Transcriptome-Wide Analysis of Low-Concentration Exposure to Bisphenol A, S, and F in Prostate Cancer Cells
Abstract
1. Introduction
2. Results
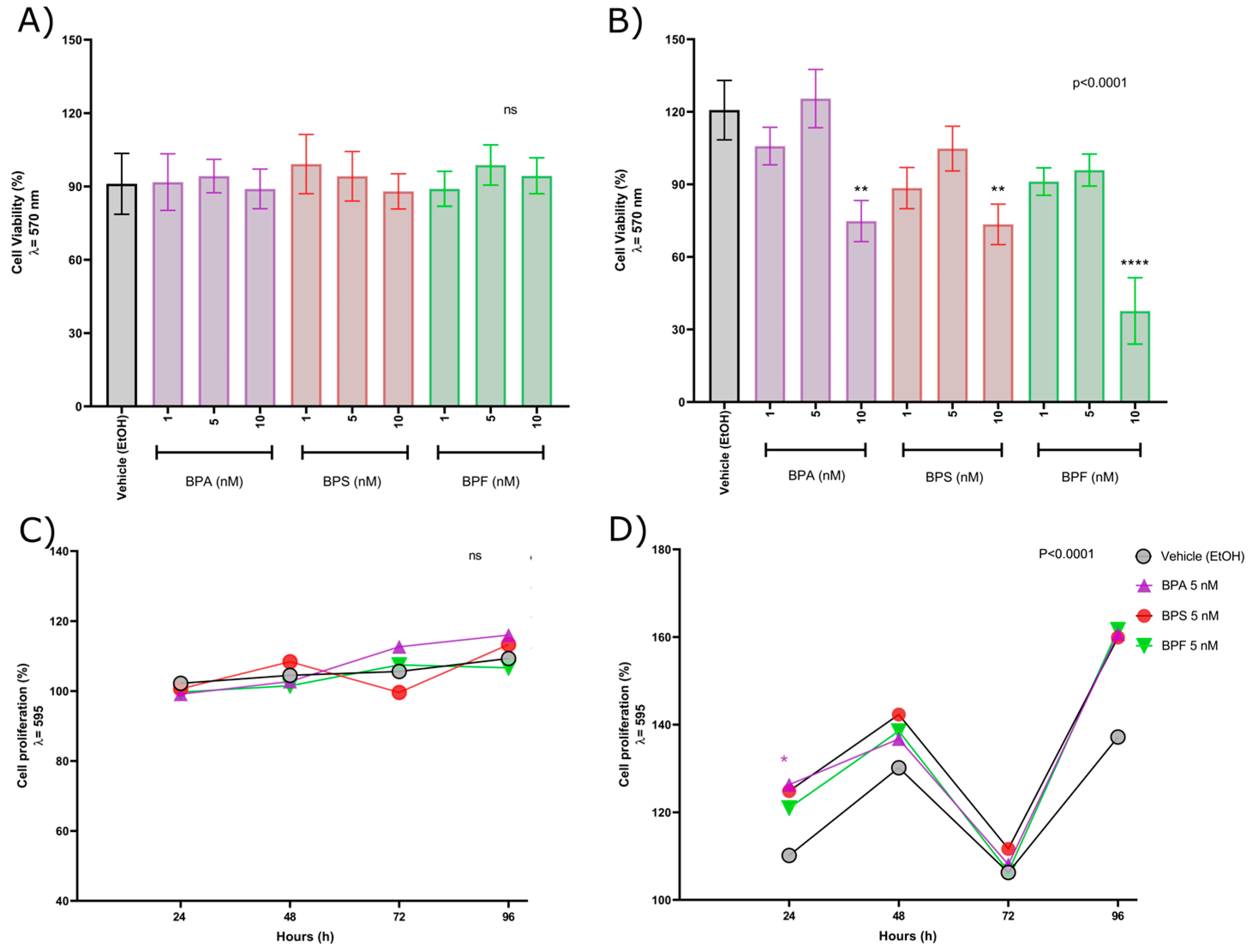
2.1. Low-Concentration Bisphenol Exposure-Induced Cytotoxicity and Produced Changes in the Cell Proliferation Pattern in Androgen-Independent PC-3 Prostate Cancer Cells
2.2. Transcriptomic Effect of Bisphenols Exposure
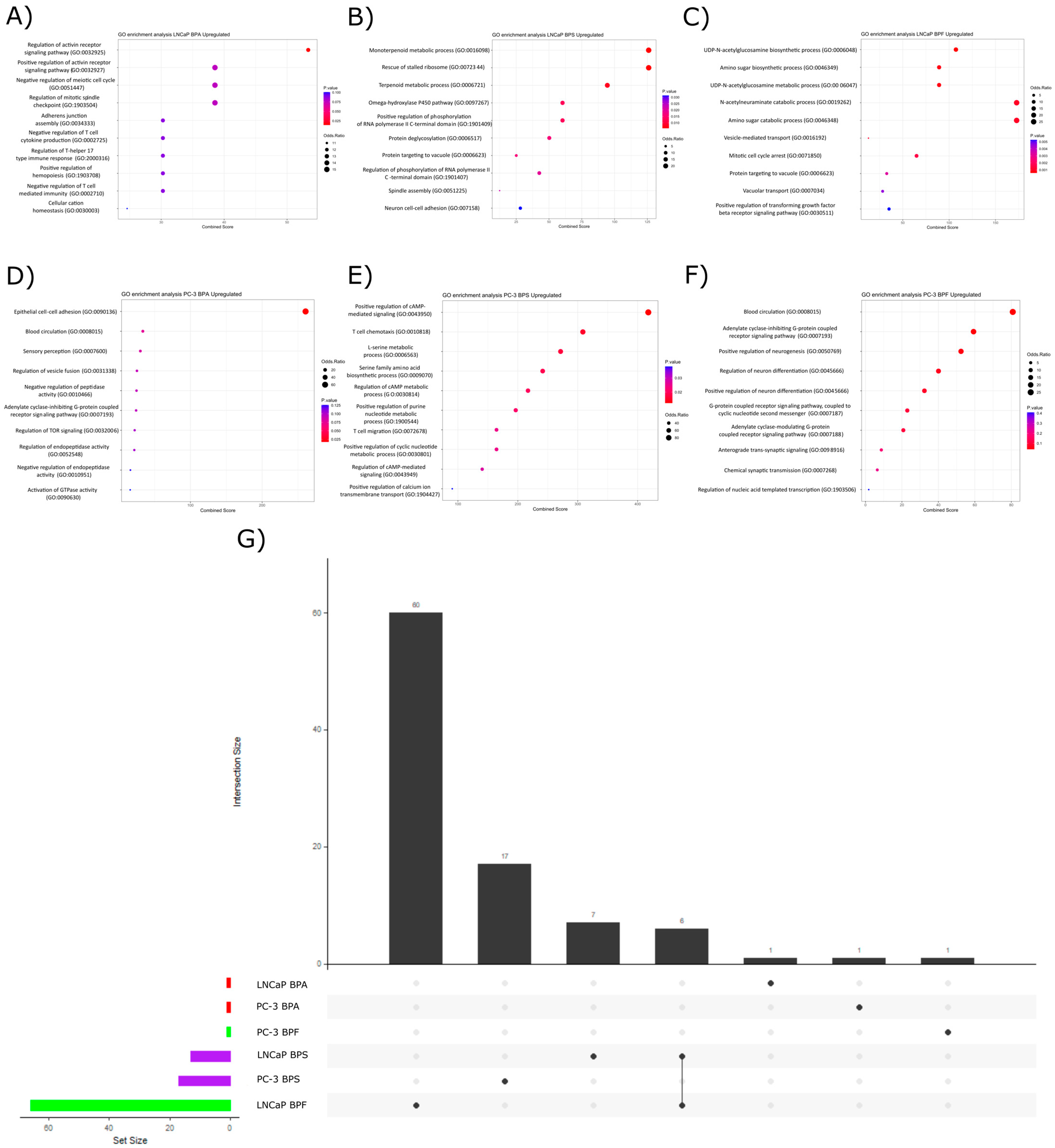
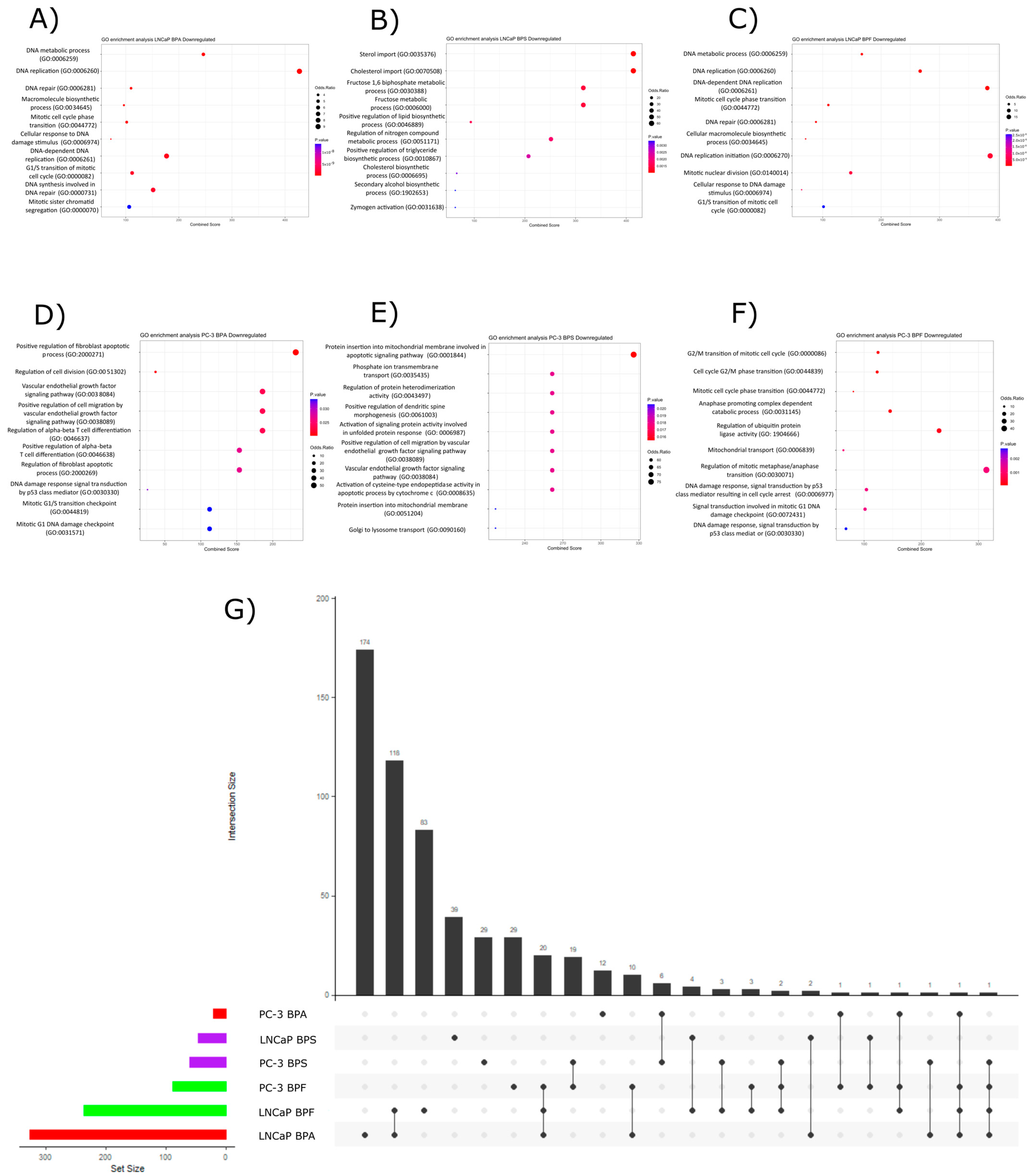
2.3. Pathway Over-Representation Analysis
2.3.1. Upregulated Pathways
2.3.2. Downregulated Pathways
2.4. DNA Damage Induction by Exposure to Bisphenols
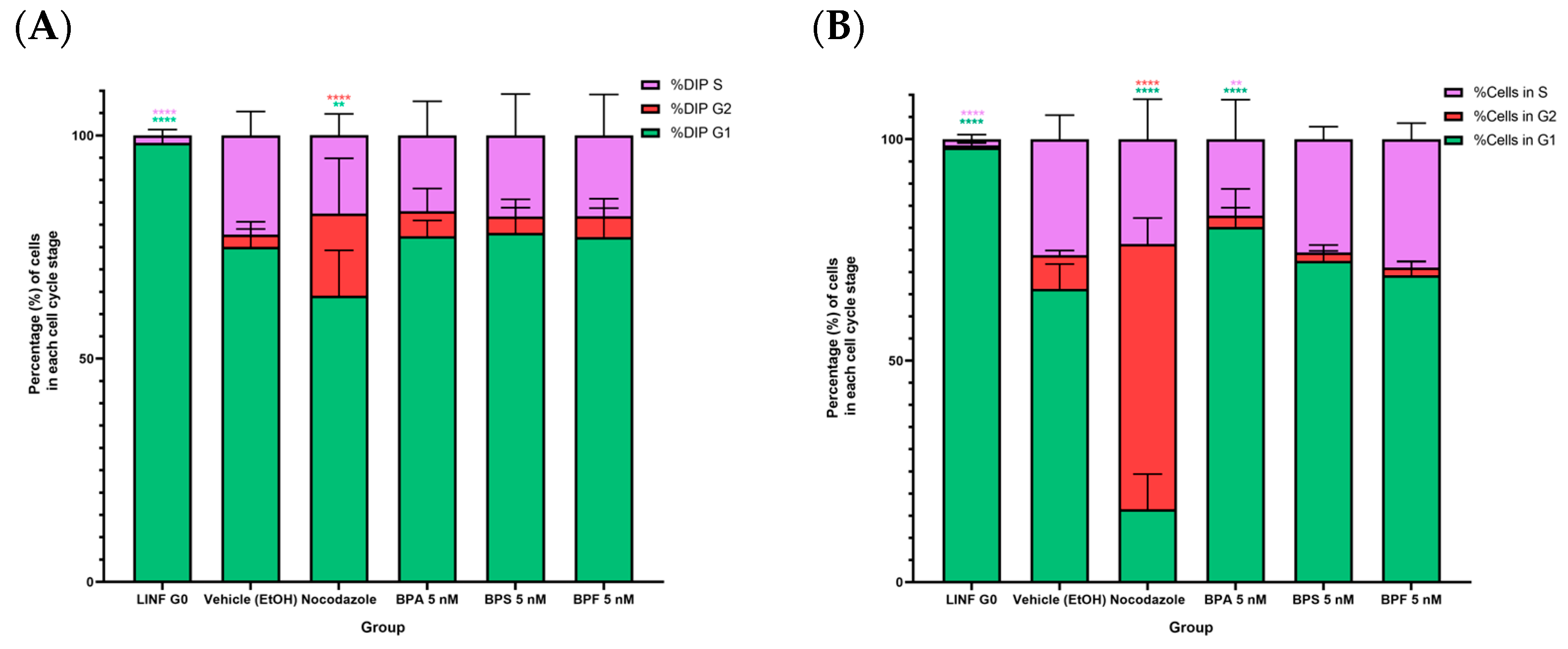
2.5. Cell Cycle Analysis
3. Discussion
4. Materials and Methods
4.1. Chemicals
4.2. Cell Culture
4.3. Cell Viability Assay
4.4. Cell Proliferation
4.5. RNA Extraction and Quality Assessment
4.6. Microarray Assay
4.7. Data Analysis
4.7.1. Differential Expression Analysis
4.7.2. Pathway Over-Representation Analysis
4.7.3. Cytokinesis-Blocked Micronuclei (CBMN) Assay
4.7.4. Alkali Comet Assay
4.7.5. Flow Cytometry and Cell Cycle Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Llevot, A.; Grau, E.; Carlotti, S.; Grelier, S.; Cramail, H. From Lignin-derived Aromatic Compounds to Novel Biobased Polymers. Macromol. Rapid Commun. 2016, 37, 9–28. [Google Scholar] [CrossRef]
- Research and Markets. Bisphenol-A—A Global Market Overview [Internet]. 2016. Available online: https://www.prnewswire.com/news-releases/global-bisphenol-a-market-overview-2016-2022---market-is-projected-to-reach-us225-billion-by-2022-up-from-156-billion-in-2016---research-and-markets-300303934.html (accessed on 31 March 2023).
- Abraham, A.; Chakraborty, P. A review on sources and health impacts of bisphenol A. Rev. Environ. Health 2020, 35, 201–210. [Google Scholar] [CrossRef]
- Sánchez-Brunete, C.; Miguel, E.; Tadeo, J.L. Determination of tetrabromobisphenol-A, tetrachlorobisphenol-A and bisphenol-A in soil by ultrasonic assisted extraction and gas chromatography–mass spectrometry. J. Chromatogr. A 2009, 1216, 5497–5503. [Google Scholar] [CrossRef] [PubMed]
- Xue, J.; Wan, Y.; Kannan, K. Occurrence of bisphenols, bisphenol A diglycidyl ethers (BADGEs), and novolac glycidyl ethers (NOGEs) in indoor air from Albany, New York, USA, and its implications for inhalation exposure. Chemosphere 2016, 151, 1–8. [Google Scholar] [CrossRef]
- Kouidhi, W.; Thannimalay, L.; Soon, C.S.; Mohd, M.A. Occupational exposure to bisphenol A (BPA) in a plastic injection molding factory in Malaysia. Int. J. Occup. Med. Environ. Health 2017, 30, 743. [Google Scholar] [CrossRef] [PubMed]
- Fromme, H.; Küchler, T.; Otto, T.; Pilz, K.; Müller, J.; Wenzel, A. Occurrence of phthalates and bisphenol A and F in the environment. Water Res. 2002, 36, 1429–1438. [Google Scholar] [CrossRef] [PubMed]
- Schecter, A.; Malik, N.; Haffner, D.; Smith, S.; Harris, T.R.; Paepke, O.; Birnbaum, L. Bisphenol A (BPA) in U.S. Food. Environ. Sci. Technol. 2010, 44, 9425–9430. [Google Scholar] [CrossRef]
- Lehmler, H.-J.; Liu, B.; Gadogbe, M.; Bao, W. Exposure to Bisphenol A, Bisphenol F, and Bisphenol S in U.S. Adults and Children: The National Health and Nutrition Examination Survey 2013–2014. ACS Omega 2018, 3, 6523–6532. [Google Scholar] [CrossRef]
- Calafat, A.M.; Kuklenyik, Z.; Reidy, J.A.; Caudill, S.P.; Ekong, J.; Needham, L.L. Urinary concentrations of bisphenol A and 4-nonylphenol in a human reference population. Environ. Health Perspect. 2005, 113, 391–395. [Google Scholar] [CrossRef]
- EPA Endocrine Disruptor Screening and Testing Advisory Committee Final Report. EDSTAC Final Rep. 1998, 1–17. Available online: http://www.epa.gov/endo/pubs/edstac/chap3v14.pdf (accessed on 27 March 2023).
- La Merrill, M.A.; Vandenberg, L.N.; Smith, M.T.; Goodson, W.; Browne, P.; Patisaul, H.B.; Guyton, K.Z.; Kortenkamp, A.; Cogliano, V.J.; Woodruff, T.J.; et al. Consensus on the key characteristics of endocrine-disrupting chemicals as a basis for hazard identification. Nat. Rev. Endocrinol. 2020, 16, 45–57. [Google Scholar] [CrossRef]
- Rancière, F.; Lyons, J.G.; Loh, V.H.Y.; Botton, J.; Galloway, T.; Wang, T.; Shaw, J.E.; Magliano, D.J. Bisphenol A and the risk of cardiometabolic disorders: A systematic review with meta-analysis of the epidemiological evidence. Environ. Health 2015, 14, 46. [Google Scholar] [CrossRef] [PubMed]
- Moon, S.; Yu, S.H.; Lee, C.B.; Park, Y.J.; Yoo, H.J.; Kim, D.S. Effects of bisphenol A on cardiovascular disease: An epidemiological study using National Health and Nutrition Examination Survey 2003–2016 and meta-analysis. Sci. Total Environ. 2021, 763, 142941. [Google Scholar] [CrossRef]
- Hwang, S.; Lim, J.-E.; Choi, Y.; Jee, S.H. Bisphenol A exposure and type 2 diabetes mellitus risk: A meta-analysis. BMC Endocr. Disord. 2018, 18, 81. [Google Scholar] [CrossRef]
- Hu, Y.; Wen, S.; Yuan, D.; Peng, L.; Zeng, R.; Yang, Z.; Liu, Q.; Xu, L.; Kang, D. The association between the environmental endocrine disruptor bisphenol A and polycystic ovary syndrome: A systematic review and meta-analysis. Gynecol. Endocrinol. 2018, 34, 370–377. [Google Scholar] [CrossRef] [PubMed]
- Abellan, A.; Mensink-Bout, S.M.; Garcia-Esteban, R.; Beneito, A.; Chatzi, L.; Duarte-Salles, T.; Fernandez, M.F.; Garcia-Aymerich, J.; Granum, B.; Iñiguez, C.; et al. In utero exposure to bisphenols and asthma, wheeze, and lung function in school-age children: A prospective meta-analysis of 8 European birth cohorts. Environ. Int. 2022, 162, 107178. [Google Scholar] [CrossRef]
- Keshavarz-Maleki, R.; Kaviani, A.; Omranipour, R.; Gholami, M.; Khoshayand, M.R.; Ostad, S.N.; Sabzevari, O. Bisphenol-A in biological samples of breast cancer mastectomy and mammoplasty patients and correlation with levels measured in urine and tissue. Sci. Rep. 2021, 11, 18411. [Google Scholar] [CrossRef]
- Nava-Castro, K.E.; Ramírez-Nieto, R.; Méndez-García, L.A.; Girón-Pérez, M.I.; Segovia-Mendoza, M.; Navidad-Murrieta, M.S.; Morales Montor, J. Environmental Pollution as a Risk Factor in Testicular Tumour Development: Focus on the Interaction between Bisphenol A and the Associated Immune Response. Int. J. Environ. Res. Public Health 2019, 16, 4113. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Guo, N.; Jin, H.; Liu, R.; Zhang, Z.; Cheng, C.; Fan, Z.; Zhang, G.; Xiao, M.; Wu, S.; et al. Bisphenol A drives di(2-ethylhexyl) phthalate promoting thyroid tumorigenesis via regulating HDAC6/PTEN and c-MYC signaling. J. Hazard. Mater. 2022, 425, 127911. [Google Scholar] [CrossRef]
- Chen, P.-P.; Yang, P.; Liu, C.; Deng, Y.-L.; Luo, Q.; Miao, Y.; Zhang, M.; Cui, F.-P.; Zeng, J.-Y.; Shi, T.; et al. Urinary concentrations of phenols, oxidative stress biomarkers and thyroid cancer: Exploring associations and mediation effects. J. Environ. Sci. 2022, 120, 30–40. [Google Scholar] [CrossRef]
- Sang, C.; Song, Y.; Jin, T.-W.; Zhang, S.; Fu, L.; Zhao, Y.; Zou, X.; Wang, Z.; Gao, H.; Liu, S. Bisphenol A induces ovarian cancer cell proliferation and metastasis through estrogen receptor-α pathways. Environ. Sci. Pollut. Res. Int. 2021, 28, 36060–36068. [Google Scholar] [CrossRef]
- Zahra, A.; Dong, Q.; Hall, M.; Jeyaneethi, J.; Silva, E.; Karteris, E.; Sisu, C. Identification of Potential Bisphenol A (BPA) Exposure Biomarkers in Ovarian Cancer. J. Clin. Med. 2021, 10, 1979. [Google Scholar] [CrossRef]
- Tse, L.A.; Lee, P.M.Y.; Ho, W.M.; Lam, A.T.; Lee, M.K.; Ng, S.S.M.; He, Y.; Leung, K.; Hartle, J.C.; Hu, H.; et al. Bisphenol A and other environmental risk factors for prostate cancer in Hong Kong. Environ. Int. 2017, 107, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Di Donato, M.; Cernera, G.; Giovannelli, P.; Galasso, G.; Bilancio, A.; Migliaccio, A.; Castoria, G. Recent advances on bisphenol-A and endocrine disruptor effects on human prostate cancer. Mol. Cell. Endocrinol. 2017, 457, 35–42. [Google Scholar] [CrossRef]
- Vandenberg, L.N.; Colborn, T.; Hayes, T.B.; Heindel, J.J.; Jacobs, D.R.J.; Lee, D.-H.; Shioda, T.; Soto, A.M.; vom Saal, F.S.; Welshons, W.V.; et al. Hormones and endocrine-disrupting chemicals: Low-dose effects and nonmonotonic dose responses. Endocr. Rev. 2012, 33, 378–455. [Google Scholar] [CrossRef] [PubMed]
- Siddique, S.; Zhang, G.; Coleman, K.; Kubwabo, C. Investigation of the migration of bisphenols from baby bottles and sippy cups. Curr. Res. Food Sci. 2021, 4, 619–626. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Kannan, K.; Tan, H.; Zheng, Z.; Feng, Y.-L.; Wu, Y.; Widelka, M. Bisphenol Analogues Other Than BPA: Environmental Occurrence, Human Exposure, and Toxicity—A Review. Environ. Sci. Technol. 2016, 50, 5438–5453. [Google Scholar] [CrossRef]
- Resnik, D.B.; Elliott, K.C. Bisphenol A and risk management ethics. Bioethics 2015, 29, 182–189. [Google Scholar] [CrossRef]
- Ullah, A.; Pirzada, M.; Jahan, S.; Ullah, H.; Khan, M.J. Bisphenol A analogues bisphenol B, bisphenol F, and bisphenol S induce oxidative stress, disrupt daily sperm production, and damage DNA in rat spermatozoa: A comparative in vitro and in vivo study. Toxicol. Ind. Health 2019, 35, 294–303. [Google Scholar] [CrossRef]
- Rosenmai, A.K.; Dybdahl, M.; Pedersen, M.; Alice van Vugt-Lussenburg, B.M.; Wedebye, E.B.; Taxvig, C.; Vinggaard, A.M. Are structural analogues to bisphenol a safe alternatives? Toxicol. Sci. 2014, 139, 35–47. [Google Scholar] [CrossRef]
- Global cancer observatory Prostate Cancer fact sheet Globocan 2020. Cancer Today 2020, 419, 119–120. Available online: https://gco.iarc.fr/today/data/factsheets/cancers/27-Prostate-fact-sheet.pdf (accessed on 27 March 2023).
- Toivanen, R.; Shen, M.M. Prostate organogenesis: Tissue induction, hormonal regulation and cell type specification. Development 2017, 144, 1382–1398. [Google Scholar] [CrossRef]
- Prins, G.S.; Korach, K.S. The role of estrogens and estrogen receptors in normal prostate growth and disease. Steroids 2008, 73, 233–244. [Google Scholar] [CrossRef]
- De Falco, M.; Laforgia, V. Combined Effects of Different Endocrine-Disrupting Chemicals (EDCs) on Prostate Gland. Int. J. Environ. Res. Public Health 2021, 18, 9772. [Google Scholar] [CrossRef] [PubMed]
- Hess-Wilson, J.K.; Knudsen, K.E. Endocrine disrupting compounds and prostate cancer. Cancer Lett. 2006, 241, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Prins, G.S.; Shu-Hua, Y.; Lynn, B.; Xiang, Z.; Ana, C.; Han, L.; Esther, C.-G.; Jacob, G.; Wen-Yang, H.; Shuk-Mei, H.; et al. Prostate Cancer Risk and DNA Methylation Signatures in Aging Rats following Developmental BPA Exposure: A Dose–Response Analysis. Environ. Health Perspect. 2022, 125, 77007. [Google Scholar] [CrossRef] [PubMed]
- Bilancio, A.; Bontempo, P.; Di Donato, M.; Conte, M.; Giovannelli, P.; Altucci, L.; Migliaccio, A.; Castoria, G. Bisphenol A induces cell cycle arrest in primary and prostate cancer cells through EGFR/ERK/p53 signaling pathway activation. Oncotarget 2017, 8, 115620–115631. [Google Scholar] [CrossRef]
- Derouiche, S.; Warnier, M.; Mariot, P.; Gosset, P.; Mauroy, B.; Bonnal, J.-L.; Slomianny, C.; Delcourt, P.; Prevarskaya, N.; Roudbaraki, M. Bisphenol A stimulates human prostate cancer cell migration via remodelling of calcium signalling. SpringerPlus 2013, 2, 54. [Google Scholar] [CrossRef]
- Gao, H.; Yang, B.-J.; Li, N.; Feng, L.-M.; Shi, X.-Y.; Zhao, W.-H.; Liu, S.-J. Bisphenol A and hormone-associated cancers: Current progress and perspectives. Medicine 2015, 94, e211. [Google Scholar] [CrossRef]
- Nadal, A.; Ropero, A.B.; Laribi, O.; Maillet, M.; Fuentes, E.; Soria, B. Nongenomic actions of estrogens and xenoestrogens by binding at a plasma membrane receptor unrelated to estrogen receptor α and estrogen receptor β. Proc. Natl. Acad. Sci. USA 2000, 97, 11603–11608. [Google Scholar] [CrossRef]
- Wetherill, Y.B.; Petre, C.E.; Monk, K.R.; Puga, A.; Knudsen, K.E. The xenoestrogen bisphenol A induces inappropriate androgen receptor activation and mitogenesis in prostatic adenocarcinoma cells. Mol. Cancer Ther. 2002, 1, 515–524. [Google Scholar]
- Wetherill, Y.B.; Hess-Wilson, J.K.; Comstock, C.E.S.; Shah, S.A.; Buncher, C.R.; Sallans, L.; Limbach, P.A.; Schwemberger, S.; Babcock, G.F.; Knudsen, K.E. Bisphenol A facilitates bypass of androgen ablation therapy in prostate cancer. Mol. Cancer Ther. 2006, 5, 3181–3190. [Google Scholar] [CrossRef]
- Wetherill, Y.B.; Fisher, N.L.; Staubach, A.; Danielsen, M.; de Vere White, R.W.; Knudsen, K.E. Xenoestrogen action in prostate cancer: Pleiotropic effects dependent on androgen receptor status. Cancer Res. 2005, 65, 54–65. [Google Scholar] [CrossRef]
- Marcoccia, D.; Pellegrini, M.; Fiocchetti, M.; Lorenzetti, S.; Marino, M. Food components and contaminants as (anti)androgenic molecules. Genes Nutr. 2017, 12, 6. [Google Scholar] [CrossRef] [PubMed]
- Hess-Wilson, J.K.; Webb, S.L.; Daly, H.K.; Leung, Y.-K.; Boldison, J.; Comstock, C.E.S.; Sartor, M.A.; Ho, S.-M.; Knudsen, K.E. Unique bisphenol A transcriptome in prostate cancer: Novel effects on ERbeta expression that correspond to androgen receptor mutation status. Environ. Health Perspect. 2007, 115, 1646–1653. [Google Scholar] [CrossRef] [PubMed]
- Acconcia, F.; Pallottini, V.; Marino, M. Molecular Mechanisms of Action of BPA. Dose-Response 2015, 13, 1559325815610582. [Google Scholar] [CrossRef] [PubMed]
- Tai, S.; Sun, Y.; Squires, J.M.; Zhang, H.; Oh, W.K.; Liang, C.-Z.; Huang, J. PC3 is a cell line characteristic of prostatic small cell carcinoma. Prostate 2011, 71, 1668–1679. [Google Scholar] [CrossRef]
- Huang, D.; Wu, J.; Su, X.; Yan, H.; Sun, Z. Effects of low dose of bisphenol A on the proliferation and mechanism of primary cultured prostate epithelial cells in rodents. Oncol. Lett. 2017, 14, 2635–2642. [Google Scholar] [CrossRef]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; et al. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016, 44, W90-7. [Google Scholar] [CrossRef] [PubMed]
- Simon, D.P.; Vadakkadath Meethal, S.; Wilson, A.C.; Gallego, M.J.; Weinecke, S.L.; Bruce, E.; Lyons, P.F.; Haasl, R.J.; Bowen, R.L.; Atwood, C.S. Activin receptor signaling regulates prostatic epithelial cell adhesion and viability. Neoplasia 2009, 11, 365–376. [Google Scholar] [CrossRef] [PubMed]
- Pu, X.; Wang, Z.; Klaunig, J.E. Alkaline comet assay for assessing DNA damage in individual cells. Curr. Protoc. Toxicol. 2015, 2015, 3.12.1–3.12.11. [Google Scholar] [CrossRef]
- Lau, K.-M.; LaSpina, M.; Long, J.; Ho, S.-M. Expression of Estrogen Receptor (ER)-α and ER-β in Normal and Malignant Prostatic Epithelial Cells: Regulation by Methylation and Involvement in Growth Regulation1. Cancer Res. 2000, 60, 3175–3182. [Google Scholar]
- Lafront, C.; Germain, L.; Weidmann, C.; Audet-Walsh, É. A Systematic Study of the Impact of Estrogens and Selective Estrogen Receptor Modulators on Prostate Cancer Cell Proliferation. Sci. Rep. 2020, 10, 4024. [Google Scholar] [CrossRef] [PubMed]
- Furic, L.; Lawrence, M.G.; Risbridger, G.P. Pro-tumorigenic role of ERα in prostate cancer cells. Aging 2015, 7, 356. [Google Scholar] [CrossRef] [PubMed]
- Hiroi, H.; Tsutsumi, O.; Momoeda, M.; Takai, Y.; Osuga, Y.; Taketani, Y. Differential Interactions of Bisphenol A and17β-estradiol with Estrogen Receptor α (ERα) and Erβ. Endocr. J. 1999, 46, 773–778. [Google Scholar] [CrossRef] [PubMed]
- Sohoni, P.; Sumpter, J.P. Several environmental oestrogens are also anti-androgens. J. Endocrinol. 1998, 158, 327–340. [Google Scholar] [CrossRef]
- Pelch, K.E.; Li, Y.; Perera, L.; Thayer, K.A.; Korach, K.S. Characterization of Estrogenic and Androgenic Activities for Bisphenol A-like Chemicals (BPs): In Vitro Estrogen and Androgen Receptors Transcriptional Activation, Gene Regulation, and Binding Profiles. Toxicol. Sci. 2019, 172, 23–37. [Google Scholar] [CrossRef]
- Chen, Y.-K.; Tan, Y.-Y.; Yao, M.; Lin, H.-C.; Tsai, M.-H.; Li, Y.-Y.; Hsu, Y.-J.; Huang, T.-T.; Chang, C.-W.; Cheng, C.-M.; et al. Bisphenol A-induced DNA damages promote to lymphoma progression in human lymphoblastoid cells through aberrant CTNNB1 signaling pathway. iScience 2021, 24, 102888. [Google Scholar] [CrossRef]
- Gassman, N.R.; Coskun, E.; Stefanick, D.F.; Horton, J.K.; Jaruga, P.; Dizdaroglu, M.; Wilson, S.H. Bisphenol A Promotes Cell Survival following Oxidative DNA Damage in Mouse Fibroblasts. PLoS ONE 2015, 10, e0118819. [Google Scholar] [CrossRef]
- Pfeifer, D.; Chung, Y.M.; Hu, M.C.-T. Effects of Low-Dose Bisphenol A on DNA Damage and Proliferation of Breast Cells: The Role of c-Myc. Environ. Health Perspect. 2015, 123, 1271–1279. [Google Scholar] [CrossRef]
- Di Pietro, P.; D’Auria, R.; Viggiano, A.; Ciaglia, E.; Meccariello, R.; Russo, R.D.; Puca, A.A.; Vecchione, C.; Nori, S.L.; Santoro, A. Bisphenol A induces DNA damage in cells exerting immune surveillance functions at peripheral and central level. Chemosphere 2020, 254, 126819. [Google Scholar] [CrossRef]
- Kose, O.; Rachidi, W.; Beal, D.; Erkekoglu, P.; Fayyad-Kazan, H.; Kocer Gumusel, B. The effects of different bisphenol derivatives on oxidative stress, DNA damage and DNA repair in RWPE-1 cells: A comparative study. J. Appl. Toxicol. 2020, 40, 643–654. [Google Scholar] [CrossRef]
- Bergholz, J.; Xiao, Z.-X. Role of p63 in Development, Tumorigenesis and Cancer Progression. Cancer Microenviron. 2012, 5, 311–322. [Google Scholar] [CrossRef]
- Ahn, C.; Jeung, E.-B. Endocrine-Disrupting Chemicals and Disease Endpoints. Int. J. Mol. Sci. 2023, 24, 5342. [Google Scholar] [CrossRef] [PubMed]
- Corti, M.; Lorenzetti, S.; Ubaldi, A.; Zilli, R.; Marcoccia, D. Endocrine Disruptors and Prostate Cancer. Int. J. Mol. Sci. 2022, 23, 1216. [Google Scholar] [CrossRef]
- Sigma-Aldrich. Bisphenol A 239658 [Internet]. 2022. Available online: https://www.sigmaaldrich.com/MX/es/product/aldrich/239658 (accessed on 5 July 2022).
- Sigma-Aldrich. Bisphenol S 103039 [Internet]. 2022. Available online: https://www.sigmaaldrich.com/MX/es/product/aldrich/103039 (accessed on 5 July 2022).
- Sigma-Aldrich. Bisphenol F B47006 [Internet]. 2022. Available online: https://www.sigmaaldrich.com/MX/es/product/aldrich/b47006 (accessed on 5 July 2022).
- Collection ATC. PC-3 CRL-1435. 2022, pp. 1–7. Available online: https://www.atcc.org/products/crl-1435 (accessed on 1 March 2023).
- American Type Culture Collection. LNCaP Clone FGC CRL-1740. 2022, pp. 1–3. Available online: https://www.atcc.org/products/crl-1740 (accessed on 1 March 2023).
- Merck KGaA. RPMI-1640 Medium Sigma-Aldrich [Internet]. 2022. Available online: https://www.sigmaaldrich.com/MX/es/product/sigma/r8758 (accessed on 5 July 2022).
- Abcam, MTT Assay Kit (Cell Proliferation) [Internet]; Abcam: Cambridge, MA, USA, 2020; Available online: https://www.abcam.com/mtt-assay-kit-cell-proliferation-ab211091.html (accessed on 15 March 2023).
- Sigma-Aldrich. TRI Reagent. 2022. Available online: https://www.sigmaaldrich.com/MX/es/product/sigma/t9424 (accessed on 15 March 2023).
- ScientificTM, T.F. NanoDropTM One/OneC [Internet]. Available online: https://www.thermofisher.com/order/catalog/product/ND-ONE-W (accessed on 15 March 2023).
- Agilent Technologies, I. 2100 Bioanalyzer Instrument [Internet]. Available online: https://www.agilent.com/en/product/automated-electrophoresis/bioanalyzer-systems/bioanalyzer-instrument/2100-bioanalyzer-instrument-228250 (accessed on 15 March 2023).
- Thermo Fisher Scientific Inc. ClariomTM D Solutions for Human, Mouse, and Rat. 2016, 122, 1–8. Available online: https://assets.thermofisher.com/TFS-Assets%2FLSG%2Fbrochures%2FEMI07313-2_DS_Clariom-D_solutions_HMR.pdf (accessed on 15 March 2023).
- Thermo Fisher Scientific Inc. GeneChipTM WT PLUS Reagent Kit Manual Target Preparation for GeneChip Whole Transcript (WT). 2020. Available online: https://assets.thermofisher.com/TFS-Assets/LSG/manuals/MAN0018137_703174_WTPlus_Reagentkit_Assay_UG.pdf (accessed on 15 March 2023).
- Affymetrix, I. Transcriptome Analysis Console (TAC) 4.0.2 User Guide. ThermoFisher Sci. 2019, 8, 219. [Google Scholar]
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis, 2nd ed.; Springer International Publishing: Cham, Switzerland, 2016; ISBN 9783319242774. [Google Scholar]
- Fenech, M.; Chang, W.P.; Kirsch-Volders, M.; Holland, N.; Bonassi, S.; Zeiger, E. HUMN project: Detailed description of the scoring criteria for the cytokinesis-block micronucleus assay using isolated human lymphocyte cultures. Mutat. Res. 2003, 534, 65–75. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Liu, Y.; Yang, C. Evaluating In Vitro DNA Damage Using Comet Assay. J. Vis. Exp. 2017, 128, e56450. [Google Scholar] [CrossRef]
- Perceptive instruments. Comet Assay IV [Internet]. 2016. Available online: http://www.perceptive.co.uk/products/comet-assay-iv/ (accessed on 5 July 2022).
- Verity Software House. ModFit LT User Guide [Internet]. 2013. Available online: https://www.vsh.com/Documentation/ModFitLT/mf40userguide.pdf (accessed on 17 March 2023).


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cortés-Ramírez, S.A.; Salazar, A.M.; Sordo, M.; Ostrosky-Wegman, P.; Morales-Pacheco, M.; Cruz-Burgos, M.; Losada-García, A.; Rodríguez-Martínez, G.; González-Ramírez, I.; Vazquez-Santillan, K.; et al. Transcriptome-Wide Analysis of Low-Concentration Exposure to Bisphenol A, S, and F in Prostate Cancer Cells. Int. J. Mol. Sci. 2023, 24, 9462. https://doi.org/10.3390/ijms24119462
Cortés-Ramírez SA, Salazar AM, Sordo M, Ostrosky-Wegman P, Morales-Pacheco M, Cruz-Burgos M, Losada-García A, Rodríguez-Martínez G, González-Ramírez I, Vazquez-Santillan K, et al. Transcriptome-Wide Analysis of Low-Concentration Exposure to Bisphenol A, S, and F in Prostate Cancer Cells. International Journal of Molecular Sciences. 2023; 24(11):9462. https://doi.org/10.3390/ijms24119462
Chicago/Turabian StyleCortés-Ramírez, Sergio A., Ana M. Salazar, Monserrat Sordo, Patricia Ostrosky-Wegman, Miguel Morales-Pacheco, Marian Cruz-Burgos, Alberto Losada-García, Griselda Rodríguez-Martínez, Imelda González-Ramírez, Karla Vazquez-Santillan, and et al. 2023. "Transcriptome-Wide Analysis of Low-Concentration Exposure to Bisphenol A, S, and F in Prostate Cancer Cells" International Journal of Molecular Sciences 24, no. 11: 9462. https://doi.org/10.3390/ijms24119462
APA StyleCortés-Ramírez, S. A., Salazar, A. M., Sordo, M., Ostrosky-Wegman, P., Morales-Pacheco, M., Cruz-Burgos, M., Losada-García, A., Rodríguez-Martínez, G., González-Ramírez, I., Vazquez-Santillan, K., González-Covarrubias, V., Maldonado-Lagunas, V., & Rodríguez-Dorantes, M. (2023). Transcriptome-Wide Analysis of Low-Concentration Exposure to Bisphenol A, S, and F in Prostate Cancer Cells. International Journal of Molecular Sciences, 24(11), 9462. https://doi.org/10.3390/ijms24119462





