Deficiency of Fam20b-Catalyzed Glycosaminoglycan Chain Synthesis in Neural Crest Leads to Cleft Palate
Abstract
1. Introduction
2. Results
2.1. The Reduced GAG Content in the Palatal Mesenchyme of Wnt1-Cre; Fam20bf/f Mice
2.2. Wnt1-Cre; Fam20bf/f Palatal Shelves Were Capable of Elevating
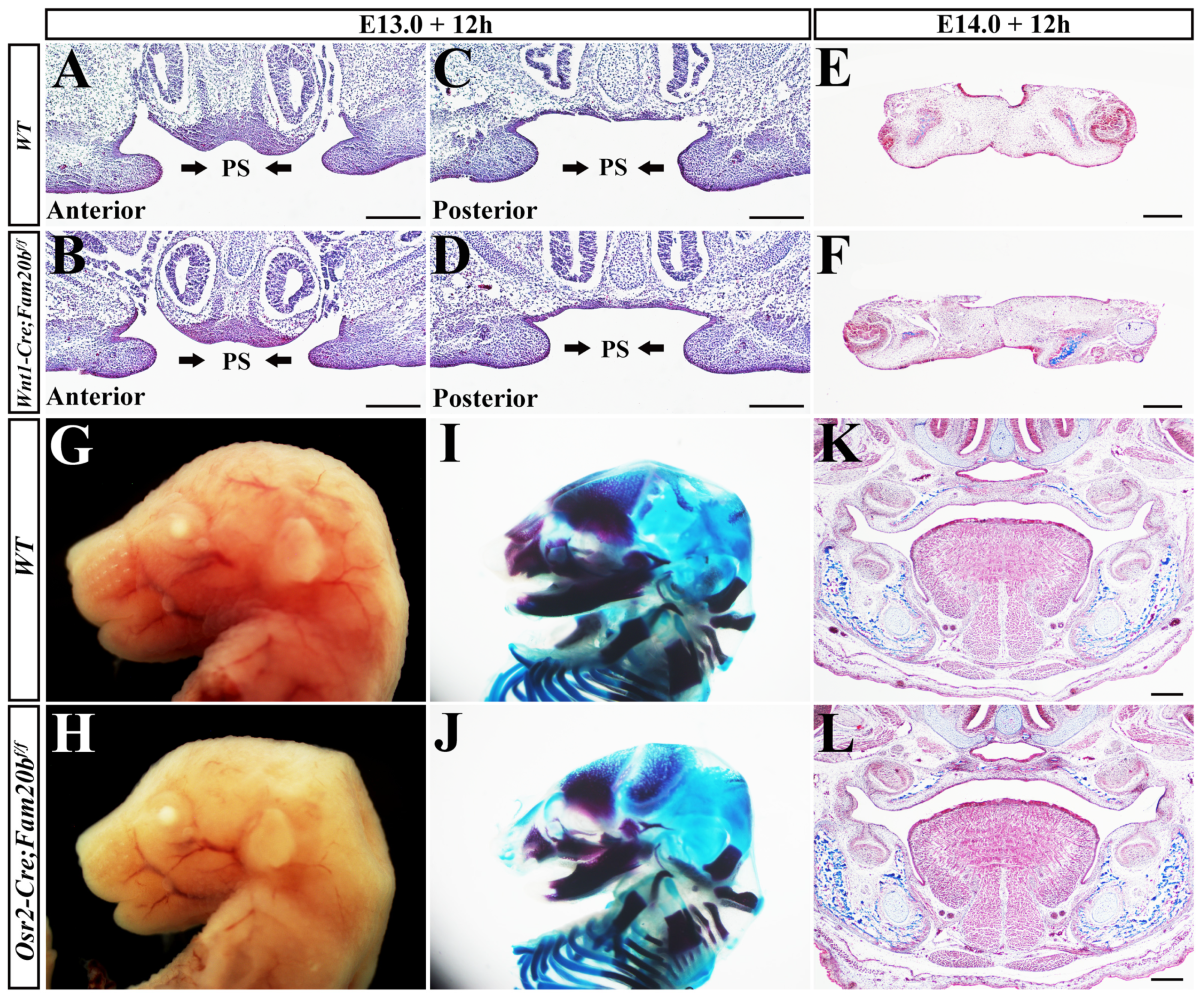
2.3. Wnt1-Cre; Fam20bf/f Mice Exhibited Human Pierre-Robin-Sequence-Like Phenotype
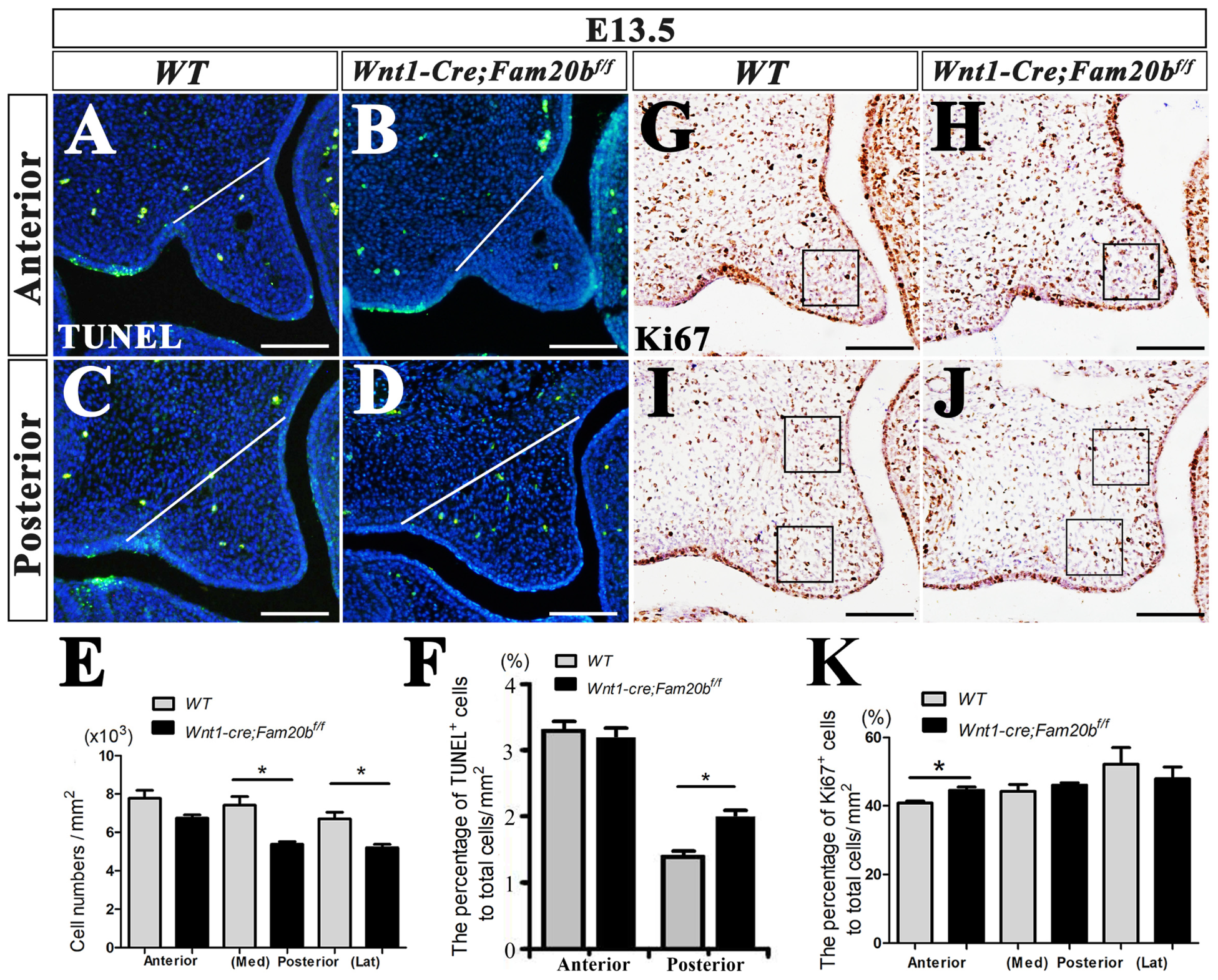
2.4. Reduced Cell Density in Wnt1-Cre; Fam20bf/f Palatal Mesenchyme
2.5. The Altered Gene Expression in Wnt1-Cre; Fam20bf/f Palatal Mesenchyme
2.6. The Palatine Osteogenesis in Wnt1-Cre; Fam20bf/f Mice Was Impaired
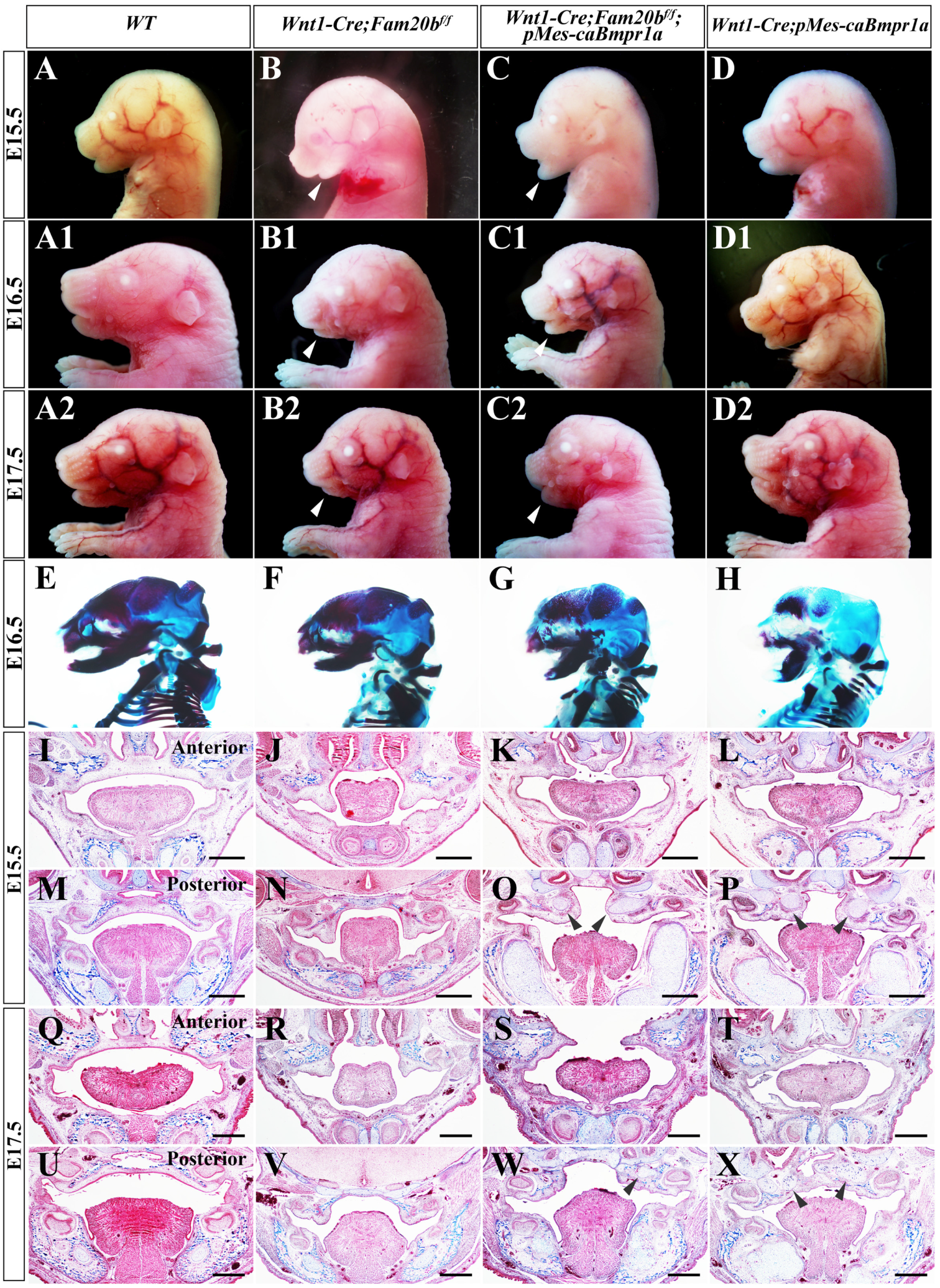
2.7. Enhanced BMP Activity in Wnt1-Cre; Fam20bf/f Mice Partially Rescued Micrognathia, Instead of Cleft Palate
3. Discussion
3.1. The Deficient Fam20b-Catalyzed Synthesis of GAG Chains Impaired Mouse Craniofacial Development
3.2. The Primary and Secondary Effects of GAG Chains in Palatal Mesenchyme on Palatogenesis
3.3. GAG Chains of Proteoglycans Were Involved in CNCC Osteogenesis by Mediating BMP Signaling
4. Materials and Methods
4.1. Mouse Lines
4.2. Alcian Blue Staining
4.3. Skeleton and Cartilage Staining
4.4. Organ Culture
4.5. Cell Proliferation and Apoptosis Analysis
4.6. In Situ Hybridization
4.7. Immunohistochemistry
4.8. Statistical Assay
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Dixon, M.J.; Marazita, M.L.; Beaty, T.H.; Murray, J.C. Cleft lip and palate: Synthesizing genetic and environmental influences. Nat. Rev. Genet. 2011, 12, 167–178. [Google Scholar] [CrossRef] [PubMed]
- Oliver, J.D.; Jia, S.; Halpern, L.R.; Graham, E.M.; Turner, E.C.; Colombo, J.S.; Grainger, D.W.; D’Souza, R.N. Innovative Molecular and Cellular Therapeutics in Cleft Palate Tissue Engineering. Tissue Eng. Part B Rev. 2021, 27, 215–237. [Google Scholar] [CrossRef] [PubMed]
- Bush, J.O.; Jiang, R. Palatogenesis: Morphogenetic and molecular mechanisms of secondary palate development. Development 2012, 139, 828. [Google Scholar] [CrossRef]
- Li, C.; Lan, Y.; Jiang, R. Molecular and Cellular Mechanisms of Palate Development. J. Dent. Res. 2017, 96, 1184–1191. [Google Scholar] [CrossRef] [PubMed]
- Parada, C.; Han, D.; Grimaldi, A.; Sarrión, P.; Park, S.S.; Pelikan, R.; Sanchez-Lara, P.A.; Chai, Y. Disruption of the ERK/MAPK pathway in neural crest cells as a potential cause of Pierre Robin sequence. Development 2015, 142, 3734–3745. [Google Scholar] [CrossRef] [PubMed]
- Song, Z.; Liu, C.; Iwata, J.; Gu, S.; Suzuki, A.; Sun, C.; He, W.; Shu, R.; Li, L.; Chai, Y.; et al. Mice with Tak1 Deficiency in Neural Crest Lineage Exhibit Cleft Palate Associated with Abnormal Tongue Development*. J. Biol. Chem. 2013, 288, 10440–10450. [Google Scholar] [CrossRef]
- Bhol, C.S.; Patil, S.; Sahu, B.B.; Patra, S.K.; Bhutia, S.K. The clinical significance and correlative signaling pathways of paired box gene 9 in development and carcinogenesis. Biochim. Biophys. Acta (BBA)-Rev. Cancer 2021, 1876, 188561. [Google Scholar] [CrossRef]
- Paradowska-Stolarz, A. MSX1 gene in the etiology orofacial deformities. Adv. Hyg. Exp. Med. 2015, 69, 1499–1504. [Google Scholar]
- Zhou, J.; Gao, Y.; Lan, Y.; Jia, S.; Jiang, R. Pax9 regulates a molecular network involving Bmp4, Fgf10, Shh signaling and the Osr2 transcription factor to control palate morphogenesis. Development 2013, 140, 4709–4718. [Google Scholar] [CrossRef]
- Gao, Y.; Lan, Y.; Ovitt, C.E.; Jiang, R. Functional equivalence of the zinc finger transcription factors Osr1 and Osr2 in mouse development. Dev. Biol. 2009, 328, 200–209. [Google Scholar] [CrossRef]
- Funato, N. Molecular basis of cleft palates in mice. World J. Biol. Chem. 2015, 6, 121. [Google Scholar] [CrossRef] [PubMed]
- Iwata, J.; Suzuki, A.; Pelikan, R.C.; Ho, T.V.; Sanchez-Lara, P.A.; Urata, M.; Dixon, M.J.; Chai, Y. Smad4-Irf6 genetic interaction and TGFbeta-mediated IRF6 signaling cascade are crucial for palatal fusion in mice. Development 2013, 140, 1220–1230. [Google Scholar] [CrossRef] [PubMed]
- Nasroen, S.L.; Maskoen, A.M.; Soedjana, H.; Hilmanto, D.; Gani, B.A. IRF6 rs2235371 as a risk factor for non-syndromic cleft palate only among the Deutero-Malay race in Indonesia and its effect on the IRF6 mRNA expression level. Dent. Med. Probl. 2022, 59, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Sun, T.; You, Y.; Wu, B.; Wang, X.; Wu, J. Proteoglycans and Glycosaminoglycans in Stem Cell Homeostasis and Bone Tissue Regeneration. Front. Cell Dev. Biol. 2021, 9, 760532. [Google Scholar] [CrossRef]
- Theocharis, A.D.; Skandalis, S.S.; Gialeli, C.; Karamanos, N.K. Extracellular matrix structure. Adv. Drug Deliv. Rev. 2016, 97, 4–27. [Google Scholar] [CrossRef]
- Paganini, C.; Costantini, R.; Superti-Furga, A.; Rossi, A. Bone and connective tissue disorders caused by defects in glycosaminoglycan biosynthesis: A panoramic view. FEBS J. 2019, 286, 3008–3032. [Google Scholar] [CrossRef]
- Renato, V.; Iozzo, L.S. Proteoglycan form and function: A comprehensive nomenclature of proteoglycans. Matrix Biol. 2015, 42, 11–55. [Google Scholar] [CrossRef]
- Koike, T.; Izumikawa, T.; Sato, B.; Kitagawa, H. Identification of Phosphatase That Dephosphorylates Xylose in the Glycosaminoglycan-Protein Linkage Region of Proteoglycans. J. Biol. Chem. 2014, 289, 6695–6708. [Google Scholar] [CrossRef]
- Tone, Y.; Pedersen, L.C.; Yamamoto, T.; Izumikawa, T.; Kitagawa, H.; Nishihara, J.; Tamura, J.; Negishi, M.; Sugahara, K. 2-O-Phosphorylation of Xylose and 6-O-Sulfation of Galactose in the Protein Linkage Region of Glycosaminoglycans Influence the Glucuronyltransferase-I Activity Involved in the Linkage Region Synthesis. J. Biol. Chem. 2008, 283, 16801–16807. [Google Scholar] [CrossRef]
- Nadanaka, S.; Kitagawa, H. EXTL2 controls liver regeneration and aortic calcification through xylose kinase-dependent regulation of glycosaminoglycan biosynthesis. Matrix Biol. 2014, 35, 18–24. [Google Scholar] [CrossRef]
- Nadanaka, S.; Zhou, S.; Kagiyama, S.; Shoji, N.; Sugahara, K.; Sugihara, K.; Asano, M.; Kitagawa, H. EXTL2, a Member of the EXT Family of Tumor Suppressors, Controls Glycosaminoglycan Biosynthesis in a Xylose Kinase-dependent Manner. J. Biol. Chem. 2013, 288, 9321–9333. [Google Scholar] [CrossRef]
- Wen, J.; Xiao, J.; Rahdar, M.; Choudhury, B.P.; Cui, J.; Taylor, G.S.; Esko, J.D.; Dixon, J.E. Xylose phosphorylation functions as a molecular switch to regulate proteoglycan biosynthesis. Proc. Natl. Acad. Sci. USA 2014, 111, 15723–15728. [Google Scholar] [CrossRef]
- Koike, T.; Izumikawa, T.; Tamura, J.; Kitagawa, H. FAM20B is a kinase that phosphorylates xylose in the glycosaminoglycan–protein linkage region. Biochem. J. 2009, 421, 157–162. [Google Scholar] [CrossRef]
- Zhang, H.; Zhu, Q.; Cui, J.; Wang, Y.; Chen, M.J.; Guo, X.; Tagliabracci, V.S.; Dixon, J.E.; Xiao, J. Structure and evolution of the Fam20 kinases. Nat. Commun. 2018, 9, 1218. [Google Scholar] [CrossRef]
- Vogel, P.; Hansen, G.M.; Read, R.W.; Vance, R.B.; Thiel, M.; Liu, J.; Wronski, T.J.; Smith, D.D.; Jeter-Jones, S.; Brommage, R. Amelogenesis Imperfecta and Other Biomineralization Defects in Fam20a and Fam20c Null Mice. Vet. Pathol. 2012, 49, 998–1017. [Google Scholar] [CrossRef]
- Wu, J.; Tian, Y.; Han, L.; Liu, C.; Sun, T.; Li, L.; Yu, Y.; Lamichhane, B.; D’Souza, R.N.; Millar, S.E.; et al. FAM20B-catalyzed glycosaminoglycans control murine tooth number by restricting FGFR2b signaling. BMC Biol. 2020, 18, 87. [Google Scholar] [CrossRef]
- Tian, Y.; Ma, P.; Liu, C.; Yang, X.; Crawford, D.M.; Yan, W.; Bai, D.; Qin, C.; Wang, X. Inactivation of Fam20B in the dental epithelium of mice leads to supernumerary incisors. Eur. J. Oral Sci. 2015, 123, 396–402. [Google Scholar] [CrossRef]
- Saiyin, W.; Li, L.; Zhang, H.; Lu, Y.; Qin, C. Inactivation of FAM20B causes cell fate changes in annulus fibrosus of mouse intervertebral disc and disc defects via the alterations of TGF-β and MAPK signaling pathways. Biochim. Biophys. Acta Mol. Basis. Dis. 2019, 1865, 165555. [Google Scholar] [CrossRef]
- Liu, X.; Li, N.; Zhang, H.; Liu, J.; Zhou, N.; Ran, C.; Chen, X.; Lu, Y.; Wang, X.; Qin, C.; et al. Inactivation of Fam20b in the neural crest-derived mesenchyme of mouse causes multiple craniofacial defects. Eur. J. Oral Sci. 2018, 126, 433–436. [Google Scholar] [CrossRef]
- Wang, X.; Li, C.; Zhu, Z.; Yuan, L.; Chan, W.Y.; Sha, O. Extracellular Matrix Remodeling During Palate Development. Organogenesis 2020, 16, 43–60. [Google Scholar] [CrossRef]
- Xu, J.; Liu, H.; Lan, Y.; Park, J.S.; Jiang, R. Genome-wide Identification of Foxf2 Target Genes in Palate Development. J. Dent. Res. 2020, 99, 463–471. [Google Scholar] [CrossRef] [PubMed]
- Hirata, A.; Katayama, K.; Tsuji, T.; Natsume, N.; Sugahara, T.; Koga, Y.; Takano, K.; Otsuki, Y.; Nakamura, H. Heparanase localization during palatogenesis in mice. Biomed Res. Int. 2013, 2013, 760236. [Google Scholar] [CrossRef] [PubMed]
- Galloway, J.L.; Jones, S.J.; Mossey, P.A.; Ellis, I.R. The control and importance of hyaluronan synthase expression in palatogenesis. Front. Physiol. 2013, 4, 10. [Google Scholar] [CrossRef] [PubMed]
- Yonemitsu, M.A.; Lin, T.; Yu, K. Hyaluronic acid is required for palatal shelf movement and its interaction with the tongue during palatal shelf elevation. Dev. Biol. 2020, 457, 57–68. [Google Scholar] [CrossRef] [PubMed]
- Lan, Y.; Qin, C.; Jiang, R. Requirement of Hyaluronan Synthase-2 in Craniofacial and Palate Development. J. Dent. Res. 2019, 98, 1367–1375. [Google Scholar] [CrossRef]
- Lan, Y.; Zhang, N.; Liu, H.; Xu, J.; Jiang, R. Golgb1 regulates protein glycosylation and is crucial for mammalian palate development. Development 2016, 143, 2344–2355. [Google Scholar] [CrossRef]
- Karempelis, P.; Hagen, M.; Morrell, N.; Roby, B.B. Associated syndromes in patients with Pierre Robin Sequence. Int. J. Pediatr. Otorhinolaryngol. 2020, 131, 109842. [Google Scholar] [CrossRef]
- Hsieh, S.T.; Woo, A.S. Pierre Robin Sequence. Clin. Plast. Surg. 2019, 46, 249–259. [Google Scholar] [CrossRef]
- Motch, P.S.; Wu, M.; Holmes, G.; Bjork, B.C.; Jabs, E.W.; Richtsmeier, J.T. Phenotypes, Developmental Basis, and Genetics of Pierre Robin Complex. J. Dev. Biol. 2020, 8, 30. [Google Scholar] [CrossRef]
- Rios, J.J.; Denton, K.; Russell, J.; Kozlitina, J.; Ferreira, C.R.; Lewanda, A.F.; Mayfield, J.E.; Moresco, E.; Ludwig, S.; Tang, M.; et al. Germline Saturation Mutagenesis Induces Skeletal Phenotypes in Mice. J. Bone Miner. Res. 2021, 36, 1548–1565. [Google Scholar] [CrossRef]
- Kuroda, Y.; Murakami, H.; Enomoto, Y.; Tsurusaki, Y.; Takahashi, K.; Mitsuzuka, K.; Ishimoto, H.; Nishimura, G.; Kurosawa, K. A novel gene (FAM20B encoding glycosaminoglycan xylosylkinase) for neonatal short limb dysplasia resembling Desbuquois dysplasia. Clin. Genet. 2019, 95, 713–717. [Google Scholar] [CrossRef]
- Ma, P.; Yan, W.; Tian, Y.; Wang, J.; Feng, J.Q.; Qin, C.; Cheng, Y.S.; Wang, X. Inactivation of Fam20B in Joint Cartilage Leads to Chondrosarcoma and Postnatal Ossification Defects. Sci. Rep. 2016, 6, 29814. [Google Scholar] [CrossRef]
- Eames, B.F.; Yan, Y.L.; Swartz, M.E.; Levic, D.S.; Knapik, E.W.; Postlethwait, J.H.; Kimmel, C.B. Mutations in fam20b and xylt1 reveal that cartilage matrix controls timing of endochondral ossification by inhibiting chondrocyte maturation. PLoS Genet. 2011, 7, e1002246. [Google Scholar] [CrossRef]
- Worby, C.A.; Mayfield, J.E.; Pollak, A.J.; Dixon, J.E.; Banerjee, S. The ABCs of the atypical Fam20 secretory pathway kinases. J. Biol. Chem. 2021, 296, 100267. [Google Scholar] [CrossRef]
- Theocharis, A.D.; Skandalis, S.S.; Tzanakakis, G.N.; Karamanos, N.K. Proteoglycans in health and disease: Novel roles for proteoglycans in malignancy and their pharmacological targeting. FEBS J. 2010, 277, 3904–3923. [Google Scholar] [CrossRef]
- Snyder-Warwick, A.K.; Perlyn, C.A.; Pan, J.; Yu, K.; Zhang, L.; Ornitz, D.M. Analysis of a gain-of-function FGFR2 Crouzon mutation provides evidence of loss of function activity in the etiology of cleft palate. Proc. Natl. Acad. Sci. USA 2010, 107, 2515–2520. [Google Scholar] [CrossRef]
- Parada, C.; Li, J.; Iwata, J.; Suzuki, A.; Chai, Y. CTGF mediates Smad-dependent transforming growth factor beta signaling to regulate mesenchymal cell proliferation during palate development. Mol. Cell. Biol. 2013, 33, 3482–3493. [Google Scholar] [CrossRef]
- Amarasekara, D.S.; Kim, S.; Rho, J. Regulation of Osteoblast Differentiation by Cytokine Networks. Int. J. Mol. Sci. 2021, 22, 2851. [Google Scholar] [CrossRef]
- Berendsen, A.D.; Olsen, B.R. Bone development. Bone 2015, 80, 14–18. [Google Scholar] [CrossRef]
- Galea, G.L.; Zein, M.R.; Allen, S.; Francis-West, P. Making and shaping endochondral and intramembranous bones. Dev. Dyn. 2021, 250, 414–449. [Google Scholar] [CrossRef]
- Baek, J.A.; Lan, Y.; Liu, H.; Maltby, K.M.; Mishina, Y.; Jiang, R. Bmpr1a signaling plays critical roles in palatal shelf growth and palatal bone formation. Dev. Biol. 2011, 350, 520–531. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Liu, J.; Liu, H.; Wang, S.; Hu, P.; Zhou, H.; Xiao, J.; Liu, C. Altered BMP-Smad4 signaling causes complete cleft palate by disturbing osteogenesis in palatal mesenchyme. J. Mol. Histol. 2021, 52, 45–61. [Google Scholar] [CrossRef] [PubMed]
- Al, K.A.; Nessib, N.; Ghachem, M.B.; Hammou, A.; Guiddana, N.; Kozlowski, K. A novel syndrome resembling Desbuquois dysplasia. Am. J. Med. Genet. A. 2005, 132A, 68–75. [Google Scholar] [CrossRef]
- Furuichi, T.; Dai, J.; Cho, T.J.; Sakazume, S.; Ikema, M.; Matsui, Y.; Baynam, G.; Nagai, T.; Miyake, N.; Matsumoto, N.; et al. CANT1 mutation is also responsible for Desbuquois dysplasia, type 2 and Kim variant. J. Med. Genet. 2011, 48, 32–37. [Google Scholar] [CrossRef]
- Takata, Y.; Lenke, L.G.; Kelly, M.P. Posterior vertebral column resection for rigid proximal thoracic kyphoscoliosis with broken growing rods in a patient with Desbuquois dysplasia. Spine Deform. 2020, 8, 135–138. [Google Scholar] [CrossRef]
- Ozdemir, M.; Kavak, R.P. Radiographic findings of Desbuquois dysplasia. BJR Case Rep. 2021, 7, 20200137. [Google Scholar] [CrossRef]
- Thomas, M.M.; Ashaat, E.A.; Otaify, G.A.; Ismail, S.; Essawi, M.L.; Abdel-Hamid, M.S.; Hassan, H.A.; Alsaiedi, S.A.; Aglan, M.; El, R.M.; et al. First Report of Two Egyptian Patients with Desbuquois Dysplasia due to Homozygous CANT1 Mutations. Mol. Syndromol. 2021, 12, 279–288. [Google Scholar] [CrossRef]
- Bui, C.; Huber, C.; Tuysuz, B.; Alanay, Y.; Bole-Feysot, C.; Leroy, J.G.; Mortier, G.; Nitschke, P.; Munnich, A.; Cormier-Daire, V. XYLT1 mutations in Desbuquois dysplasia type 2. Am. J. Hum. Genet. 2014, 94, 405–414. [Google Scholar] [CrossRef]
- Huber, C.; Oules, B.; Bertoli, M.; Chami, M.; Fradin, M.; Alanay, Y.; Al-Gazali, L.I.; Ausems, M.G.; Bitoun, P.; Cavalcanti, D.P.; et al. Identification of CANT1 mutations in Desbuquois dysplasia. Am. J. Hum. Genet. 2009, 85, 706–710. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, X.; Li, N.; Hu, P.; Li, L.; Li, D.; Liu, H.; Zhu, L.; Xiao, J.; Liu, C. Deficiency of Fam20b-Catalyzed Glycosaminoglycan Chain Synthesis in Neural Crest Leads to Cleft Palate. Int. J. Mol. Sci. 2023, 24, 9634. https://doi.org/10.3390/ijms24119634
Chen X, Li N, Hu P, Li L, Li D, Liu H, Zhu L, Xiao J, Liu C. Deficiency of Fam20b-Catalyzed Glycosaminoglycan Chain Synthesis in Neural Crest Leads to Cleft Palate. International Journal of Molecular Sciences. 2023; 24(11):9634. https://doi.org/10.3390/ijms24119634
Chicago/Turabian StyleChen, Xiaoyan, Nan Li, Ping Hu, Leilei Li, Danya Li, Han Liu, Lei Zhu, Jing Xiao, and Chao Liu. 2023. "Deficiency of Fam20b-Catalyzed Glycosaminoglycan Chain Synthesis in Neural Crest Leads to Cleft Palate" International Journal of Molecular Sciences 24, no. 11: 9634. https://doi.org/10.3390/ijms24119634
APA StyleChen, X., Li, N., Hu, P., Li, L., Li, D., Liu, H., Zhu, L., Xiao, J., & Liu, C. (2023). Deficiency of Fam20b-Catalyzed Glycosaminoglycan Chain Synthesis in Neural Crest Leads to Cleft Palate. International Journal of Molecular Sciences, 24(11), 9634. https://doi.org/10.3390/ijms24119634







