Genome-Wide Identification of Detoxification Genes in Wild Silkworm Antheraea pernyi and Transcriptional Response to Coumaphos
Abstract
1. Introduction
2. Results
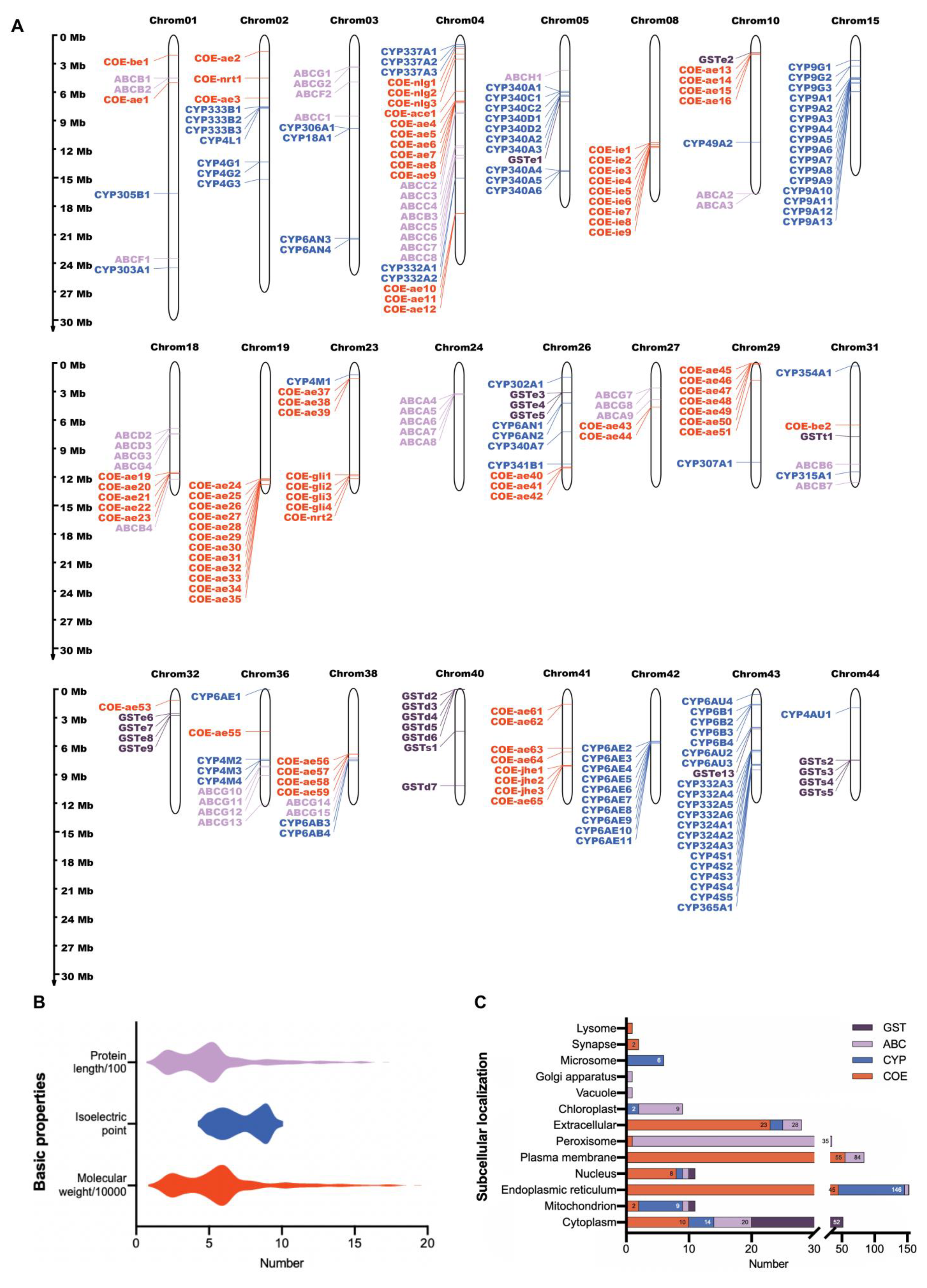
2.1. Genome-Wide Identification of Detoxification Genes in A. pernyi
2.2. Glutathione S-Transferase (GST) Genes
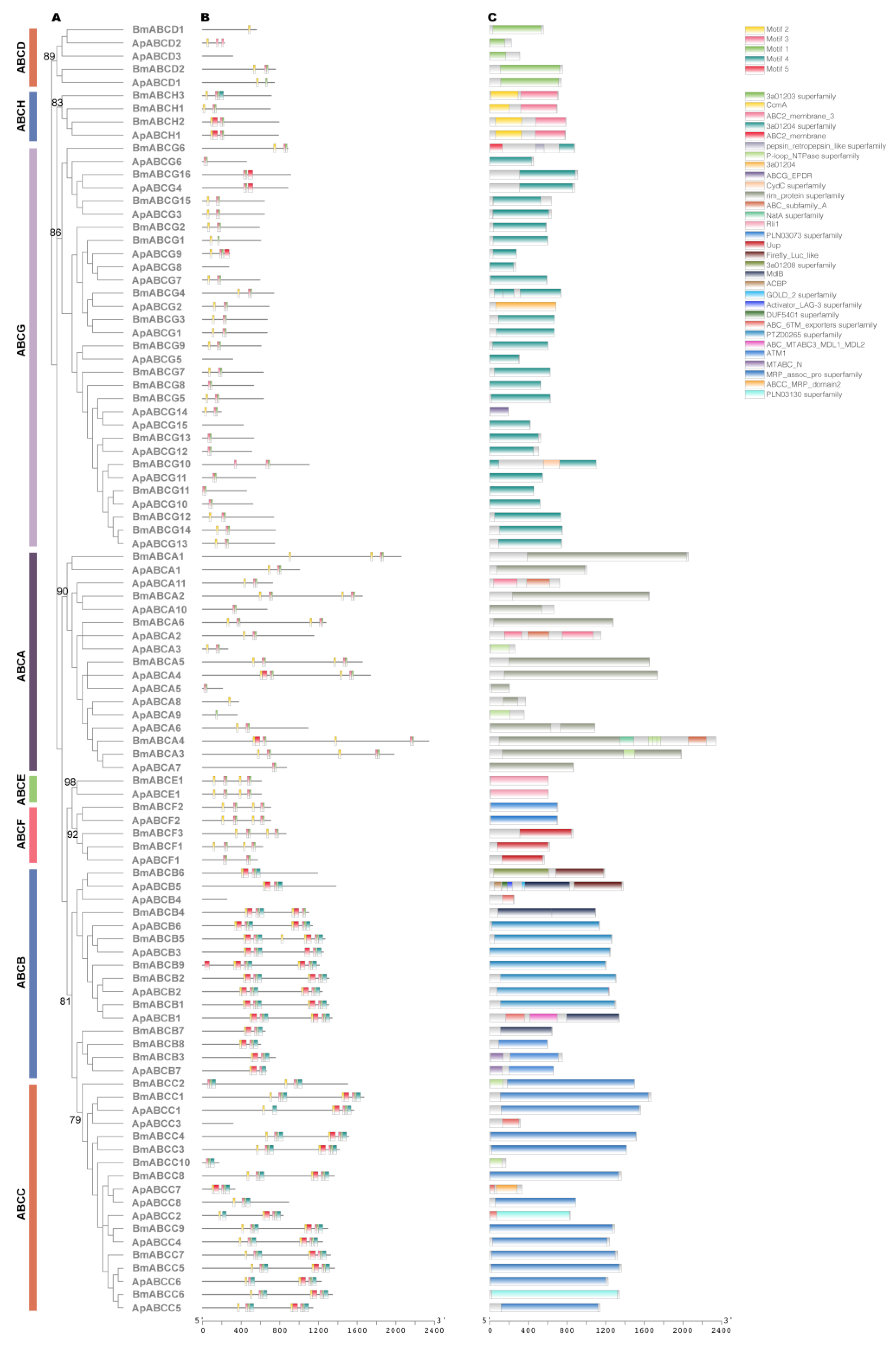
2.3. ABC Transporter Genes
2.4. Cytochrome P450 (CYP) Genes
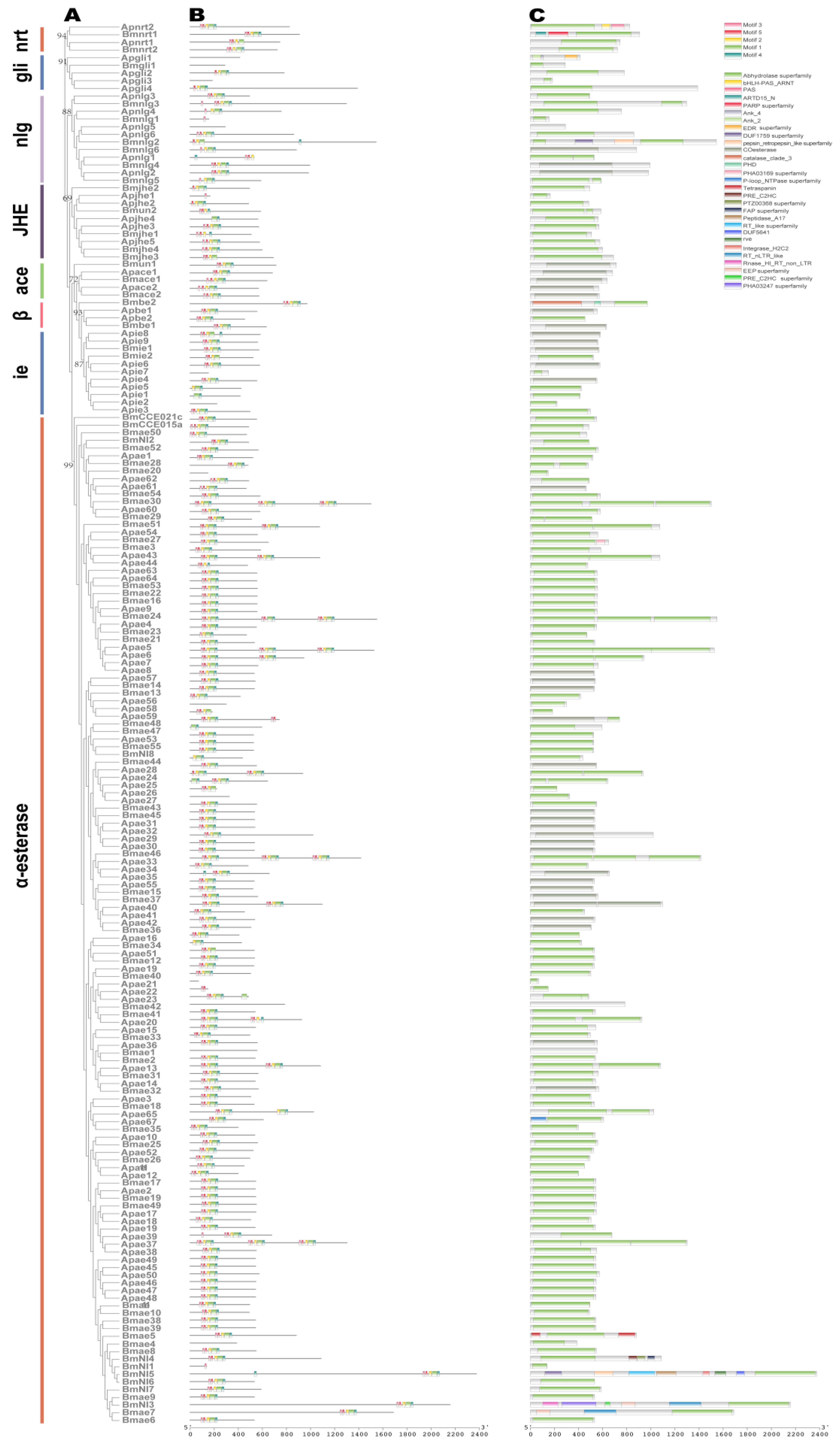
2.5. Carboxylesterase (COE) Genes
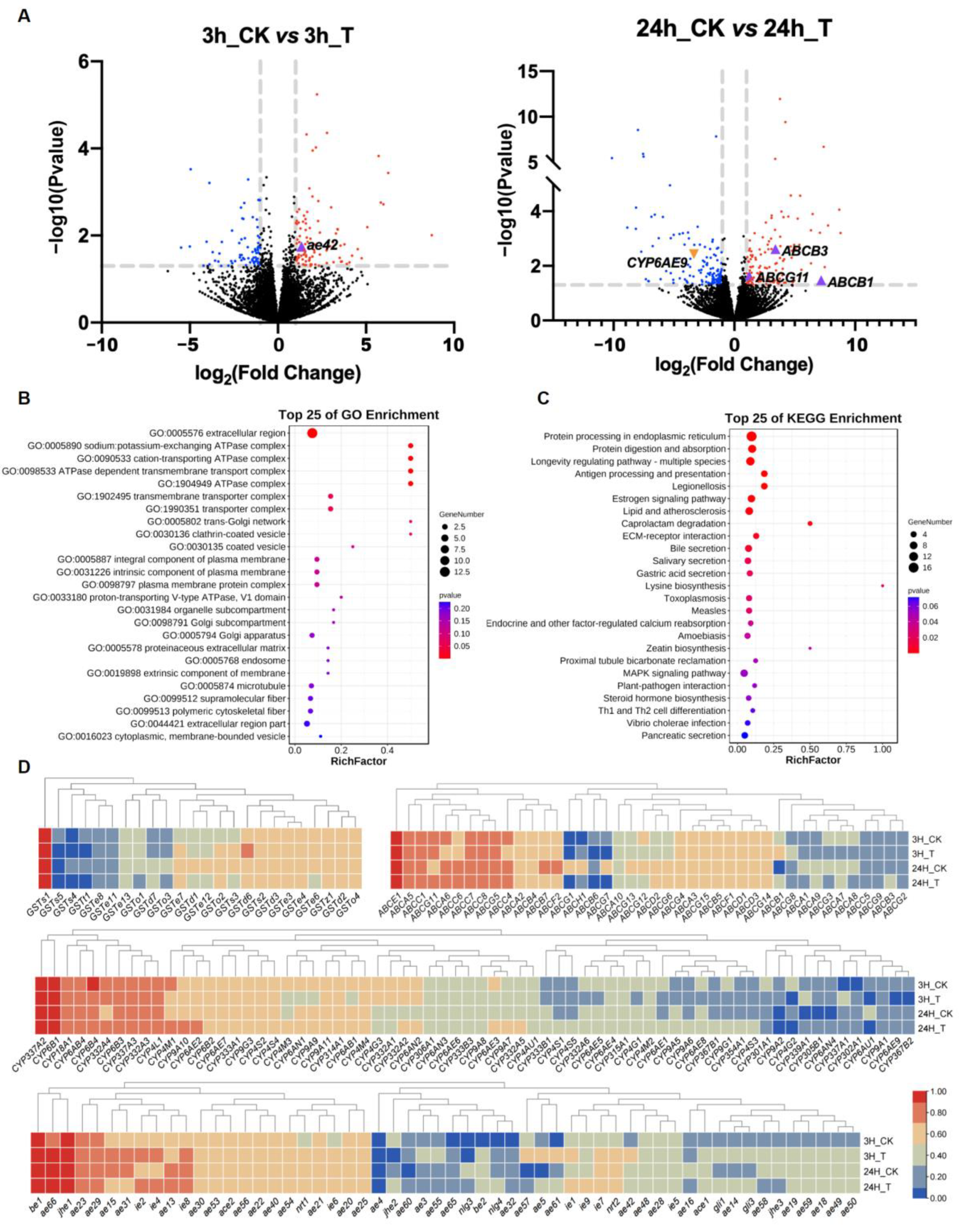
2.6. Expression Patterns of Detoxification Genes Associated with Coumaphos Stress
3. Discussion
4. Materials and Methods
4.1. Identification of A. pernyi Detoxification Genes
4.2. Phylogenetic Reconstruction
4.3. Analysis of the Conserved Domains and Motifs
4.4. RNA-Seq Sample Collection and Illumina Sequencing
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Liu, Y.Q.; Li, Y.P.; Li, X.S.; Qin, L. The origin and dispersal of the domesticated Chinese oak silkworm, Antheraea pernyi, in China: A reconstruction based on ancient texts. J. Insect Sci. 2010, 10, 180. [Google Scholar] [CrossRef] [PubMed]
- Li, W.L.; Zhang, Z.Y.; Lin, L.; Terenius, O. Antheraea pernyi (Lepidoptera: Saturniidae) and its importance in sericulture, food consumption, and traditional Chinese medicine. J. Econ. Entomol. 2017, 110, 1404–1411. [Google Scholar] [CrossRef] [PubMed]
- Li, X.Y.; Liu, Y.C.; Zhang, R.S.; Chen, D.B.; Chen, M.M.; Li, Y.P.; Liu, Y.Q.; Qin, L. The mitochondrial genome of Qinghuang_1, the first modern improved strain of Chinese oak silkworm, Antheraea pernyi (Lepidoptera: Saturniidae). J. Insects Food Feed 2021, 7, 233–243. [Google Scholar] [CrossRef]
- Liu, Q.N.; Zhu, B.J.; Wang, L.; Wei, G.Q.; Dai, L.S.; Lin, K.Z.; Sun, Y.; Qiu, J.F.; Fu, W.W.; Liu, C.L. Identification of immune response-related genes in the Chinese oak silkworm, Antheraea pernyi by suppression subtractive hybridization. J. Invertebr. Pathol. 2013, 114, 313–323. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Chen, M.M.; Yao, R.; Li, Y.; Wang, H.; Li, Y.P.; Liu, Y.Q. Molecular cloning and characterization of two 12 kDa FK506-binding protein genes in the Chinese oak silkworm, Antheraea pernyi. J. Agric. Food Chem. 2013, 61, 4599–4605. [Google Scholar] [CrossRef]
- Xin, Z.Z.; Liu, Q.N.; Liu, Y.; Zhang, D.Z.; Wang, Z.F.; Zhang, H.B.; Ge, B.M.; Zhou, C.L.; Chai, X.Y.; Tang, B.P. Transcriptome-wide identification of differentially expressed genes in Chinese oak silkworm Antheraea pernyi in response to lead challenge. J. Agric. Food Chem. 2017, 65, 9305–9314. [Google Scholar] [CrossRef]
- Jiang, Y.R.; Qin, L. Prevention and control of the parasitic fly, Blepharipa tibialis Chao. Modern Agric. 2012, 6, 53. (In Chinese) [Google Scholar]
- Qin, L.; Li, S.Y. Sericulture of Chinese Oak Silkworm; China Agricultural Press: Beijing, China, 2017. [Google Scholar]
- Li, H.D.; Jia, P.; Shi, S.L.; Dong, X.G.; Zhao, S.W.; Li, S.Y.; Li, X.S.; Chen, Z.L. Influences of coumaphos on the activity of several physiological metabolic enzymes in Antheraea pernyi. Sci. Seric. 2015, 41, 486–490. (In Chinese) [Google Scholar]
- Cheng, T.C.; Wu, J.Q.; Wu, Y.Q.; Chilukuri, R.V.; Huang, L.H.; Yamamoto, K.; Feng, L.; Li, W.S.; Chen, Z.W.; Guo, H.Z.; et al. Genomic adaptation to polyphagy and insecticides in a major East Asian noctuid pest. Nat. Ecol. Evol. 2017, 1, 1747–1756. [Google Scholar] [CrossRef]
- Pearce, S.L.; Clarke, D.F.; East, P.D.; Elfekih, S.; Gordon, K.H.J.; Jermiin, L.S.; McGaughran, A.; Oakeshott, J.G.; Papanicolaou, A.; Perera, O.P.; et al. Genomic innovations, transcriptional plasticity and gene loss underlying the evolution and divergence of two highly polyphagous and invasive Helicoverpa pest species. BMC Biol. 2017, 15, 63. [Google Scholar] [CrossRef]
- Gouin, A.; Bretaudeau, A.; Nam, K.; Gimenez, S.; Aury, J.M.; Duvic, B.; Hilliou, F.; Durand, N.; Montagné, N.; Darboux, I.; et al. Two genomes of highly polyphagous lepidopteran pests (Spodoptera frugiperda, Noctuidae) with different host-plant ranges. Sci. Rep. 2017, 7, 11816. [Google Scholar] [CrossRef] [PubMed]
- You, M.S.; Yue, Z.; He, W.Y.; Yang, X.H.; Yang, G.; Xie, M.; Zhan, D.L.; Baxter, S.W.; Vasseur, L.; Gurr, G.M.; et al. A heterozygous moth genome provides insights into herbivory and detoxification. Nat. Genet. 2013, 45, 220–225. [Google Scholar] [CrossRef] [PubMed]
- Kawamoto, M.; Jouraku, A.; Toyoda, A.; Yokoi, K.; Minakuchi, Y.; Katsuma, S.; Fujiyama, A.; Kiuchi, T.; Yamamoto, K.; Shimada, T. High-quality genome assembly of the silkworm, Bombyx mori. Insect Biochem. Mol. Biol. 2019, 107, 53–62. [Google Scholar] [CrossRef] [PubMed]
- Yu, Q.Y.; Lu, C.; Li, W.L.; Xiang, Z.H.; Zhang, Z. Annotation and expression of carboxylesterases in the silkworm, Bombyx mori. BMC Genomics 2009, 10, 553. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.M.; Zhou, S.; Tian, L.; Guo, E.E.; Luan, Y.X.; Zhang, J.Z.; Li, S. Genome-wide identification and characterization of ATP-binding cassette transporters in the silkworm, Bombyx mori. BMC Genomics 2011, 12, 491. [Google Scholar] [CrossRef]
- Xie, X.D.; Cheng, T.C.; Wang, G.H.; Duan, J.; Niu, W.H.; Xia, Q.Y. Genome-wide analysis of the ATP-binding cassette (ABC) transporter gene family in the silkworm, Bombyx mori. Mol. Biol. Rep. 2012, 39, 7281–7291. [Google Scholar] [CrossRef]
- Yu, Q.Y.; Lu, C.; Li, B.; Fang, S.M.; Zuo, W.D.; Dai, F.Y.; Zhang, Z.; Xiang, Z.H. Identification, genomic organization and expression pattern of glutathione S-transferase in the silkworm, Bombyx mori. Insect Biochem. Mol. Biol. 2008, 38, 1158–1164. [Google Scholar] [CrossRef]
- Duan, J.P.; Li, Y.; Du, J.; Duan, E.Z.; Lei, Y.Y.; Liang, S.M.; Zhang, X.; Zhao, X.; Kan, Y.C.; Yao, L.G.; et al. A chromosome-scale genome assembly of Antheraea pernyi (Saturniidae, Lepidoptera). Mol. Ecol. Resour. 2020, 20, 1372–1383. [Google Scholar] [CrossRef]
- Mirth, C.K.; Riddiford, L.M. Size assessment and growth control: How adult size is determined in insects. Bioessays 2007, 29, 344–355. [Google Scholar] [CrossRef]
- Peng, J.; Li, Z.; Yang, Y.; Wang, P.; Zhou, X.; Zhao, T.J.; Guo, M.P.; Meng, M.; Zhang, T.L.; Qian, W.L.; et al. Comparative transcriptome analysis provides novel insight into morphologic and metabolic changes in the fat body during silkworm metamorphosis. Int. J. Mol. Sci. 2018, 19, 3523. [Google Scholar] [CrossRef]
- Enayati, A.A.; Ranson, H.; Hemingway, J. Insect glutathione transferases and insecticide resistance. Insect Mol. Biol. 2005, 14, 3–8. [Google Scholar] [CrossRef]
- Sheehan, D.; Meade, G.; Foley, V.M.; Dowd, C.A. Structure, function and evolution of glutathione transferases: Implications for classification of non-mammalian members of an ancient enzyme superfamily. Biochem. J. 2001, 360, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Schneider, E.; Hunke, S. ATP-binding-cassette (ABC) transport systems: Functional and structural aspects of the ATP-hydrolyzing subunits/domains. FEMS Microbiol. Rev. 1998, 22, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Nauen, R.; Bass, C.; Feyereisen, R.; Vontas, J. The role of cytochrome P450s in insect toxicology and resistance. Annu. Rev. Entomol. 2022, 67, 105–124. [Google Scholar] [CrossRef] [PubMed]
- Dermauw, W.; Van, L.T.; Feyereisen, R. Diversity and evolution of the P450 family in arthropods. Insect Biochem. Mol. Biol. 2020, 127, 103490. [Google Scholar] [CrossRef] [PubMed]
- Nelson, D.R. Cytochrome P450 diversity in the tree of life. Biochim. Biophys. Acta Proteins Proteom. 2018, 1866, 141–154. [Google Scholar] [CrossRef]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef]
- Satoh, T.; Hosokawa, M. The mammalian carboxylesterases: From molecules to functions. Annu. Rev. Pharmacol. Toxicol. 1998, 38, 257–288. [Google Scholar] [CrossRef]
- Bornscheuer, U.T. Microbial carboxyl esterases: Classification, properties and application in biocatalysis. FEMS Microbiol. Rev. 2002, 26, 73–81. [Google Scholar] [CrossRef]
- Marshall, S.D.G.; Putterill, J.J.; Plummer, K.M.; Newcomb, R.D. The carboxylesterase gene family from Arabidopsis thaliana. J. Mol. Evol. 2003, 57, 487–500. [Google Scholar]
- Ranson, H.; Claudianos, C.; Ortelli, F.; Abgrall, C.; Hemingway, J.; Sharakhova, M.V.; Unger, M.F.; Collins, F.H.; Feyereisen, R. Evolution of supergene families associated with insecticide resistance. Science 2002, 298, 179–181. [Google Scholar] [CrossRef]
- Sun, H.; Pu, J.; Chen, F.; Wang, J.; Han, Z. Multiple ATP-binding cassette transporters are involved in insecticide resistance in the small brown planthopper, Laodelphax striatellus. Insect Mol. Biol. 2017, 26, 343–355. [Google Scholar] [CrossRef] [PubMed]
- Li, J.Y.; Lv, Y.T.; Yan, K.P.; Yang, F.T.; Chen, X.W.; Gao, X.W.; Wen, S.Y.; Xu, H.F.; Pan, Y.O.; Shang, Q.L. Functional analysis of cyantraniliprole tolerance ability mediated by ATP-binding cassette transporters in Aphis gossypii glover. Pestic. Biochem. Physiol. 2022, 184, 105104. [Google Scholar] [CrossRef]
- Han, H.; Yang, Y.Y.; Hu, J.; Wang, Y.X.; Zhao, Z.G.; Ma, R.Y.; Gao, L.L.; Guo, Y.Q. Identification and characterization of CYP6 family genes from the oriental fruit moth (Grapholita molesta) and their responses to insecticides. Insects 2022, 13, 300. [Google Scholar] [CrossRef] [PubMed]
- Kostaropoulos, I.; Papadopoulos, A.I.; Metaxakis, A.; Boukouvala, E.; Papadopoulou-Mourkidou, E. The role of glutathione S-transferases in the detoxification of some organophosphorus insecticides in larvae and pupae of the yellow mealworm, Tenebrio molitor (Coleoptera: Tenebrionidae). Pest Manag. Sci. 2001, 57, 501–508. [Google Scholar] [CrossRef]
- Zhou, L.; Fang, S.M.; Huang, K.; Yu, Q.Y.; Zhang, Z. Characterization of an epsilon-class glutathione S-transferase involved in tolerance in the silkworm larvae after long term exposure to insecticides. Ecotoxicol. Environ. Saf. 2015, 120, 20–26. [Google Scholar] [CrossRef]
- Hu, C.; Liu, J.Y.; Wang, W.; Mota-Sanchez, D.; He, S.; Shi, Y.; Yang, X.Q. Glutathione S-transferase genes are involved in lambda-cyhalothrin resistance in Cydia pomonella via sequestration. J. Agric. Food Chem. 2022, 70, 2265–2279. [Google Scholar] [CrossRef] [PubMed]
- Ranson, H.; Rossiter, L.; Ortelli, F.; Jensen, B.; Wang, X.; Roth, C.W.; Collins, F.H.; Hemingway, J. Identification of a novel class of insect glutathione S-transferases involved in resistance to DDT in the malaria vector Anopheles gambiae. Biochem. J. 2002, 359, 295–304. [Google Scholar] [CrossRef]
- Mao, W.F.; Schuler, M.A.; Berenbaum, M.R. Honey constituents up-regulate detoxification and immunity genes in the western honey bee Apis mellifera. Proc. Natl. Acad. Sci. USA 2013, 110, 8842–8846. [Google Scholar] [CrossRef]
- Siebert, S.C.; Kanga, L.H.B.; Basha, S.M.; Legaspi, J.C. Molecular assessment of genes linked to immune response traits of honey bees in conventional and organically managed apiaries. Insects 2020, 11, 637. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinformatics 2019, 10, 421. [Google Scholar] [CrossRef] [PubMed]
- Mistry, J.; Finn, R.D.; Eddy, S.R.; Bateman, A.; Punta, M. Challenges in homology search: HMMER3 and convergent evolution of coiled-coil regions. Nucleic Acids Res. 2013, 41, e121. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Bateman, A.; Clements, J.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Heger, A.; Hetherington, K.; Holm, L.; Mistry, J.; et al. Pfam: The protein families database. Nucleic Acids Res. 2014, 42, D222–D230. [Google Scholar] [CrossRef] [PubMed]
- Wilkins, M.R.; Gasteiger, E.; Bairoch, A.; Sanchez, J.Z.; Williams, K.L.; Appel, R.D.; Hochstrasser, D.F. Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. 1999, 112, 531–552. [Google Scholar] [PubMed]
- Madeira, F.; Park, Y.M.; Lee, J.; Buso, N.; Gur, T.; Madhusoodanan, N.; Basutkar, P.; Tivey, A.R.N.; Potter, S.C.; Finn, R.D.; et al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019, 47, W636–W641. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Yang, M.Z.; Derbyshire, M.K.; Yamashita, R.A.; Marchler-Bauer, A. NCBI’s conserved domain database and tools for protein domain analysis. Curr. Protoc. Bioinformatics 2020, 69, e90. [Google Scholar] [CrossRef]
- Chen, C.J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.H.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq-a Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2022; Available online: https://www.R-project.org/ (accessed on 26 February 2023).


| Species | GSTs | ABCs | CYPs | COEs | Total | References |
|---|---|---|---|---|---|---|
| Antheraea pernyi | 32 | 48 | 104 | 97 | 281 | This study |
| Bombyx mori | 23 | 52 | 83 | 87 | 245 | [14] |
| Spodoptera litura | 47 | 54 | 138 | 110 | 349 | [10] |
| Spodoptera frugiperda (corn/rice strain) | 46/45 | NA/NA | 117/135 | 93/90 | 256/270 | [12] |
| Helicoverpa armigera | 42 | 54 | 114 | 97 | 307 | [11] |
| Helicoverpa zea | 40 | 54 | 108 | 93 | 295 | [11] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, D.-B.; Xia, R.-X.; Li, Q.; Li, Y.-P.; Cao, H.-Y.; Liu, Y.-Q. Genome-Wide Identification of Detoxification Genes in Wild Silkworm Antheraea pernyi and Transcriptional Response to Coumaphos. Int. J. Mol. Sci. 2023, 24, 9775. https://doi.org/10.3390/ijms24119775
Chen D-B, Xia R-X, Li Q, Li Y-P, Cao H-Y, Liu Y-Q. Genome-Wide Identification of Detoxification Genes in Wild Silkworm Antheraea pernyi and Transcriptional Response to Coumaphos. International Journal of Molecular Sciences. 2023; 24(11):9775. https://doi.org/10.3390/ijms24119775
Chicago/Turabian StyleChen, Dong-Bin, Run-Xi Xia, Qun Li, Yu-Ping Li, Hui-Ying Cao, and Yan-Qun Liu. 2023. "Genome-Wide Identification of Detoxification Genes in Wild Silkworm Antheraea pernyi and Transcriptional Response to Coumaphos" International Journal of Molecular Sciences 24, no. 11: 9775. https://doi.org/10.3390/ijms24119775
APA StyleChen, D.-B., Xia, R.-X., Li, Q., Li, Y.-P., Cao, H.-Y., & Liu, Y.-Q. (2023). Genome-Wide Identification of Detoxification Genes in Wild Silkworm Antheraea pernyi and Transcriptional Response to Coumaphos. International Journal of Molecular Sciences, 24(11), 9775. https://doi.org/10.3390/ijms24119775






