J.S.; Lee, O.J.; Lee, H.; Park, H.S.; Choi, K.Y.; et al. 3D bioprinted silk fibroin hydrogels for tissue engineering. Nat. Protoc. 2021, 16, 5484–5532. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Li, H.B. Polyprotein of GB1 is an ideal artificial elastomeric protein. Nat. Mater. 2007, 6, 109–114. [Google Scholar] [CrossRef]
- Lei, H.; Guo, Y.; Hu, X.; Hu, C.G.; Hu, X.T.; Li, H.B. Reversible Unfolding and Folding of the Metalloprotein Ferredoxin Revealed by Single-Molecule Atomic Force Microscopy. J. Am. Chem. Soc. 2017, 139, 1538–1544. [Google Scholar] [CrossRef]
- Neuman, K.C.; Nagy, A. Single-molecule force spectroscopy: Optical tweezers, magnetic tweezers and atomic force microscopy. Nat. Methods 2008, 5, 491–505. [Google Scholar] [CrossRef]
- Tunn, I.; de Leon, A.S.; Blank, K.G.; Harrington, M.J. Tuning coiled coil stability with histidine-metal coordination. Nanoscale 2018, 10, 22725–22729. [Google Scholar] [CrossRef]
- Kwon, H.; Squire, C.J.; Young, P.G.; Baker, E.N. Autocatalytically generated Thr-Gln ester bond cross-links stabilize the repetitive Ig-domain shaft of a bacterial cell surface adhesin. Proc. Natl. Acad. Sci. USA 2014, 111, 1367–1372. [Google Scholar] [CrossRef]
- Lei, H.; Ma, Q.; Li, W.F.; Wen, J.; Ma, H.B.; Qin, M.; Wang, W.; Cao, Y. An ester bond underlies the mechanical strength of a pathogen surface protein. Nat. Commun. 2021, 12, 5082. [Google Scholar] [CrossRef]
- Zheng, P.; Wang, Y.Y.; Li, H.B. Reversible Unfolding-Refolding of Rubredoxin: A Single-Molecule Force Spectroscopy Study. Angew. Chem. Int. Ed. 2014, 53, 14060–14063. [Google Scholar] [CrossRef]
- Fairbanks, B.B.D.; Schwartz, M.P.; Halevi, A.E.; Nuttelman, C.R.; Bowman, C.N.; Anseth, K.S. A Versatile Synthetic Extracellular Matrix Mimic via Thiol-Norbornene Photopolymerization. Adv. Mater. 2009, 21, 5005–5010. [Google Scholar] [CrossRef] [PubMed]
- Gramlich, W.M.; Kim, I.L.; Burdick, J.A. Synthesis and orthogonal photopatterning of hyaluronic acid hydrogels with thiol-norbornene chemistry. Biomaterials 2013, 34, 9803–9811. [Google Scholar] [CrossRef]
- Wu, J.H.; Li, P.F.; Dong, C.L.; Jiang, H.T.; Xue, B.; Gao, X.; Qin, M.; Wang, W.; Chen, B.; Cao, Y. Rationally designed synthetic protein hydrogels with predictable mechanical properties. Nat. Commun. 2018, 9, 620. [Google Scholar] [CrossRef] [PubMed]
- Fang, J.; Mehlich, A.; Koga, N.; Huang, J.Q.; Koga, R.; Gao, X.Y.; Hu, C.G.; Jin, C.; Rief, M.; Kast, J.; et al. Forced protein unfolding leads to highly elastic and tough protein hydrogels. Nat. Commun. 2013, 4, 2974. [Google Scholar] [CrossRef] [PubMed]
- Lake, G.J.; Thomas, A.G. The strength of highly elastic materials. Proc. Math. Phys. Eng. Sci. 1967, 300, 108–119. [Google Scholar] [CrossRef]
- Lin, S.T.; Zhao, X.H. Fracture of polymer networks with diverse topological defects. Phys. Rev. E 2020, 102, 052503. [Google Scholar] [CrossRef] [PubMed]
- Slootman, J.; Waltz, V.; Yeh, C.J.; Baumann, C.; Göstl, R.; Comtet, J.; Creton, C. Quantifying Rate- and Temperature-Dependent Molecular Damage in Elastomer Fracture. Phys. Rev. X 2020, 10, 041045. [Google Scholar] [CrossRef]
- Wang, S.; Panyukov, S.; Rubinstein, M.; Craig, S.L. Quantitative Adjustment to the Molecular Energy Parameter in the Lake–Thomas Theory of Polymer Fracture Energy. Macromolecules 2019, 52, 2772–2777. [Google Scholar] [CrossRef]
- Arora, A.; Lin, T.S.; Beech, H.K.; Mochigase, H.; Wang, R.; Olsen, B.D. Fracture of Polymer Networks Containing Topological Defects. Macromolecules 2020, 53, 7346–7355. [Google Scholar] [CrossRef]
- Xie, J.B.; Qin, M.; Cao, Y.; Wang, W. Mechanistic insight of photo-induced aggregation of chicken egg white lysozyme: The interplay between hydrophobic interactions and formation of intermolecular disulfide bonds. Proteins 2011, 79, 2505–2516. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, D.; Li, L.; Fang, Y.; Ma, Q.; Cao, Y.; Lei, H. Ester Bonds for Modulation of the Mechanical Properties of Protein Hydrogels. Int. J. Mol. Sci. 2023, 24, 10778. https://doi.org/10.3390/ijms241310778
Zhang D, Li L, Fang Y, Ma Q, Cao Y, Lei H. Ester Bonds for Modulation of the Mechanical Properties of Protein Hydrogels. International Journal of Molecular Sciences. 2023; 24(13):10778. https://doi.org/10.3390/ijms241310778
Chicago/Turabian StyleZhang, Di, Luofei Li, Yizhou Fang, Quan Ma, Yi Cao, and Hai Lei. 2023. "Ester Bonds for Modulation of the Mechanical Properties of Protein Hydrogels" International Journal of Molecular Sciences 24, no. 13: 10778. https://doi.org/10.3390/ijms241310778
APA StyleZhang, D., Li, L., Fang, Y., Ma, Q., Cao, Y., & Lei, H. (2023). Ester Bonds for Modulation of the Mechanical Properties of Protein Hydrogels. International Journal of Molecular Sciences, 24(13), 10778. https://doi.org/10.3390/ijms241310778





