Resistance to Resveratrol Treatment in Experimental PTSD Is Associated with Abnormalities in Hepatic Metabolism of Glucocorticoids
Abstract
1. Introduction
2. Results
2.1. Effects of Trans-Resveratrol Treatment on EPM Test Scores in PTSD Rats
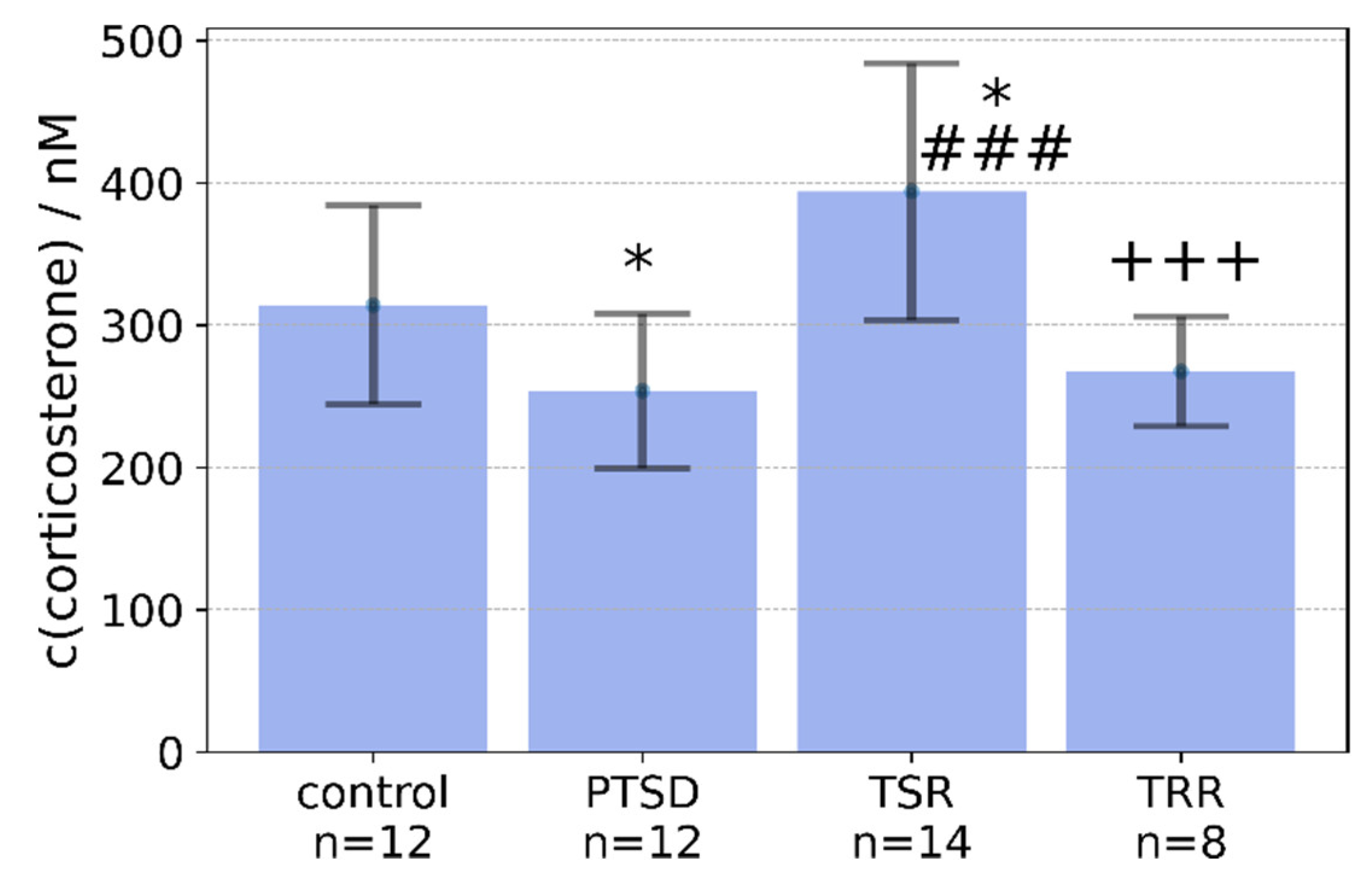
2.2. Effects of Trans-Resveratrol Treatment on Plasma Concentrations of Corticosterone in Rats with PTSD
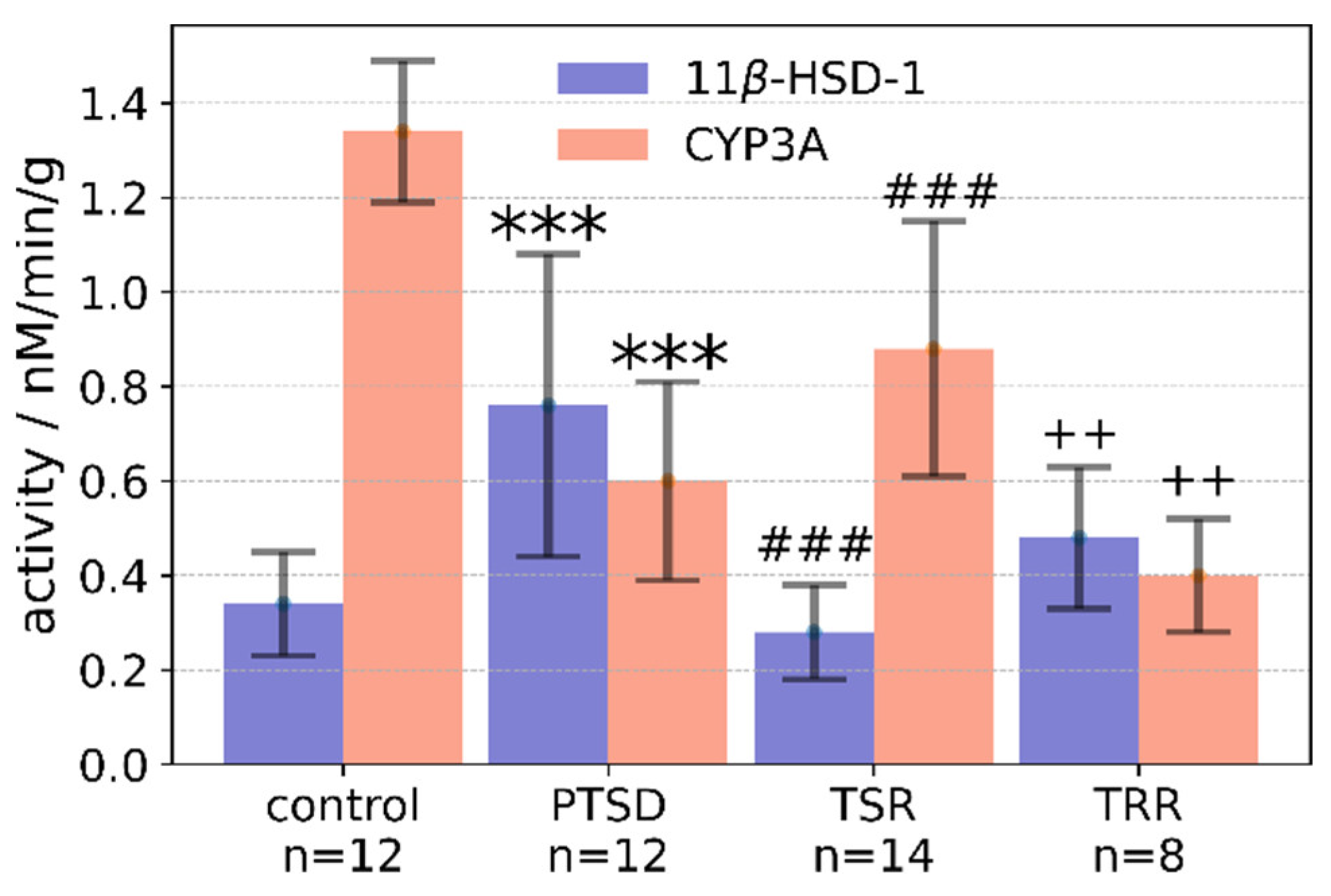
2.3. Effects of Trans-Resveratrol Treatment on Liver 11β-HSD-1 Activity
2.4. Effects of Trans-Resveratrol Treatment on CYP3A Activity in the Liver
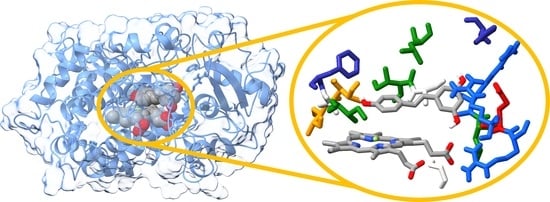
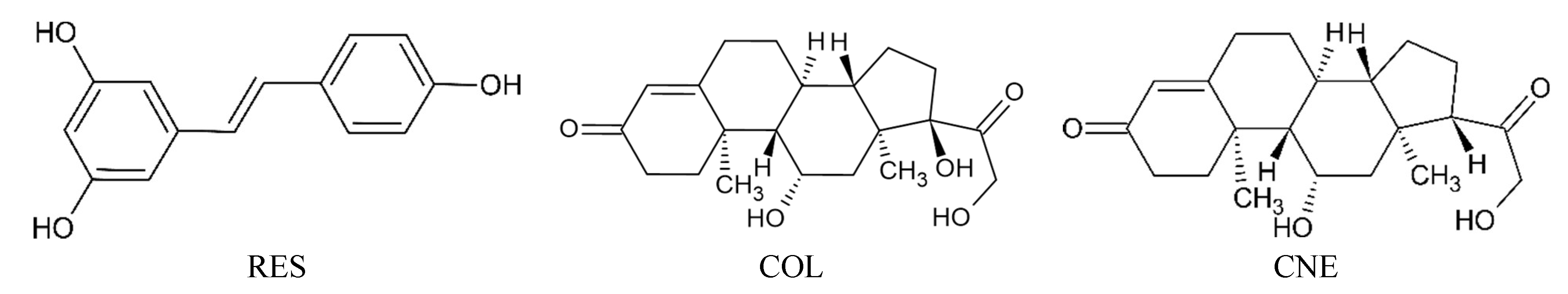
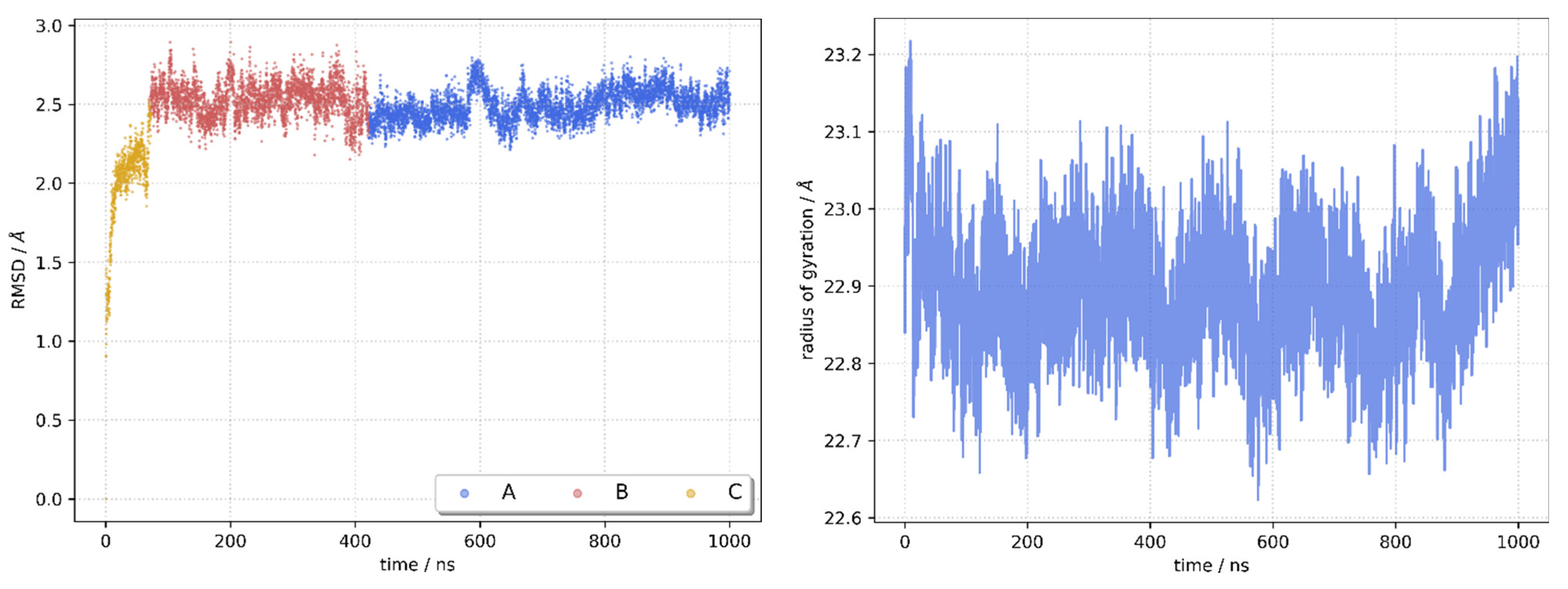
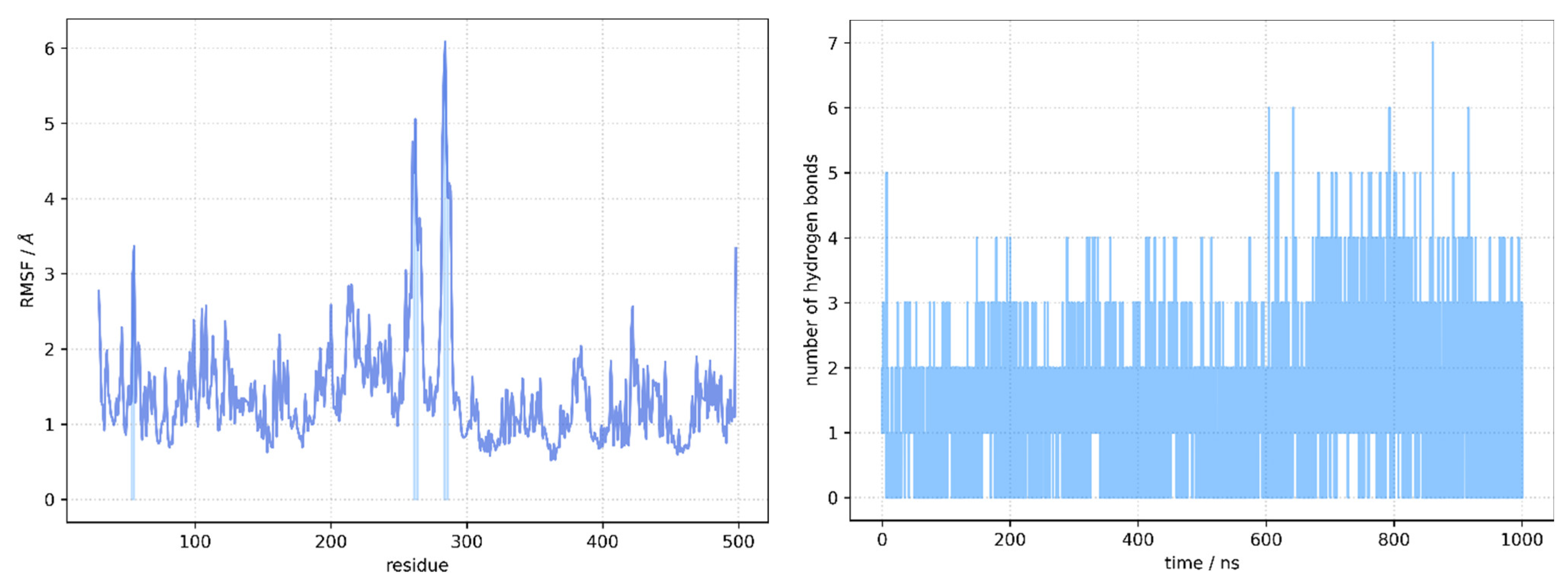
2.5. Molecular Docking and Molecular Dynamics Simulations
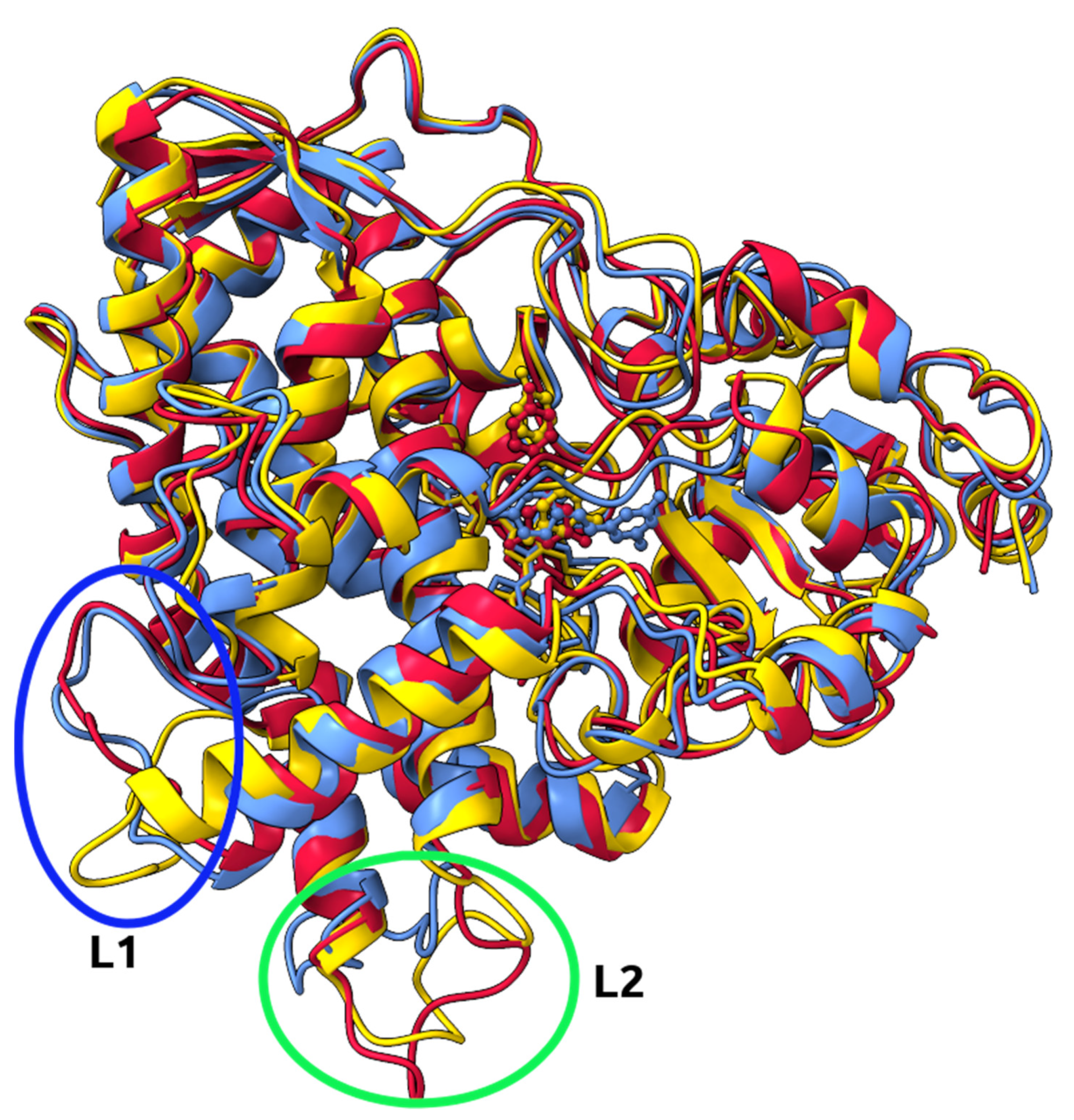
2.6. Identification of Three Conformations
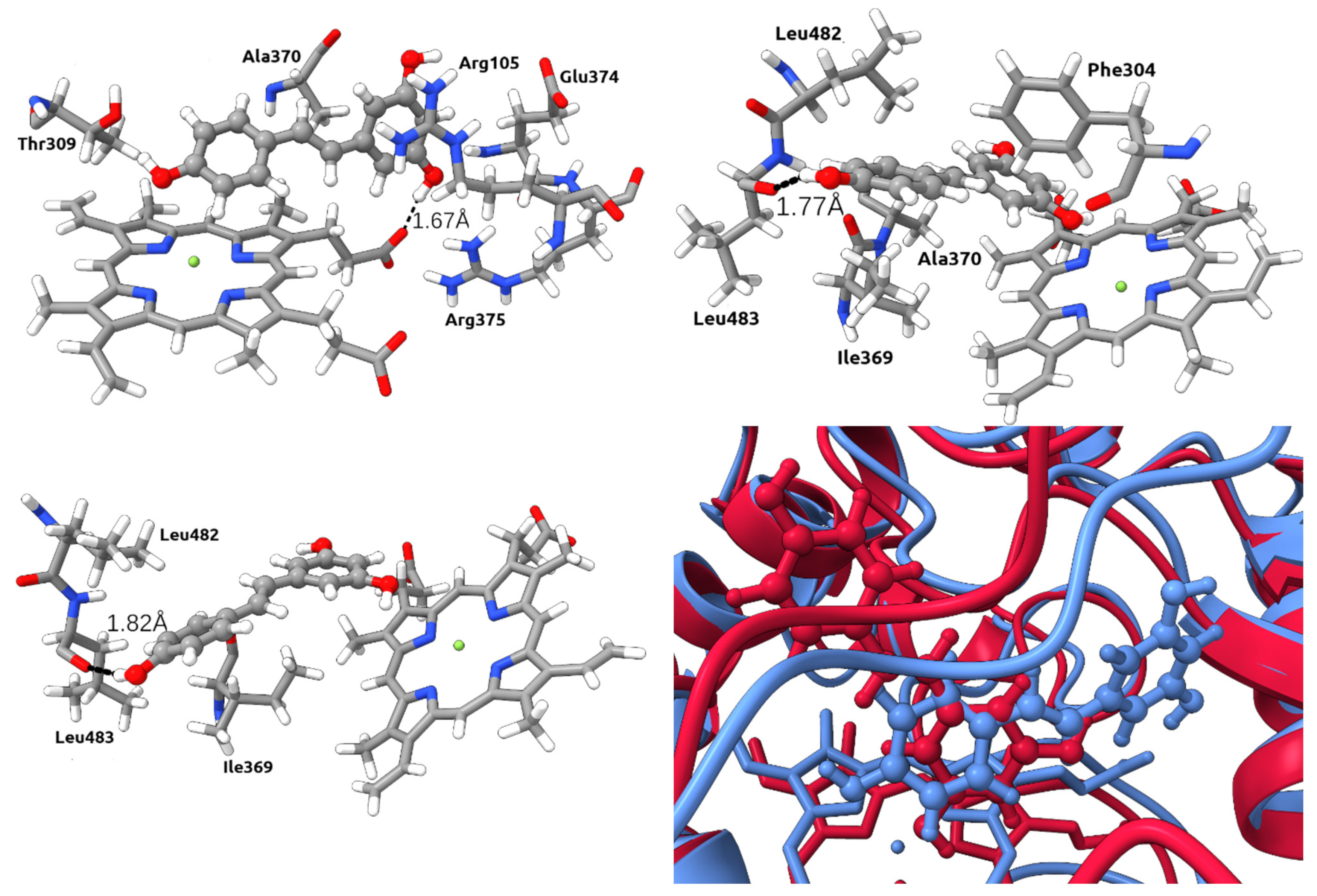
2.7. Binding Free Energy Calculation
3. Discussion
4. Materials and Methods
- Control rats (treated with vehicle only for 10 days, n = 12);
- PTSD (rats previously exposed to chronic predator stress followed by a two-week rest, n = 12);
- RES + PTSD (an effective dose of resveratrol was administered to rats via a tube one hour before the onset of predatory stress; n = 22). The effective dose of resveratrol was determined based on data presented in [11]. After performing a behavioral test in the elevated-plus maze (EPM), animals were classified into two phenotypes based on AI value: treatment-sensitive Rats (TSR; AI < 0.8) based on AI value, whereas the second phenotype was defined as treatment-resistant rats (TRR; AI > 0.8).
4.1. Behavioral Assessment
4.2. Statistical Analysis
4.3. Molecular Docking
4.4. Molecular Dynamics Simulations
4.5. Binding Free Energy Calculation
4.6. Dynamics Cross-Correlation Map (DCCM) Analysis
4.7. Cluster Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wang, X.; Li, X.; Qi, M.; Hu, X.; Zhu, H.; Shi, X. Incidence of post-traumatic stress disorder in survivors of traumatic fracture: A systematic review and meta-analysis. Psychol. Health Med. 2022, 27, 902–916. [Google Scholar] [CrossRef] [PubMed]
- Manukhina, E.B.; Tseilikman, V.E.; Karpenko, M.N.; Pestereva, N.S.; Tseilikman, O.B.; Komelkova, M.V.; Kondashevskaya, M.V.; Goryacheva, A.V.; Lapshin, M.S.; Platkovskii, P.O.; et al. Intermittent Hypoxic Conditioning Alleviates Post-Traumatic Stress Disorder-Induced Damage and Dysfunction of Rat Visceral Organs and Brain. Int. J. Mol. Sci. 2020, 21, 345. [Google Scholar] [CrossRef] [PubMed]
- Hoskins, M.D.; Bridges, J.; Sinnerton, R.; Nakamura, A.; Underwood, J.F.G.; Slater, A.; Lee, M.R.D.; Clarke, L.; Lewis, C.; Roberts, N.P.; et al. Pharmacological therapy for post-traumatic stress disorder: A systematic review and meta-analysis of monotherapy, augmentation and head-to-head approaches. Eur. J. Psychotraumatol. 2021, 12, 1802920. [Google Scholar] [CrossRef] [PubMed]
- Costa, G.D.M.; Zanatta, F.B.; Ziegelmann, P.K.; Barros, A.J.S.; Mello, C.F. Pharmacological treatments for adults with post-traumatic stress disorder: A network meta-analysis of comparative efficacy and acceptability. J. Psychiatr. Res. 2020, 130, 412–420. [Google Scholar] [CrossRef]
- Bertolini, F.; Robertson, L.; Bisson, J.I.; Meader, N.; Churchill, R.; Ostuzzi, G.; Stein, D.J.; Williams, T.; Barbui, C. Early pharmacological interventions for universal prevention of post-traumatic stress disorder (PTSD). Cochrane Database Syst. Rev. 2022, 2, CD013443. [Google Scholar] [CrossRef]
- Locher, C.; Koechlin, H.; Zion, S.R.; Werner, C.; Pine, D.S.; Kirsch, I.; Kessler, R.C.; Kossowsky, J. Efficacy and Safety of Selective Serotonin Reuptake Inhibitors, Serotonin-Norepinephrine Reuptake Inhibitors, and Placebo for Common Psychiatric Disorders Among Children and Adolescents. JAMA Psychiatry 2017, 74, 1011–1020. [Google Scholar] [CrossRef]
- Rezaeiamiri, E.; Bahramsoltani, R.; Rahimi, R. Plant-derived natural agents as dietary supplements for the regulation of glycosylated hemoglobin: A review of clinical trials. Clin. Nutr. 2020, 39, 331–342. [Google Scholar] [CrossRef]
- Gambini, J.; Inglés, M.; Olaso, G.; Lopez-Grueso, R.; Bonet-Costa, V.; Gimeno-Mallench, L.; Mas-Bargues, C.; Abdelaziz, K.M.; Gomez-Cabrera, M.C.; Vina, J.; et al. Properties of Resveratrol: In Vitro and In Vivo Studies about Metabolism, Bioavailability, and Biological Effects in Animal Models and Humans. Oxid. Med. Cell Longev. 2015, 2015, 837042. [Google Scholar] [CrossRef]
- Li, I.-H.; Shih, J.-H.; Jhao, Y.-T.; Chen, H.-C.; Chiu, C.-H.; Chen, C.-F.F.; Huang, Y.-S.; Shiue, C.-Y.; Ma, K.-H. Regulation of Noise-Induced Loss of Serotonin Transporters with Resveratrol in a Rat Model Using 4-[18F]-ADAM/Small-Animal Positron Emission Tomography. Molecules 2019, 24, 1344. [Google Scholar] [CrossRef]
- Li, G.; Wang, G.; Shi, J.; Xie, X.; Fei, N.; Chen, L.; Liu, N.; Yang, M.; Pan, J.; Huang, W.; et al. trans-Resveratrol ameliorates anxiety-like behaviors and fear memory deficits in a rat model of post-traumatic stress disorder. Neuropharmacology 2018, 133, 181–188. [Google Scholar] [CrossRef]
- Novak, J.; Tseilikman, V.E.; Tseilikman, O.B.; Lazuko, S.S.; Belyeva, L.E.; Rahmani, A.; Fedotova, J. Can Resveratrol Influence the Activity of 11β-Hydroxysteroid Dehydrogenase Type 1? A Combined In Silico and In Vivo Study. Pharmaceuticals 2023, 16, 251. [Google Scholar] [CrossRef]
- Basheer, L.; Schultz, K.; Guttman, Y.; Kerem, Z. In silico and in vitro inhibition of cytochrome P450 3A by synthetic stilbenoids. Food Chem. 2017, 237, 895–903. [Google Scholar] [CrossRef] [PubMed]
- Tseilikman, V.; Dremencov, E.; Tseilikman, O.; Pavlovicova, M.; Lacinova, L.; Jezova, D. Role of glucocorticoid- and monoamine-metabolizing enzymes in stress-related psychopathological processes. Stress 2020, 23, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-C. Beware of docking! Trends Pharmacol. Sci. 2015, 36, 78–95. [Google Scholar] [CrossRef]
- Salentin, S.; Schreiber, S.; Haupt, V.J.; Adasme, M.F.; Schroeder, M. PLIP: Fully automated protein–ligand interaction profiler. Nucleic Acids Res. 2015, 43, W443–W447. [Google Scholar] [CrossRef]
- Kabsch, W.; Sander, C. Dictionary of protein secondary structure: Pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 1983, 22, 2577–2637. [Google Scholar] [CrossRef]
- Novak, J.; Rimac, H.; Kandagalla, S.; Grishina, M.A.; Potemkin, V.A. Can natural products stop the SARS-CoV-2 virus? A docking and molecular dynamics study of a natural product database. Futur. Med. Chem. 2021, 13, 363–378. [Google Scholar] [CrossRef]
- Kandagalla, S.; Novak, J.; Shekarappa, S.B.; Grishina, M.A.; Potemkin, V.A.; Kumbar, B. Exploring potential inhibitors against Kyasanur forest disease by utilizing molecular dynamics simulations and ensemble docking. J. Biomol. Struct. Dyn. 2022, 40, 13547–13563. [Google Scholar] [CrossRef]
- Basheer, L.; Schultz, K.; Fichman, M.; Kerem, Z. Use of In Vitro and Predictive In Silico Models to Study the Inhibition of Cytochrome P4503A by Stilbenes. PLoS ONE 2015, 10, e0141061. [Google Scholar] [CrossRef]
- Basheer, L.; Schultz, K.; Kerem, Z. Inhibition of cytochrome P450 3A by acetoxylated analogues of resveratrol in in vitro and in silico models. Sci. Rep. 2016, 6, 31557. [Google Scholar] [CrossRef]
- Wheeler, S.E.; Bloom, J.W.G. Toward a More Complete Understanding of Noncovalent Interactions Involving Aromatic Rings. J. Phys. Chem. A 2014, 118, 6133–6147. [Google Scholar] [CrossRef] [PubMed]
- Cohen, H.; Zohar, J. An Animal Model of Posttraumatic Stress Disorder: The Use of Cut-Off Behavioral Criteria. Ann. N. Y. Acad. Sci. 2004, 1032, 167–178. [Google Scholar] [CrossRef] [PubMed]
- Manukhina, E.B.; Tseilikman, V.E.; Komelkova, M.V.; Lapshin, M.S.; Goryacheva, A.V.; Kondashevskaya, M.V.; Mkhitarov, V.A.; Lazuko, S.S.; Tseilikman, O.B.; Sarapultsev, A.P.; et al. Cardiac injury in rats with experimental posttraumatic stress disorder and mechanisms of its limitation in experimental posttraumatic stress disorder-resistant rats. J. Appl. Physiol. 2021, 130, 759–771. [Google Scholar] [CrossRef] [PubMed]
- Tseilikman, V.E.; Tseilikman, O.B.; Pashkov, A.A.; Ivleva, I.S.; Karpenko, M.N.; Shatilov, V.A.; Zhukov, M.S.; Fedotova, J.O.; Kondashevskaya, M.V.; Downey, H.F.; et al. Mechanisms of Susceptibility and Resilience to PTSD: Role of Dopamine Metabolism and BDNF Expression in the Hippocampus. Int. J. Mol. Sci. 2022, 23, 14575. [Google Scholar] [CrossRef] [PubMed]
- Komelkova, M.; Manukhina, E.; Downey, H.F.; Sarapultsev, A.; Cherkasova, O.; Kotomtsev, V.; Platkovskiy, P.; Fedorov, S.; Sarapultsev, P.; Tseilikman, O.; et al. Hexobarbital Sleep Test for Predicting the Susceptibility or Resistance to Experimental Posttraumatic Stress Disorder. Int. J. Mol. Sci. 2020, 21, 5900. [Google Scholar] [CrossRef]
- Tseilikman, V.; Lapshin, M.; Klebanov, I.; Chrousos, G.; Vasilieva, M.; Pashkov, A.; Fedotova, J.; Tseilikman, D.; Shatilov, V.; Manukhina, E.; et al. The Link between Activities of Hepatic 11beta-Hydroxysteroid Dehydrogenase-1 and Monoamine Oxidase-A in the Brain Following Repeated Predator Stress: Focus on Heightened Anxiety. Int. J. Mol. Sci. 2022, 23, 4881. [Google Scholar] [CrossRef] [PubMed]
- Sarapultsev, A.; Sarapultsev, P.; Dremencov, E.; Komelkova, M.; Tseilikman, O.; Tseilikman, V. Low glucocorticoids in stress-related disorders: The role of inflammation. Stress 2020, 23, 651–661. [Google Scholar] [CrossRef]
- Lazuko, S.S.; Kuzhel, O.P.; Belyaeva, L.E.; Manukhina, E.B.; Downey, H.F.; Tseilikman, O.B.; Komelkova, M.V.; Tseilikman, V.E. Posttraumatic Stress Disorder Disturbs Coronary Tone and Its Regulatory Mechanisms. Cell. Mol. Neurobiol. 2018, 38, 209–217. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. PubChem in 2021: New data content and improved web interfaces. Nucleic Acids Res. 2021, 49, D1388–D1395. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wang, W.; Kollman, P.A.; Case, D.A. Automatic atom type and bond type perception in molecular mechanical calculations. J. Mol. Graph. Model. 2006, 25, 247–260. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wolf, R.M.; Caldwell, J.W.; Kollman, P.A.; Case, D.A. Development and testing of a general amber force field. J. Comput. Chem. 2004, 25, 1157–1174. [Google Scholar] [CrossRef]
- Shahrokh, K.; Orendt, A.; Yost, G.S.; Cheatham, T.E. Quantum mechanically derived AMBER-compatible heme parameters for various states of the cytochrome P450 catalytic cycle. J. Comput. Chem. 2012, 33, 119–133. [Google Scholar] [CrossRef]
- Tian, C.; Kasavajhala, K.; Belfon, K.A.A.; Raguette, L.; Huang, H.; Migues, A.N.; Bickel, J.; Wang, Y.; Pincay, J.; Wu, Q.; et al. ff19SB: Amino-Acid-Specific Protein Backbone Parameters Trained against Quantum Mechanics Energy Surfaces in Solution. J. Chem. Theory Comput. 2020, 16, 528–552. [Google Scholar] [CrossRef]
- Dolinsky, T.J.; Czodrowski, P.; Li, H.; Nielsen, J.E.; Jensen, J.H.; Klebe, G.; Baker, N.A. PDB2PQR: Expanding and upgrading automated preparation of biomolecular structures for molecular simulations. Nucleic Acids Res. 2007, 35, W522–W525. [Google Scholar] [CrossRef]
- Machado, M.R.; Pantano, S. Split the Charge Difference in Two! A Rule of Thumb for Adding Proper Amounts of Ions in MD Simulations. J. Chem. Theory Comput. 2020, 16, 1367–1372. [Google Scholar] [CrossRef]
- Darden, T.; York, D.; Pedersen, L. Particle mesh Ewald: An N log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993, 98, 10089–10092. [Google Scholar] [CrossRef]
- Case, D.A.; Betz, R.M.; Cerutti, D.S.; Cheatham, T.E., III; Darden, T.A.; Duke, R.E.; Giese, T.J.; Gohlke, H.; Goetz, A.W.; Homeyer, N.; et al. Amber 2016; University of California: San Francisco, CA, USA, 2016. [Google Scholar]
- Genheden, S.; Ryde, U. The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expert Opin. Drug Discov. 2015, 10, 449–461. [Google Scholar] [CrossRef]
- Rastelli, G.; Del Rio, A.; Degliesposti, G.; Sgobba, M. Fast and accurate predictions of binding free energies using MM-PBSA and MM-GBSA. J. Comput. Chem. 2010, 31, 797–810. [Google Scholar] [CrossRef] [PubMed]
- Hünenberger, P.; Mark, A.; Van Gunsteren, W. Fluctuation and Cross-correlation Analysis of Protein Motions Observed in Nanosecond Molecular Dynamics Simulations. J. Mol. Biol. 1995, 252, 492–503. [Google Scholar] [CrossRef] [PubMed]
- McCammon, J.A. Protein dynamics. Rep. Prog. Phys. 1984, 47, 1–46. [Google Scholar] [CrossRef]
- Roe, D.R.; Cheatham, T.E., III. PTRAJ and CPPTRAJ: Software for Processing and Analysis of Molecular Dynamics Trajectory Data. J. Chem. Theory Comput. 2013, 9, 3084–3095. [Google Scholar] [CrossRef] [PubMed]
- Waskom, M.L. seaborn: Statistical data visualization. J. Open Source Softw. 2021, 6, 3021. [Google Scholar] [CrossRef]
- Laskowski, R.A.; Jabłońska, J.; Pravda, L.; Vařeková, R.S.; Thornton, J. PDBsum: Structural summaries of PDB entries. Protein Sci. 2018, 27, 129–134. [Google Scholar] [CrossRef]

| Variable | Control | PTSD | TSR | TRR |
|---|---|---|---|---|
| n | 12 | 12 | 14 | 8 |
| Anxiety index | 0.71 ± 0.07 | 0.83 ± 0.059 *** | 0.67 ± 0.14 | 0.84 ± 0.06 |
| Number of entries into the closed arms | 3 ± 1.5 | 7 ± 1.5 | 6 ± 2.81 | 6 ± 0.7 |
| Number of entries into the open arms | 3 ± 1.1 | 2 ± 0.5 | 3 ± 2.8 | 2 ± 0.3 |
| Time in the closed arms | 415.83 ± 108.2 | 524.16 ± 38.4 * | 423 ± 17.6 ## | 563.7 ± 17.5 ++ |
| Time in the open arms | 184.17 ± 108.2 | 75.84 ± 38.4 * | 177 ± 17.6 ## | 36.3 ± 17.6 ++ |
| Cluster | Conformation | Cluster Population | d a | csd b | RMSD against A |
|---|---|---|---|---|---|
| 1 | A | 0.579 | 2.306 | 0.218 | 0 |
| 2 | B | 0.350 | 2.400 | 0.230 | 1.728 |
| 3 | C | 0.071 | 2.287 | 0.358 | 2.221 |
| ΔGbind | ΔEvdW | ΔEelectrostatic | ΔGGB | ΔGSA | TΔS | |
|---|---|---|---|---|---|---|
| RES | −21.4 ± 2.4 | −30.8 ± 1.3 | −18.3 ± 3.6 | 32.0 ± 2.3 | −4.2 ± 0.2 | −18.7 |
| COL | −31.0 ± 2.6 | −43.3 ± 2.3 | −19.7 ± 5.3 | 37.4 ± 4.9 | −5.5 ± 0.3 | −24.1 |
| CNE | −37.6 ± 3.0 | −42.2 ± 2.5 | −23.5 ± 6.3 | 33.4 ± 2.7 | −5.3 ± 0.3 | −22.9 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tseilikman, V.E.; Fedotova, J.O.; Tseilikman, O.B.; Novak, J.; Karpenko, M.N.; Maistrenko, V.A.; Lazuko, S.S.; Belyeva, L.E.; Kamel, M.; Buhler, A.V.; et al. Resistance to Resveratrol Treatment in Experimental PTSD Is Associated with Abnormalities in Hepatic Metabolism of Glucocorticoids. Int. J. Mol. Sci. 2023, 24, 9333. https://doi.org/10.3390/ijms24119333
Tseilikman VE, Fedotova JO, Tseilikman OB, Novak J, Karpenko MN, Maistrenko VA, Lazuko SS, Belyeva LE, Kamel M, Buhler AV, et al. Resistance to Resveratrol Treatment in Experimental PTSD Is Associated with Abnormalities in Hepatic Metabolism of Glucocorticoids. International Journal of Molecular Sciences. 2023; 24(11):9333. https://doi.org/10.3390/ijms24119333
Chicago/Turabian StyleTseilikman, Vadim E., Julia O. Fedotova, Olga B. Tseilikman, Jurica Novak, Marina N. Karpenko, Victoria A. Maistrenko, Svetlana S. Lazuko, Lyudmila E. Belyeva, Mustapha Kamel, Alexey V. Buhler, and et al. 2023. "Resistance to Resveratrol Treatment in Experimental PTSD Is Associated with Abnormalities in Hepatic Metabolism of Glucocorticoids" International Journal of Molecular Sciences 24, no. 11: 9333. https://doi.org/10.3390/ijms24119333
APA StyleTseilikman, V. E., Fedotova, J. O., Tseilikman, O. B., Novak, J., Karpenko, M. N., Maistrenko, V. A., Lazuko, S. S., Belyeva, L. E., Kamel, M., Buhler, A. V., & Kovaleva, E. G. (2023). Resistance to Resveratrol Treatment in Experimental PTSD Is Associated with Abnormalities in Hepatic Metabolism of Glucocorticoids. International Journal of Molecular Sciences, 24(11), 9333. https://doi.org/10.3390/ijms24119333