Inhibition of Pertussis Toxin by Human α-Defensins-1 and -5: Differential Mechanisms of Action
Abstract
1. Introduction
2. Results
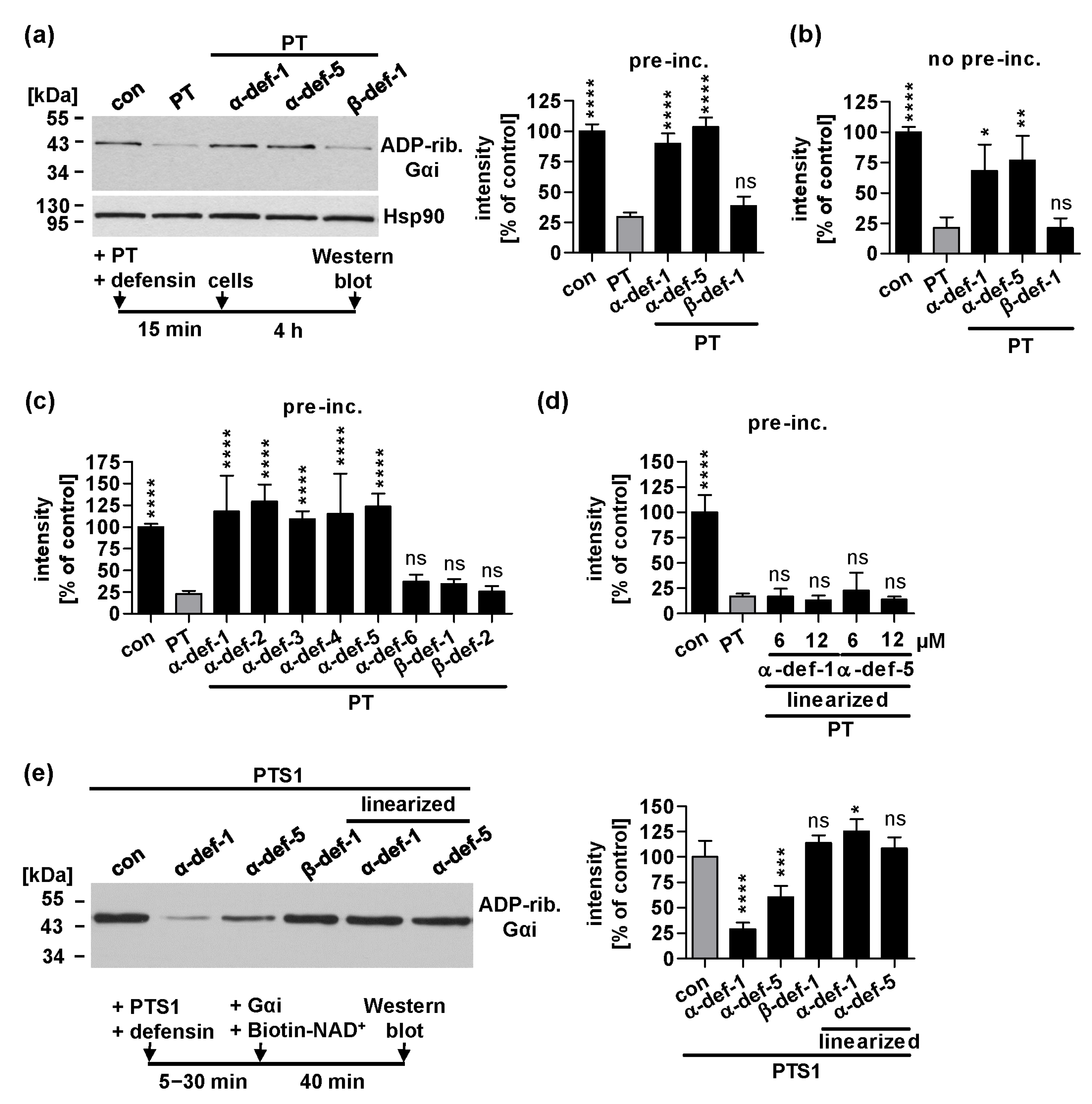
2.1. Human α-Defensin-1 and -5 Inhibit ADP-Ribosylation of Gαi in PT-Treated Cells and Enzyme Activity of PTS1 In Vitro
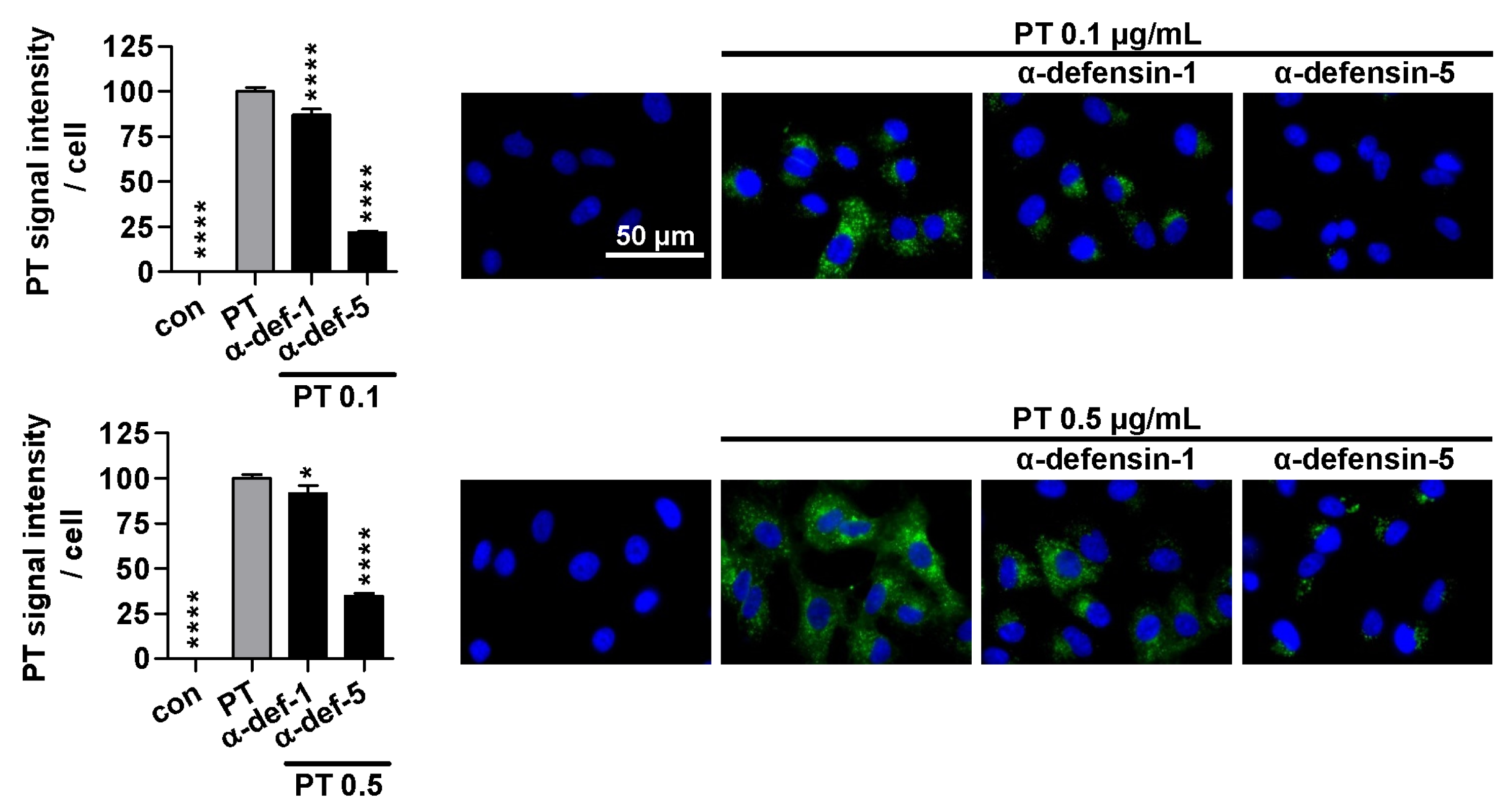
2.2. α-Defensin-5 Reduces the Amount of Cell-Bound PT Stronger Than α-Defensin-1
2.3. Treatment with α-Defensin-5 Results in a Reduction in the Amount of PT in Cells
2.4. α-Defensin-1 Is Internalized into Cells
2.5. α-Defensin-1 but Not α-Defensin-5 Is Detected in Close Proximity to PTS1 in Cells
2.6. Computational Modeling of the PTS1/α-Defensin-1 and PTS1/α-Defensin-5 Binding Interface
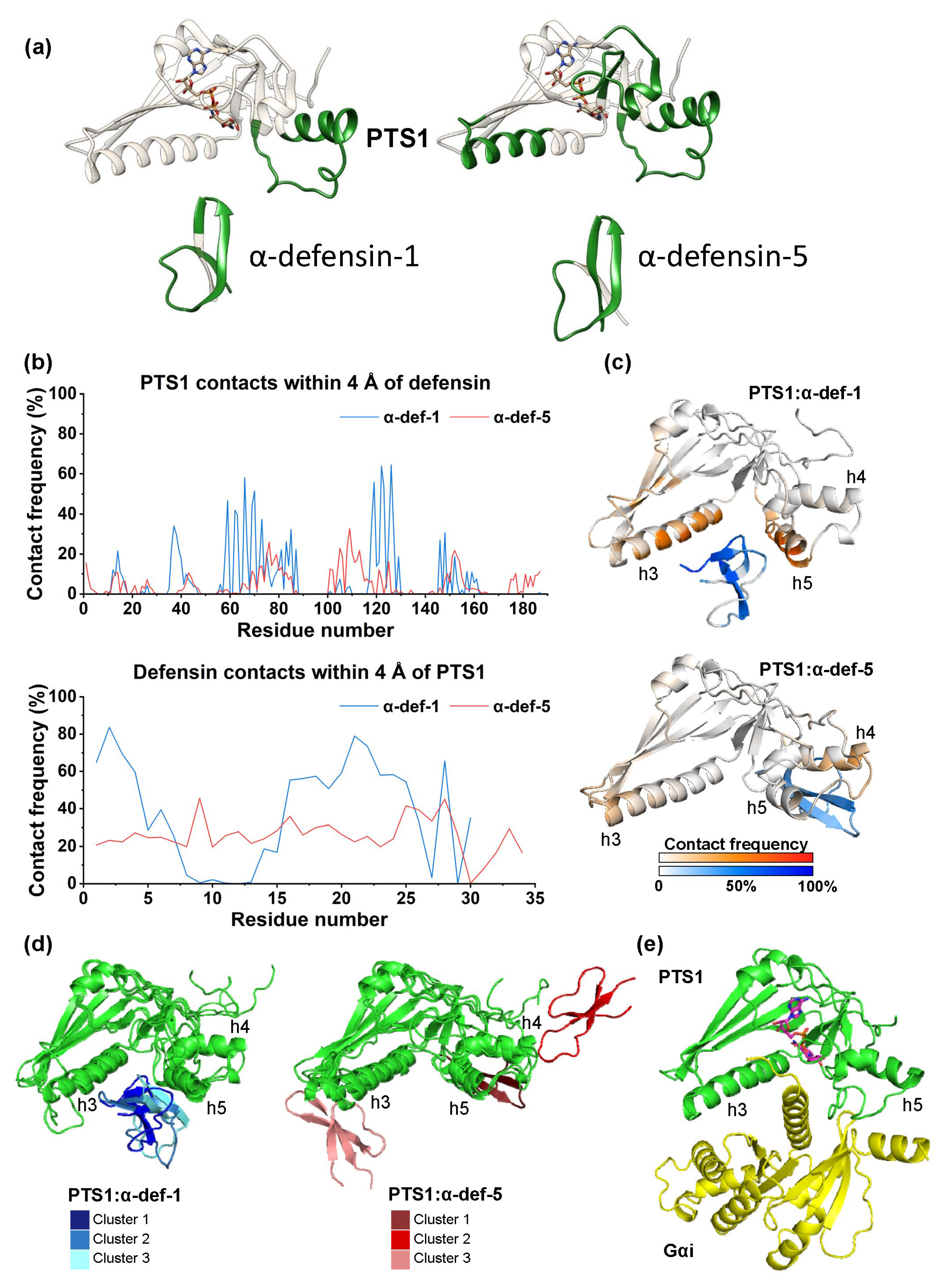
2.7. α-Defensin-1 Directly Binds to PTS1 and Gαi In Vitro
3. Discussion
4. Materials and Methods
4.1. Compounds and Reagents
4.2. Cell Lines
4.3. Sequential ADP-Ribosylation of Gαi in Lysates from Toxin-Treated Cells
4.4. In Vitro Enzyme Activity of PTS1
4.5. Flow Cytometry Binding Assay
4.6. Immunostaining and Fluorescence Microscopy
4.7. Cell Viability Assay
4.8. Protein Interaction Analysis in Cells Using Proximity Ligation Assay
4.9. In Silico Modeling of PTS1/Defensin Complexes
4.10. Protein Interaction Analysis In Vitro Using Dot Blot System
4.11. NADase Activity Assay
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Stein, P.E.; Boodhoo, A.; Armstrong, G.D.; Cockle, S.A.; Klein, M.H.; Read, R.J. The Crystal Structure of Pertussis Toxin. Structure 1994, 2, 45–57. [Google Scholar] [PubMed]
- Tamura, M.; Nogimori, K.; Murai, S.; Yajima, M.; Ito, K.; Katada, T.; Ui, M.; Ishii, S. Subunit Structure of Islet-Activating Protein, Pertussis Toxin, in Conformity with the A-B Model. Biochemistry 1982, 21, 5516–5522. [Google Scholar] [CrossRef] [PubMed]
- Weiss, A.A.; Johnson, F.D.; Burns, D.L. Molecular Characterization of an Operon Required for Pertussis Toxin Secretion. Proc. Natl. Acad. Sci. USA 1993, 90, 2970–2974. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, G.D.; Howard, L.A.; Peppler, M.S. Use of Glycosyltransferases to Restore Pertussis Toxin Receptor Activity to Asialoagalactofetuin. J. Biol. Chem. 1988, 263, 8677–8684. [Google Scholar] [PubMed]
- Witvliet, M.H.; Burns, D.L.; Brennan, M.J.; Poolman, J.T.; Manclark, C.R. Binding of Pertussis Toxin to Eucaryotic Cells and Glycoproteins. Infect. Immun. 1989, 57, 3324–3330. [Google Scholar] [CrossRef]
- Plaut, R.D.; Carbonetti, N.H. Retrograde Transport of Pertussis Toxin in the Mammalian Cell. Cell. Microbiol. 2008, 10, 1130–1139. [Google Scholar] [CrossRef]
- el Bayâ, A.; Linnemann, R.; von Olleschik-Elbheim, L.; Robenek, H.; Schmidt, M.A. Endocytosis and Retrograde Transport of Pertussis Toxin to the Golgi Complex as a Prerequisite for Cellular Intoxication. Eur. J. Cell Biol. 1997, 73, 40–48. [Google Scholar]
- Burns, D.L.; Manclark, C.R. Adenine Nucleotides Promote Dissociation of Pertussis Toxin Subunits. J. Biol. Chem. 1986, 261, 4324–4327. [Google Scholar]
- Hazes, B.; Boodhoo, A.; Cockle, S.A.; Read, R.J. Crystal Structure of the Pertussis Toxin-ATP Complex: A Molecular Sensor. J. Mol. Biol. 1996, 258, 661–671. [Google Scholar] [CrossRef]
- Plaut, R.D.; Scanlon, K.M.; Taylor, M.; Teter, K.; Carbonetti, N.H. Intracellular Disassembly and Activity of Pertussis Toxin Require Interaction with ATP. Pathog Dis 2016, 74, ftw065. [Google Scholar] [CrossRef]
- Banerjee, T.; Cilenti, L.; Taylor, M.; Showman, A.; Tatulian, S.A.; Teter, K. Thermal Unfolding of the Pertussis Toxin S1 Subunit Facilitates Toxin Translocation to the Cytosol by the Mechanism of Endoplasmic Reticulum-Associated Degradation. Infect. Immun. 2016, 84, 3388–3398. [Google Scholar] [CrossRef]
- Pande, A.H.; Moe, D.; Jamnadas, M.; Tatulian, S.A.; Teter, K. The Pertussis Toxin S1 Subunit Is a Thermally Unstable Protein Susceptible to Degradation by the 20S Proteasome. Biochemistry 2006, 45, 13734–13740. [Google Scholar] [CrossRef]
- Worthington, Z.E.V.; Carbonetti, N.H. Evading the Proteasome: Absence of Lysine Residues Contributes to Pertussis Toxin Activity by Evasion of Proteasome Degradation. Infect. Immun. 2007, 75, 2946–2953. [Google Scholar] [CrossRef]
- Ernst, K.; Eberhardt, N.; Mittler, A.-K.; Sonnabend, M.; Anastasia, A.; Freisinger, S.; Schiene-Fischer, C.; Malešević, M.; Barth, H. Pharmacological Cyclophilin Inhibitors Prevent Intoxication of Mammalian Cells with Bordetella Pertussis Toxin. Toxins 2018, 10, 181. [Google Scholar] [CrossRef]
- Ernst, K.; Mittler, A.-K.; Winkelmann, V.; Kling, C.; Eberhardt, N.; Anastasia, A.; Sonnabend, M.; Lochbaum, R.; Wirsching, J.; Sakari, M.; et al. Pharmacological Targeting of Host Chaperones Protects from Pertussis Toxin in Vitro and in Vivo. Sci. Rep. 2021, 11, 5429. [Google Scholar] [CrossRef]
- Kellner, A.; Taylor, M.; Banerjee, T.; Britt, C.B.T.; Teter, K. A Binding Motif for Hsp90 in the A Chains of ADP-Ribosylating Toxins That Move from the Endoplasmic Reticulum to the Cytosol. Cell. Microbiol. 2019, 21, e13074. [Google Scholar] [CrossRef]
- Kellner, A.; Cherubin, P.; Harper, J.K.; Teter, K. Proline Isomerization as a Key Determinant for Hsp90-Toxin Interactions. Front. Cell. Infect. Microbiol. 2021, 11, 771653. [Google Scholar] [CrossRef]
- Ernst, K. Requirement of Peptidyl-Prolyl Cis/Trans Isomerases and Chaperones for Cellular Uptake of Bacterial AB-Type Toxins. Front. Cell. Infect. Microbiol. 2022, 12, 938015. [Google Scholar] [CrossRef]
- Bokoch, G.M.; Katada, T.; Northup, J.K.; Hewlett, E.L.; Gilman, A.G. Identification of the Predominant Substrate for ADP-Ribosylation by Islet Activating Protein. J. Biol. Chem. 1983, 258, 2072–2075. [Google Scholar]
- Katada, T.; Ui, M. Direct Modification of the Membrane Adenylate Cyclase System by Islet-Activating Protein Due to ADP-Ribosylation of a Membrane Protein. Proc. Natl. Acad. Sci. USA 1982, 79, 3129–3133. [Google Scholar]
- Scanlon, K.; Skerry, C.; Carbonetti, N. Association of Pertussis Toxin with Severe Pertussis Disease. Toxins 2019, 11, 373. [Google Scholar] [CrossRef] [PubMed]
- Toyota, T.; Kakizaki, M.; Kimura, K.; Yajima, M.; Okamoto, T.; Ui, M. Islet Activating Protein (IAP) Derived from the Culture Supernatant Fluid of Bordetella Pertussis: Effect on Spontaneous Diabetic Rats. Diabetologia 1978, 14, 319–323. [Google Scholar] [CrossRef] [PubMed]
- Andreasen, C.; Carbonetti, N.H. Pertussis Toxin Inhibits Early Chemokine Production to Delay Neutrophil Recruitment in Response to Bordetella Pertussis Respiratory Tract Infection in Mice. Infect. Immun. 2008, 76, 5139–5148. [Google Scholar] [CrossRef]
- Kirimanjeswara, G.S.; Agosto, L.M.; Kennett, M.J.; Bjornstad, O.N.; Harvill, E.T. Pertussis Toxin Inhibits Neutrophil Recruitment to Delay Antibody-Mediated Clearance of Bordetella Pertussis. J. Clin. Investig. 2005, 115, 3594–3601. [Google Scholar] [CrossRef]
- Spangrude, G.J.; Sacchi, F.; Hill, H.R.; Van Epps, D.E.; Daynes, R.A. Inhibition of Lymphocyte and Neutrophil Chemotaxis by Pertussis Toxin. J. Immunol. 1985, 135, 4135–4143. [Google Scholar] [CrossRef] [PubMed]
- Warnock, R.A.; Askari, S.; Butcher, E.C.; Andrian, U.H. von Molecular Mechanisms of Lymphocyte Homing to Peripheral Lymph Nodes. J. Exp. Med. 1998, 187, 205–216. [Google Scholar] [CrossRef]
- Schenkel, A.R.; Pauza, C.D. Pertussis Toxin Treatment In Vivo Reduces Surface Expression of the Adhesion Integrin Leukocyte Function Antigen-1 (LFA-1). Cell Adhes. Commun. 1999, 7, 183–193. [Google Scholar] [CrossRef]
- Mattoo, S.; Cherry, J.D. Molecular Pathogenesis, Epidemiology, and Clinical Manifestations of Respiratory Infections Due to Bordetella Pertussis and Other Bordetella Subspecies. Clin. Microbiol. Rev. 2005, 18, 326–382. [Google Scholar] [CrossRef]
- Carbonetti, N.H. Contribution of Pertussis Toxin to the Pathogenesis of Pertussis Disease. Pathog. Dis. 2015, 73, ftv073. [Google Scholar] [CrossRef]
- Surridge, J.; Segedin, E.R.; Grant, C.C. Pertussis Requiring Intensive Care. Arch. Dis. Child 2007, 92, 970–975. [Google Scholar] [CrossRef]
- Yeung, K.H.T.; Duclos, P.; Nelson, E.A.S.; Hutubessy, R.C.W. An Update of the Global Burden of Pertussis in Children Younger than 5 Years: A Modelling Study. Lancet Infect. Dis. 2017, 17, 974–980. [Google Scholar] [CrossRef]
- WHO Immunization Coverage. Available online: https://www.who.int/news-room/fact-sheets/detail/immunization-coverage (accessed on 15 June 2023).
- Locht, C.; Antoine, R. The History of Pertussis Toxin. Toxins 2021, 13, 623. [Google Scholar] [CrossRef]
- Connelly, C.E.; Sun, Y.; Carbonetti, N.H. Pertussis Toxin Exacerbates and Prolongs Airway Inflammatory Responses during Bordetella Pertussis Infection. Infect. Immun. 2012, 80, 4317–4332. [Google Scholar] [CrossRef]
- Kling, C.; Pulliainen, A.T.; Barth, H.; Ernst, K. Human Peptides α-Defensin-1 and -5 Inhibit Pertussis Toxin. Toxins 2021, 13, 480. [Google Scholar] [CrossRef]
- Cederlund, A.; Gudmundsson, G.H.; Agerberth, B. Antimicrobial Peptides Important in Innate Immunity. FEBS J. 2011, 278, 3942–3951. [Google Scholar] [CrossRef]
- Romero-Molina, S.; Ruiz-Blanco, Y.B.; Harms, M.; Münch, J.; Sanchez-Garcia, E. PPI-Detect: A Support Vector Machine Model for Sequence-Based Prediction of Protein–Protein Interactions. J. Comput. Chem. 2019, 40, 1233–1242. [Google Scholar] [CrossRef]
- Evans, R.; O’Neill, M.; Pritzel, A.; Antropova, N.; Senior, A.; Green, T.; Žídek, A.; Bates, R.; Blackwell, S.; Yim, J.; et al. Protein Complex Prediction with AlphaFold-Multimer. bioRxiv 2021, arXiv:2021.10.04.463034. [Google Scholar]
- Sakari, M.; Tran, M.T.; Rossjohn, J.; Pulliainen, A.T.; Beddoe, T.; Littler, D.R. Crystal Structures of Pertussis Toxin with NAD+ and Analogs Provide Structural Insights into the Mechanism of Its Cytosolic ADP-Ribosylation Activity. J. Biol. Chem. 2022, 298, 101892. [Google Scholar] [CrossRef]
- Ashok, Y.; Miettinen, M.; de Oliveira, D.K.H.; Tamirat, M.Z.; Näreoja, K.; Tiwari, A.; Hottiger, M.O.; Johnson, M.S.; Lehtiö, L.; Pulliainen, A.T. Discovery of Compounds Inhibiting the ADP-Ribosyltransferase Activity of Pertussis Toxin. ACS Infect. Dis. 2020, 6, 588–602. [Google Scholar] [CrossRef]
- Ernst, K. Novel Strategies to Inhibit Pertussis Toxin. Toxins 2022, 14, 187. [Google Scholar] [CrossRef]
- Altunaiji, S.; Kukuruzovic, R.; Curtis, N.; Massie, J. Antibiotics for Whooping Cough (Pertussis). Cochrane Database Syst. Rev. 2007, 3, CD004404. [Google Scholar] [CrossRef]
- De Graaf, H.; Ibrahim, M.; Hill, A.R.; Gbesemete, D.; Vaughan, A.T.; Gorringe, A.; Preston, A.; Buisman, A.M.; Faust, S.N.; Kester, K.E.; et al. Controlled Human Infection with Bordetella Pertussis Induces Asymptomatic, Immunizing Colonization. Clin. Infect. Dis. 2020, 71, 403–411. [Google Scholar] [CrossRef] [PubMed]
- Lehrer, R.I.; Barton, A.; Daher, K.A.; Harwig, S.S.; Ganz, T.; Selsted, M.E. Interaction of Human Defensins with Escherichia Coli. Mechanism of Bactericidal Activity. J. Clin. Investig. 1989, 84, 553–561. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.; Gajendran, N.; Mittrücker, H.-W.; Weiwad, M.; Song, Y.-H.; Hurwitz, R.; Wilmanns, M.; Fischer, G.; Kaufmann, S.H.E. Human α-Defensins Neutralize Anthrax Lethal Toxin and Protect against Its Fatal Consequences. Proc. Natl. Acad. Sci. USA 2005, 102, 4830–4835. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.; Slavinskaya, Z.; Merrill, A.R.; Kaufmann, S.H.E. Human α-Defensins Neutralize Toxins of the Mono-ADP-Ribosyltransferase Family. Biochem. J. 2006, 399, 225–229. [Google Scholar] [CrossRef] [PubMed]
- Korbmacher, M.; Fischer, S.; Landenberger, M.; Papatheodorou, P.; Aktories, K.; Barth, H. Human α-Defensin-5 Efficiently Neutralizes Clostridioides Difficile Toxins TcdA, TcdB, and CDT. Front. Pharmacol. 2020, 11, 1204. [Google Scholar] [CrossRef]
- Fischer, S.; Ückert, A.-K.; Landenberger, M.; Papatheodorou, P.; Hoffmann-Richter, C.; Mittler, A.-K.; Ziener, U.; Hägele, M.; Schwan, C.; Müller, M.; et al. Human Peptide α-Defensin-1 Interferes with Clostridioides Difficile Toxins TcdA, TcdB, and CDT. FASEB J. 2020, 34, 6244–6261. [Google Scholar] [CrossRef]
- Fischer, S.; Popoff, M.R.; Barth, H. Human Alpha-Defensin-1 Protects Cells from Intoxication with Clostridium Perfringens Iota Toxin. Pathog. Dis. 2018, 76, fty022. [Google Scholar] [CrossRef]
- Kudryashova, E.; Quintyn, R.; Seveau, S.; Lu, W.; Wysocki, V.H.; Kudryashov, D.S. Human Defensins Facilitate Local Unfolding of Thermodynamically Unstable Regions of Bacterial Protein Toxins. Immunity 2014, 41, 709–721. [Google Scholar] [CrossRef]
- Castagnini, M.; Picchianti, M.; Talluri, E.; Biagini, M.; Del Vecchio, M.; Di Procolo, P.; Norais, N.; Nardi-Dei, V.; Balducci, E. Arginine-Specific Mono ADP-Ribosylation in Vitro of Antimicrobial Peptides by ADP-Ribosylating Toxins. PLoS ONE 2012, 7, e41417. [Google Scholar] [CrossRef]
- Paone, G.; Wada, A.; Stevens, L.A.; Matin, A.; Hirayama, T.; Levine, R.L.; Moss, J. ADP Ribosylation of Human Neutrophil Peptide-1 Regulates Its Biological Properties. Proc. Natl. Acad. Sci. USA 2002, 99, 8231–8235. [Google Scholar] [CrossRef]
- Lee, A.C.-L.; Harris, J.L.; Khanna, K.K.; Hong, J.-H. A Comprehensive Review on Current Advances in Peptide Drug Development and Design. Int. J. Mol. Sci. 2019, 20, E2383. [Google Scholar] [CrossRef]
- Bosso, M.; Ständker, L.; Kirchhoff, F.; Münch, J. Exploiting the Human Peptidome for Novel Antimicrobial and Anticancer Agents. Bioorg. Med. Chem. 2018, 26, 2719–2726. [Google Scholar] [CrossRef]
- Hazrati, E.; Galen, B.; Lu, W.; Wang, W.; Ouyang, Y.; Keller, M.J.; Lehrer, R.I.; Herold, B.C. Human α- and β-Defensins Block Multiple Steps in Herpes Simplex Virus Infection1. J. Immunol. 2006, 177, 8658–8666. [Google Scholar] [CrossRef]
- Arnett, E.; Lehrer, R.I.; Pratikhya, P.; Lu, W.; Seveau, S. Defensins Enable Macrophages to Inhibit the Intracellular Proliferation of Listeria Monocytogenes. Cell. Microbiol. 2011, 13, 635–651. [Google Scholar] [CrossRef]
- Romero-Molina, S.; Ruiz-Blanco, Y.B.; Green, J.R.; Sanchez-Garcia, E. ProtDCal-Suite: A Web Server for the Numerical Codification and Functional Analysis of Proteins. Protein Sci. 2019, 28, 1734–1743. [Google Scholar] [CrossRef]
- Wei, G.; de Leeuw, E.; Pazgier, M.; Yuan, W.; Zou, G.; Wang, J.; Ericksen, B.; Lu, W.-Y.; Lehrer, R.I.; Lu, W. Through the Looking Glass, Mechanistic Insights from Enantiomeric Human Defensins. J. Biol. Chem. 2009, 284, 29180–29192. [Google Scholar] [CrossRef]
- Szyk, A.; Wu, Z.; Tucker, K.; Yang, D.; Lu, W.; Lubkowski, J. Crystal Structures of Human α-Defensins HNP4, HD5, and HD6. Protein Sci. 2006, 15, 2749–2760. [Google Scholar] [CrossRef]
- MacKerell, A.D., Jr.; Bashford, D.; Bellott, M.; Dunbrack, R.L., Jr.; Evanseck, J.D.; Field, M.J.; Fischer, S.; Gao, J.; Guo, H.; Ha, S.; et al. All-Atom Empirical Potential for Molecular Modeling and Dynamics Studies of Proteins. J. Phys. Chem. B 1998, 102, 3586–3616. [Google Scholar] [CrossRef]
- Huang, J.; MacKerell Jr, A.D. CHARMM36 All-Atom Additive Protein Force Field: Validation Based on Comparison to NMR Data. J. Comput. Chem. 2013, 34, 2135–2145. [Google Scholar] [CrossRef]
- Haug, E.J.; Arora, J.S.; Matsui, K. A Steepest-Descent Method for Optimization of Mechanical Systems. J. Optim. Theory Appl. 1976, 19, 401–424. [Google Scholar] [CrossRef]
- Verlet, L. Computer “Experiments” on Classical Fluids. I. Thermodynamical Properties of Lennard-Jones Molecules. Phys. Rev. 1967, 159, 98–103. [Google Scholar] [CrossRef]
- Bussi, G.; Donadio, D.; Parrinello, M. Canonical Sampling through Velocity Rescaling. J. Chem. Phys. 2007, 126, 014101. [Google Scholar] [CrossRef] [PubMed]
- Bernetti, M.; Bussi, G. Pressure Control Using Stochastic Cell Rescaling. J. Chem. Phys. 2020, 153, 114107. [Google Scholar] [CrossRef]
- Hess, B.; Bekker, H.; Berendsen, H.J.C.; Fraaije, J.G.E.M. LINCS: A Linear Constraint Solver for Molecular Simulations. J. Comput. Chem. 1997, 18, 1463–1472. [Google Scholar] [CrossRef]
- Darden, T.; York, D.; Pedersen, L. Particle Mesh Ewald: An N⋅log(N) Method for Ewald Sums in Large Systems. J. Chem. Phys. 1993, 98, 10089–10092. [Google Scholar] [CrossRef]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High Performance Molecular Simulations through Multi-Level Parallelism from Laptops to Supercomputers. SoftwareX 2015, 1–2, 19–25. [Google Scholar] [CrossRef]
- Daura, X.; Gademann, K.; Jaun, B.; Seebach, D.; van Gunsteren, W.F.; Mark, A.E. Peptide Folding: When Simulation Meets Experiment. Angew. Chem. Int. Ed. 1999, 38, 236–240. [Google Scholar] [CrossRef]
- Vicente, F.E.M.; González-Garcia, M.; Diaz Pico, E.; Moreno-Castillo, E.; Garay, H.E.; Rosi, P.E.; Jimenez, A.M.; Campos-Delgado, J.A.; Rivera, D.G.; Chinea, G.; et al. Design of a Helical-Stabilized, Cyclic, and Nontoxic Analogue of the Peptide Cm-P5 with Improved Antifungal Activity. ACS Omega 2019, 4, 19081–19095. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kling, C.; Sommer, A.; Almeida-Hernandez, Y.; Rodríguez, A.; Perez-Erviti, J.A.; Bhadane, R.; Ständker, L.; Wiese, S.; Barth, H.; Pupo-Meriño, M.; et al. Inhibition of Pertussis Toxin by Human α-Defensins-1 and -5: Differential Mechanisms of Action. Int. J. Mol. Sci. 2023, 24, 10557. https://doi.org/10.3390/ijms241310557
Kling C, Sommer A, Almeida-Hernandez Y, Rodríguez A, Perez-Erviti JA, Bhadane R, Ständker L, Wiese S, Barth H, Pupo-Meriño M, et al. Inhibition of Pertussis Toxin by Human α-Defensins-1 and -5: Differential Mechanisms of Action. International Journal of Molecular Sciences. 2023; 24(13):10557. https://doi.org/10.3390/ijms241310557
Chicago/Turabian StyleKling, Carolin, Anja Sommer, Yasser Almeida-Hernandez, Armando Rodríguez, Julio A. Perez-Erviti, Rajendra Bhadane, Ludger Ständker, Sebastian Wiese, Holger Barth, Mario Pupo-Meriño, and et al. 2023. "Inhibition of Pertussis Toxin by Human α-Defensins-1 and -5: Differential Mechanisms of Action" International Journal of Molecular Sciences 24, no. 13: 10557. https://doi.org/10.3390/ijms241310557
APA StyleKling, C., Sommer, A., Almeida-Hernandez, Y., Rodríguez, A., Perez-Erviti, J. A., Bhadane, R., Ständker, L., Wiese, S., Barth, H., Pupo-Meriño, M., Pulliainen, A. T., Sánchez-García, E., & Ernst, K. (2023). Inhibition of Pertussis Toxin by Human α-Defensins-1 and -5: Differential Mechanisms of Action. International Journal of Molecular Sciences, 24(13), 10557. https://doi.org/10.3390/ijms241310557









