DNA Polymerases for Whole Genome Amplification: Considerations and Future Directions
Abstract
1. Introduction
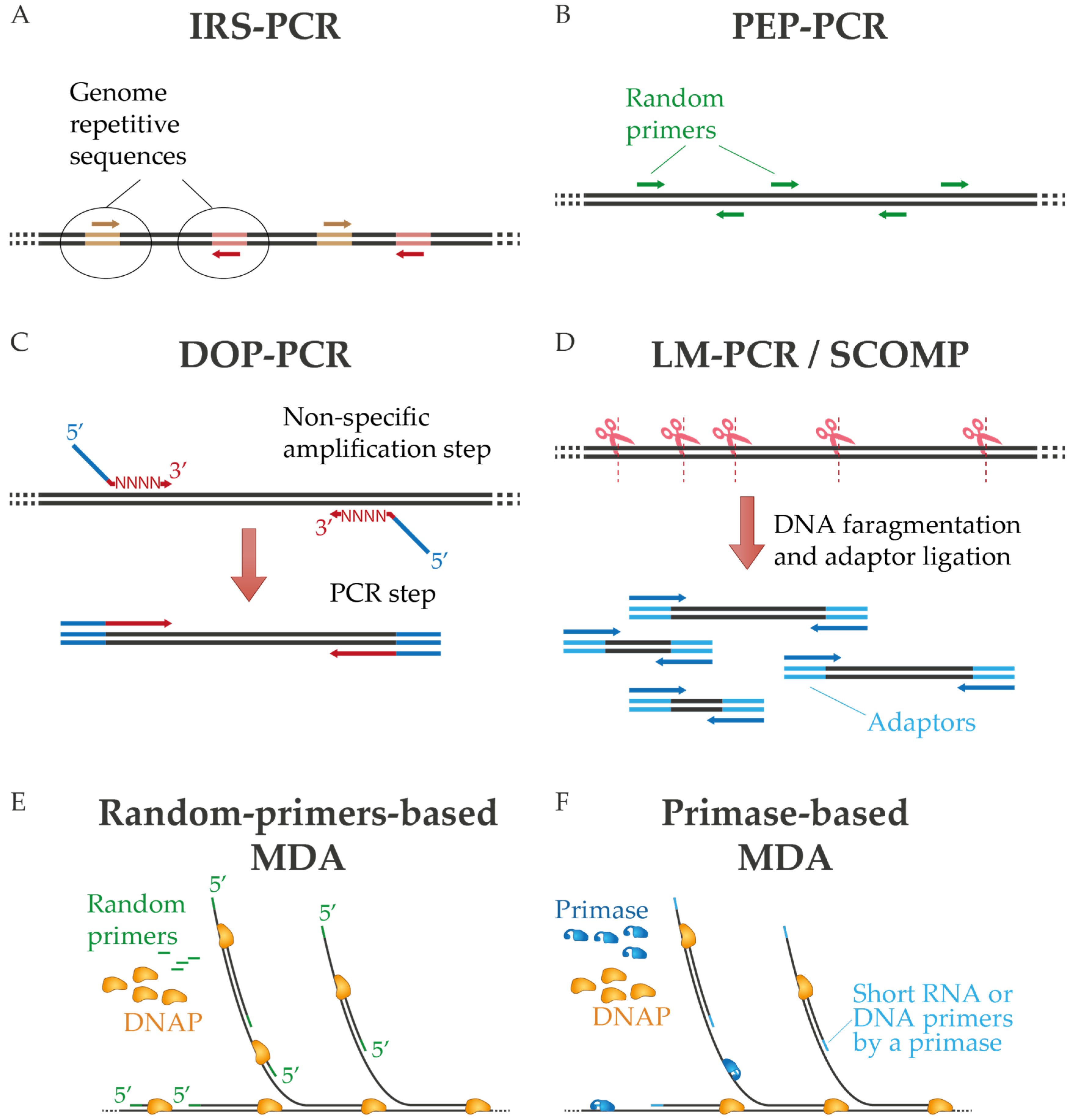
2. Overview of Whole Genome Amplification Methods
2.1. WGA Protocols Based on PCR
2.2. WGA Protocols Based on Multiple Displacement Amplification (MDA)
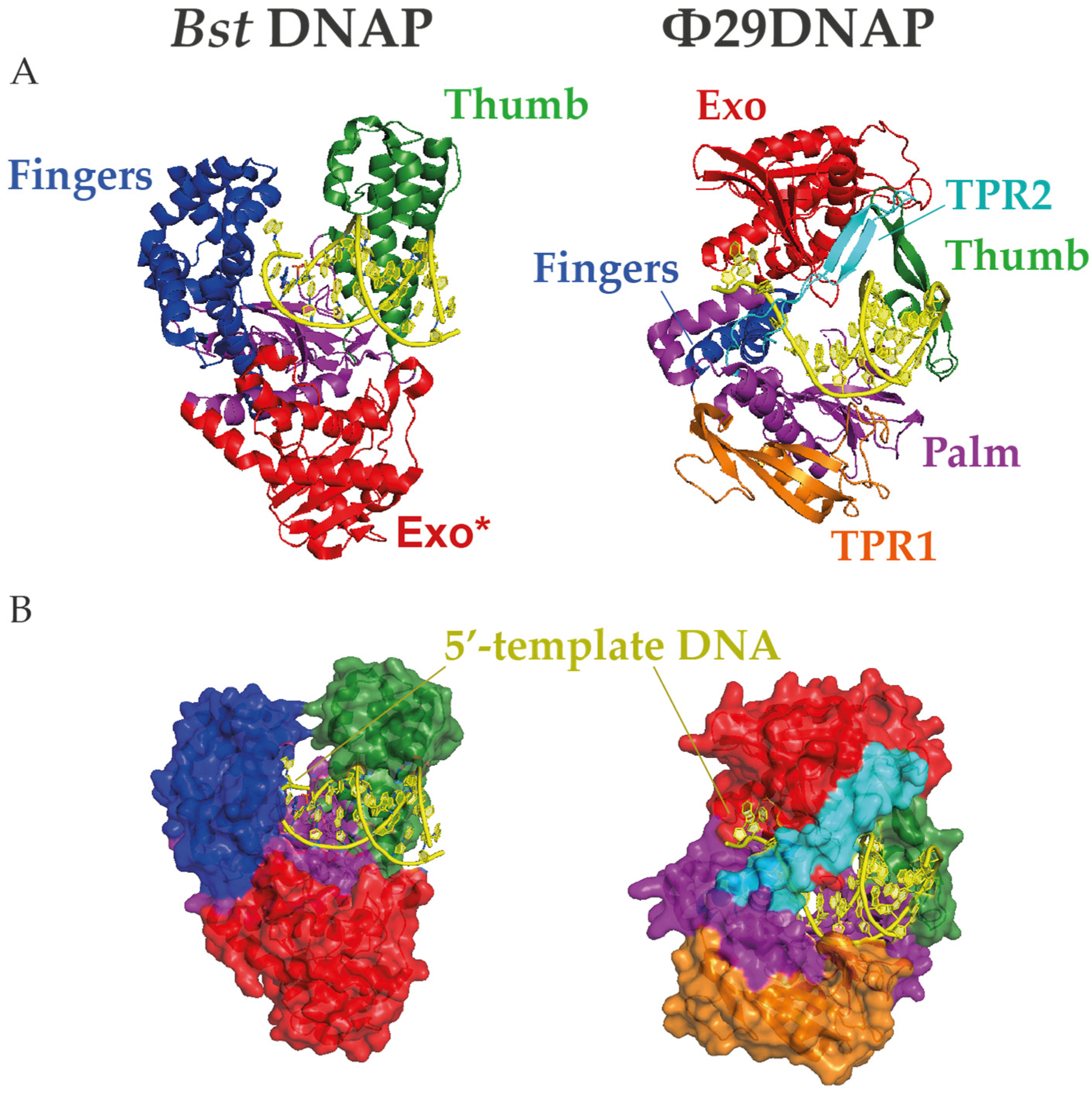
3. Main DNA Polymerases in WGA
4. Key Features of DNA Polymerase for Accurate WGA
4.1. Processivity of DNA Synthesis
4.2. Strand Displacement Capacity
| DNA Polymerase | Family | Processivity | 5′-3′ Exo | 3′-5′ Exo | Strand Displacement | Average Error Rate | TLS | Optimum Temp. (°C) |
|---|---|---|---|---|---|---|---|---|
| Taq | A | 20–50 nt [78,79] | +/− c | − | − | 1–2 × 10−4 | THF [80] | 70–75 |
| Pwo | B | <Taq [81] a | − | + | − | 2.4 × 10−6 | ND | 72 b |
| Bst | A | <Φ29DNAP [73] a | +/− c | − | + | 1.5 × 10−5 | ND | 65 |
| Φ29DNAP | B | >40 Kb [33,35,73] | − | + | + | 5 × 10−6 | [74] | 30 |
| B35DNAP | B | >23 Kb [74] | − | + | + | 5 × 10−6 | THF, Tg, T:T [74,82] | 37 |
4.3. Fidelity and Accuracy in DNA Replication and Amplification
4.4. Damage Bypass and Translesion DNA Synthesis
4.5. Thermoresistance
5. The Impact of DNA Polymerases on Limitations and Weaknesses of WGA
6. Improvement of WGA by DNA Polymerases Engineering
6.1. Targeted Mutagenesis
6.2. Directed Evolution
6.3. Fusion or Chimeric Enzymes
7. Concluding Remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Aschenbrenner, J.; Marx, A. DNA Polymerases and Biotechnological Applications. Curr. Opin. Biotechnol. 2017, 48, 187–195. [Google Scholar] [CrossRef]
- Garcia-Heredia, I.; Bhattacharjee, A.S.; Fornas, O.; Gomez, M.L.; Martínez, J.M.; Martinez-Garcia, M. Benchmarking of Single-Virus Genomics: A New Tool for Uncovering the Virosphere. Environ. Microbiol. 2021, 23, 1584–1593. [Google Scholar] [CrossRef] [PubMed]
- Khan, T.; Becker, T.M.; Po, J.W.; Chua, W.; Ma, Y. Single-Circulating Tumor Cell Whole Genome Amplification to Unravel Cancer Heterogeneity and Actionable Biomarkers. Int. J. Mol. Sci. 2022, 23, 8386. [Google Scholar] [CrossRef] [PubMed]
- Simon, C.; Daniel, R. Construction of Small-Insert and Large-Insert Metagenomic Libraries. Methods Mol. Biol. Clifton NJ 2023, 2555, 1–12. [Google Scholar] [CrossRef]
- Czyz, Z.T.; Kirsch, S.; Polzer, B. Principles of Whole-Genome Amplification. Methods Mol. Biol. 2015, 1347, 1–14. [Google Scholar] [CrossRef]
- Hawkins, T.L.; Detter, J.C.; Richardson, P.M. Whole Genome Amplification—Applications and Advances. Curr. Opin. Biotechnol. 2002, 13, 65–67. [Google Scholar] [CrossRef]
- Sims, D.; Sudbery, I.; Ilott, N.E.; Heger, A.; Ponting, C.P. Sequencing Depth and Coverage: Key Considerations in Genomic Analyses. Nat. Rev. Genet. 2014, 15, 121–132. [Google Scholar] [CrossRef]
- Wang, X.; Liu, Y.; Liu, H.; Pan, W.; Ren, J.; Zheng, X.; Tan, Y.; Chen, Z.; Deng, Y.; He, N.; et al. Recent Advances and Application of Whole Genome Amplification in Molecular Diagnosis and Medicine. MedComm 2022, 3, e116. [Google Scholar] [CrossRef]
- Huang, L.; Ma, F.; Chapman, A.; Lu, S.; Xie, X.S. Single-Cell Whole-Genome Amplification and Sequencing: Methodology and Applications. Annu. Rev. Genom. Hum. Genet. 2015, 16, 79–102. [Google Scholar] [CrossRef]
- Saiki, R.K.; Scharf, S.; Faloona, F.; Mullis, K.B.; Horn, G.T.; Erlich, H.A.; Arnheim, N. Enzymatic Amplification of β-Globin Genomic Sequences and Restriction Site Analysis for Diagnosis of Sickle Cell Anemia. Science 1985, 230, 1350–1354. [Google Scholar] [CrossRef] [PubMed]
- Mullis, K.; Faloona, F.; Scharf, S.; Saiki, R.; Horn, G.; Erlich, H. Specific Enzymatic Amplification of DNA In Vitro: The Polymerase Chain Reaction. Cold Spring Harb. Symp. Quant. Biol. 1986, 51, 263–273. [Google Scholar] [CrossRef]
- McIver, C.J.; Jacques, C.F.H.; Chow, S.S.W.; Munro, S.C.; Scott, G.M.; Roberts, J.A.; Craig, M.E.; Rawlinson, W.D. Development of Multiplex PCRs for Detection of Common Viral Pathogens and Agents of Congenital Infections. J. Clin. Microbiol. 2005, 43, 5102–5110. [Google Scholar] [CrossRef]
- Wang, W.; Xu, Y.; Gao, R.; Lu, R.; Han, K.; Wu, G.; Tan, W. Detection of SARS-CoV-2 in Different Types of Clinical Specimens. JAMA-J. Am. Med. Assoc. 2020, 323, 1843–1844. [Google Scholar] [CrossRef]
- Śpibida, M.; Krawczyk, B.; Olszewski, M.; Kur, J. Modified DNA Polymerases for PCR Troubleshooting. J. Appl. Genet. 2017, 58, 133–142. [Google Scholar] [CrossRef]
- Himmelbauer, H.; Schalkwyk, L.C.; Lehrach, H. Interspersed Repetitive Sequence (IRS)-PCR for Typing of Whole Genome Radiation Hybrid Panels. Nucleic Acids Res. 2000, 28, e7. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Nelson, D.L. Interspersed Repetitive Sequence Polymerase Chain Reaction (IRS PCR) for Generation of Human DNA Fragments from Complex Sources. Methods 1991, 2, 60–74. [Google Scholar] [CrossRef]
- Reyes, G.R.; Kim, J.P. Sequence-Independent, Single-Primer Amplification (SISPA) of Complex DNA Populations. Mol. Cell. Probes 1991, 5, 473–481. [Google Scholar] [CrossRef]
- Froussard, P. RPCR: A Powerful Tool for Random Amplification of Whole RNA Sequences. Genome Res. 1993, 2, 185–190. [Google Scholar] [CrossRef]
- Zou, N.; Ditty, S.; Li, B.; Lo, S.C. Random Priming PCR Strategy to Amplify and Clone Trace Amounts of DNA. BioTechniques 2003, 35, 758–765. [Google Scholar] [CrossRef]
- Brinkmann, A.; Uddin, S.; Krause, E.; Surtees, R.; Dinçer, E.; Kar, S.; Hacıoğlu, S.; Özkul, A.; Ergünay, K.; Nitsche, A. Utility of a Sequence-Independent, Single-Primer-Amplification (SISPA) and Nanopore Sequencing Approach for Detection and Characterization of Tick-Borne Viral Pathogens. Viruses 2021, 13, 203. [Google Scholar] [CrossRef] [PubMed]
- Fouts, D.E. Amplification for Whole Genome Sequencing of Bacteriophages from Single Isolated Plaques Using SISPA. Bacteriophages 2018, 3, 165–178. [Google Scholar] [CrossRef]
- Blagodatskikh, K.A.; Kramarov, V.M.; Barsova, E.V.; Garkovenko, A.V.; Shcherbo, D.S.; Shelenkov, A.A.; Ustinova, V.V.; Tokarenko, M.R.; Baker, S.C.; Kramarova, T.V.; et al. Improved DOP-PCR (IDOP-PCR): A Robust and Simple WGA Method for Efficient Amplification of Low Copy Number Genomic DNA. PLoS ONE 2017, 12, e0184507. [Google Scholar] [CrossRef]
- Telenius, H.; Carter, N.P.; Bebb, C.E.; Nordenskjöld, M.; Ponder, B.A.J.; Tunnacliffe, A. Degenerate Oligonucleotide-Primed PCR: General Amplification of Target DNA by a Single Degenerate Primer. Genomics 1992, 13, 718–725. [Google Scholar] [CrossRef]
- Arneson, N.; Hughes, S.; Houlston, R.; Done, S. Whole-Genome Amplification by Improved Primer Extension Preamplification PCR (I-PEP-PCR). Cold Spring Harb. Protoc. 2008, 3, pdb.prot4921. [Google Scholar] [CrossRef][Green Version]
- Moghaddaszadeh-Ahrabi, S.; Farajnia, S.; Rahimi-Mianji, G.; Nejati-Javaremi, A. A Short and Simple Improved-Primer Extension Preamplification (I-PEP) Procedure for Whole Genome Amplification (WGA) of Bovine Cells. Anim. Biotechnol. 2012, 23, 24–42. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Cui, X.; Schmitt, K.; Hubert, R.; Navidi, W.; Arnheim, N. Whole Genome Amplification from a Single Cell: Implications for Genetic Analysis. Proc. Natl. Acad. Sci. USA 1992, 89, 5847–5851. [Google Scholar] [CrossRef] [PubMed]
- Tagoh, H.; Cockerill, P.N.; Bonifer, C. In Vivo Genomic Footprinting Using LM–PCR Methods. In Nuclear Reprogramming: Methods and Protocols; Pells, S., Ed.; Methods in Molecular BiologyTM; Humana Press: Totowa, NJ, USA, 2006; pp. 285–314. ISBN 978-1-59745-005-8. [Google Scholar]
- Arneson, N.; Hughes, S.; Houlston, R.; Done, S. Whole-Genome Amplification by Single-Cell Comparative Genomic Hybridization PCR (SCOMP). Cold Spring Harb. Protoc. 2008, 3, pdb.prot4923. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Stoecklein, N.H.; Erbersdobler, A.; Schmidt-Kittler, O.; Diebold, J.; Schardt, J.A.; Izbicki, J.R.; Klein, C.A. SCOMP Is Superior to Degenerated Oligonucleotide Primed-Polymerase Chain Reaction for Global Amplification of Minute Amounts of DNA from Microdissected Archival Tissue Samples. Am. J. Pathol. 2002, 161, 43–51. [Google Scholar] [CrossRef]
- Jäger, R. New Perspectives for Whole Genome Amplification in Forensic STR Analysis. Int. J. Mol. Sci. 2022, 23, 7090. [Google Scholar] [CrossRef]
- Volozonoka, L.; Miskova, A.; Gailite, L. Whole Genome Amplification in Preimplantation Genetic Testing in the Era of Massively Parallel Sequencing. Int. J. Mol. Sci. 2022, 23, 4819. [Google Scholar] [CrossRef]
- Bodulev, O.L.; Sakharov, I.Y. Isothermal Nucleic Acid Amplification Techniques and Their Use in Bioanalysis. Biochem. Mosc. 2020, 85, 147–166. [Google Scholar] [CrossRef] [PubMed]
- Dean, F.B.; Nelson, J.R.; Giesler, T.L.; Lasken, R.S. Rapid Amplification of Plasmid and Phage DNA Using Phi29 DNA Polymerase and Multiply-Primed Rolling Circle Amplification. Genome Res. 2001, 11, 1095–1099. [Google Scholar] [CrossRef] [PubMed]
- Salas, M.; Blanco, L.; Lázaro, J.M.; De Vega, M. The Bacteriophage Φ29 DNA Polymerase. IUBMB Life 2008, 60, 82–85. [Google Scholar] [CrossRef]
- Blanco, L.; Bernad, A.; Lázaro, J.M.; Martín, G.; Garmendia, C.; Salas, M. Highly Efficient DNA Synthesis by the Phage Φ29 DNA Polymerase: Symmetrical Mode of DNA Replication. J. Biol. Chem. 1989, 264, 8935–8940. [Google Scholar] [CrossRef]
- Asadi, R.; Mollasalehi, H. The Mechanism and Improvements to the Isothermal Amplification of Nucleic Acids, at a Glance. Anal. Biochem. 2021, 631, 114260. [Google Scholar] [CrossRef] [PubMed]
- Nelson, J.R. Random-Primed, Phi29 DNA Polymerase-Based Whole Genome Amplification. Curr. Protoc. Mol. Biol. 2014, 105, 15.13. [Google Scholar] [CrossRef] [PubMed]
- Lage, J.M.; Leamon, J.H.; Pejovic, T.; Hamann, S.; Lacey, M.; Dillon, D.; Segraves, R.; Vossbrinck, B.; González, A.; Pinkel, D.; et al. Whole Genome Analysis of Genetic Alterations in Small DNA Samples Using Hyperbranched Strand Displacement Amplification and Array–CGH. Genome Res. 2003, 13, 294. [Google Scholar] [CrossRef]
- Sabina, J.; Leamon, J.H. Bias in Whole Genome Amplification: Causes and Considerations. Methods Mol. Biol. 2015, 1347, 15–41. [Google Scholar] [CrossRef]
- Li, Y.; Kim, H.-J.; Zheng, C.; Chow, W.H.A.; Lim, J.; Keenan, B.; Pan, X.; Lemieux, B.; Kong, H. Primase-Based Whole Genome Amplification. Nucleic Acids Res. 2008, 36, e79. [Google Scholar] [CrossRef][Green Version]
- Schaerli, Y.; Stein, V.; Spiering, M.M.; Benkovic, S.J.; Abell, C.; Hollfelder, F. Isothermal DNA Amplification Using the T4 Replisome: Circular Nicking Endonuclease-Dependent Amplification and Primase-Based Whole-Genome Amplification. Nucleic Acids Res. 2010, 38, e201. [Google Scholar] [CrossRef][Green Version]
- Picher, Á.J.; Budeus, B.; Wafzig, O.; Krüger, C.; García-Gómez, S.; Martínez-Jiménez, M.I.; Díaz-Talavera, A.; Weber, D.; Blanco, L.; Schneider, A. TruePrime Is a Novel Method for Whole-Genome Amplification from Single Cells Based on TthPrimPol. Nat. Commun. 2016, 7, 13296. [Google Scholar] [CrossRef]
- Martínez-Jiménez, M.I.; García-Gómez, S.; Bebenek, K.; Sastre-Moreno, G.; Calvo, P.A.; Díaz-Talavera, A.; Kunkel, T.A.; Blanco, L. Alternative Solutions and New Scenarios for Translesion DNA Synthesis by Human PrimPol. DNA Repair 2015, 29, 127–138. [Google Scholar] [CrossRef]
- Direito, S.O.L.L.; Zaura, E.; Little, M.; Ehrenfreund, P.; Röling, W.F.M.M. Systematic Evaluation of Bias in Microbial Community Profiles Induced by Whole Genome Amplification. Environ. Microbiol. 2014, 16, 643–657. [Google Scholar] [CrossRef] [PubMed]
- Parras-Moltó, M.; Rodríguez-Galet, A.; Suárez-Rodríguez, P.; López-Bueno, A. Evaluation of Bias Induced by Viral Enrichment and Random Amplification Protocols in Metagenomic Surveys of Saliva DNA Viruses. Microbiome 2018, 6, 119. [Google Scholar] [CrossRef]
- Deleye, L.; De Coninck, D.; Dheedene, A.; De Sutter, P.; Menten, B.; Deforce, D.; Van Nieuwerburgh, F. Performance of a TthPrimPol-Based Whole Genome Amplification Kit for Copy Number Alteration Detection Using Massively Parallel Sequencing. Sci. Rep. 2016, 6, 31825. [Google Scholar] [CrossRef]
- Ordóñez, C.D.; Mayoral-Campos, C.; Egas, C.; Redrejo-Rodríguez, M. A Primer-Independent DNA Polymerase-Based Method for Competent Whole-(Meta)Genome Amplification of Intermediate to High GC Sequences. bioRxiv 2023. [Google Scholar] [CrossRef]
- Redrejo-Rodríguez, M.; Ordóñez, C.D.; Berjón-Otero, M.; Moreno-González, J.; Aparicio-Maldonado, C.; Forterre, P.; Salas, M.; Krupovic, M. Primer-Independent DNA Synthesis by a Family B DNA Polymerase from Self-Replicating Mobile Genetic Elements. Cell Rep. 2017, 21, 1574–1587. [Google Scholar] [CrossRef] [PubMed]
- Zong, C.; Lu, S.; Chapman, A.R.; Xie, X.S. Genome-Wide Detection of Single Nucleotide and Copy Number Variations of a Single Human Cell. Science 2012, 338, 1622–1626. [Google Scholar] [CrossRef]
- Bessman, M.J.; Kornberg, A.; Lehman, I.R.; Simms, E.S. Enzymic Synthesis of Deoxyribonucleic Acid. Biochim. Biophys. Acta 1956, 21, 197–198. [Google Scholar] [CrossRef]
- Kornberg, A.; Baker, T.A. DNA Replication; Wh Freeman: New York, NY, USA, 1992; Volume 3, ISBN 978-0-7167-2003-4. [Google Scholar]
- Bebenek, K.; Kunkel, T.A. Functions of DNA Polymerases. Adv. Protein Chem. 2004, 69, 137–165. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Diaz, M.; Bebenek, K. Multiple Functions of DNA Polymerases. Crit. Rev. Plant Sci. 2007, 26, 105–122. [Google Scholar] [CrossRef] [PubMed]
- Zarrouk, K.; Piret, J.; Boivin, G. Herpesvirus DNA Polymerases: Structures, Functions and Inhibitors. Virus Res. 2017, 234, 177–192. [Google Scholar] [CrossRef]
- Yang, W.; Gao, Y. Translesion and Repair DNA Polymerases: Diverse Structure and Mechanism. Annu. Rev. Biochem. 2018, 87, 239–261. [Google Scholar] [CrossRef]
- Ito, J.; Braithwaite, D.K. Compilation and Alignment of DNA Polymerase Sequences. Nucleic Acids Res. 1991, 19, 4045–4057. [Google Scholar] [CrossRef]
- Ohmori, H.; Friedberg, E.C.; Fuchs, R.P.P.; Goodman, M.F.; Hanaoka, F.; Hinkle, D.; Kunkel, T.A.; Lawrence, C.W.; Livneh, Z.; Nohmi, T.; et al. The Y-Family of DNA Polymerases. Mol. Cell 2001, 8, 7–8. [Google Scholar] [CrossRef]
- Temin, H.M.; Mizutani, S. Viral RNA-Dependent DNA Polymerase: RNA-Dependent DNA Polymerase in Virions of Rous Sarcoma Virus. Nature 1970, 226, 1211–1213. [Google Scholar] [CrossRef] [PubMed]
- Baltimore, D. Viral RNA-Dependent DNA Polymerase: RNA-Dependent DNA Polymerase in Virions of RNA Tumour Viruses. Nature 1970, 226, 1209–1211. [Google Scholar] [CrossRef]
- Blanco, L.; Calvo, P.A.; Diaz-Talavera, A.; Carvalho, G.; Calero, N.; Martínez-Carrón, A.; Velázquez-Ruiz, C.; Villadangos, S.; Guerra, S.; Martínez-Jim Enez, M.I. Mechanism of DNA Primer Synthesis by Human PrimPol. Enzymes 2019, 45, 289–310. [Google Scholar]
- Iyer, L.M. Origin and Evolution of the Archaeo-Eukaryotic Primase Superfamily and Related Palm-Domain Proteins: Structural Insights and New Members. Nucleic Acids Res. 2005, 33, 3875–3896. [Google Scholar] [CrossRef]
- Rudd, S.G.; Bianchi, J.; Doherty, A.J. PrimPol-A New Polymerase on the Block. Mol. Cell. Oncol. 2014, 1, e960754. [Google Scholar] [CrossRef]
- Ishino, S.; Ishino, Y. DNA Polymerases as Useful Reagents for Biotechnology—The History of Developmental Research in the Field. Front. Microbiol. 2014, 5, 465. [Google Scholar] [CrossRef] [PubMed]
- Ghasemi, A.; Salmanian, A.H.; Sadeghifard, N.; Salarian, A.A.; Gholi, M.K. Cloning, Expression and Purification of Pwo Polymerase from Pyrococcus woesei. Iran. J. Microbiol. 2011, 3, 118–122. [Google Scholar]
- Kim, S.W.; Kim, D.U.; Kim, J.K.; Kang, L.W.; Cho, H.S. Crystal Structure of Pfu, the High Fidelity DNA Polymerase from Pyrococcus furiosus. Int. J. Biol. Macromol. 2008, 42, 356–361. [Google Scholar] [CrossRef] [PubMed]
- Perler, F.B.; Comb, D.G.; Jack, W.E.; Moran, L.S.; Qiang, B.; Kucera, R.B.; Benner, J.; Slatko, B.E.; Nwankwo, D.O.; Hempstead, S.K. Intervening Sequences in an Archaea DNA Polymerase Gene. Proc. Natl. Acad. Sci. USA 1992, 89, 5577–5581. [Google Scholar] [CrossRef] [PubMed]
- de Vega, M.; Salas, M. Bacteriophage Φ29 DNA Polymerase: An Outstanding Replicase. In Bacterial DNA, DNA Polymerase and DNA Helicases; Knudsen, W.D., Bruns, S.S., Eds.; Nova Science Publishers, Inc.: New York, NY, USA, 2009; ISBN 978-1-60741-094-2. [Google Scholar]
- Estévez-Gómez, N.; Prieto, T.; Guillaumet-Adkins, A.; Heyn, H.; Prado-López, S.; Posada, D. Comparison of Single-Cell Whole-Genome Amplification Strategies. bioRxiv 2018, 443754. [Google Scholar] [CrossRef]
- Lechuga, A. Disclosing Bacillus Virus BAM35 and Its Host. Identification and Characterization of the Viral SSB, Host Genomic Characterization and Phage-Bacteria Interactome. Ph.D. Thesis, Universidad Autónoma de Madrid, Madrid, Spain, 2020. [Google Scholar]
- Soengas, M.S.; Gutiérrez, C.; Salas, M. Helix-Destabilizing Activity of Φ29 Single-Stranded DNA Binding Protein: Effect on the Elongation Rate During Strand Displacement DNA Replication. J. Mol. Biol. 1995, 253, 517–529. [Google Scholar] [CrossRef]
- Inoue, J.; Shigemori, Y.; Mikawa, T. Improvements of Rolling Circle Amplification (RCA) Efficiency and Accuracy Using Thermus Thermophilus SSB Mutant Protein. Nucleic Acids Res. 2006, 34, e69. [Google Scholar] [CrossRef]
- Tate, C.M.; Nuñez, A.N.; Goldstein, C.A.; Gomes, I.; Robertson, J.M.; Kavlick, M.F.; Budowle, B. Evaluation of Circular DNA Substrates for Whole Genome Amplification Prior to Forensic Analysis. Forensic Sci. Int. Genet. 2012, 6, 185–190. [Google Scholar] [CrossRef]
- Chen, J.; Baker, Y.R.; Brown, A.; El-Sagheer, A.H.; Brown, T. Enzyme-Free Synthesis of Cyclic Single-Stranded DNA Constructs Containing a Single Triazole, Amide or Phosphoramidate Backbone Linkage and Their Use as Templates for Rolling Circle Amplification and Nanoflower Formation. Chem. Sci. 2018, 9, 8110–8120. [Google Scholar] [CrossRef]
- Berjon-Otero, M.; Villar, L.; de Vega, M.; Salas, M.; Redrejo-Rodriguez, M. DNA Polymerase from Temperate Phage Bam35 Is Endowed with Processive Polymerization and Abasic Sites Translesion Synthesis Capacity. Proc. Natl. Acad. Sci. USA 2015, 112, E3476–E3484. [Google Scholar] [CrossRef]
- Longás, E.; de Vega, M.; Lázaro, J.M.; Salas, M. Functional Characterization of Highly Processive Protein-Primed DNA Polymerases from Phages Nf and GA-1, Endowed with a Potent Strand Displacement Capacity. Nucleic Acids Res. 2006, 34, 6051–6063. [Google Scholar] [CrossRef] [PubMed]
- Ignatov, K.B.; Barsova, E.V.; Fradkov, A.F.; Blagodatskikh, K.A.; Kramarova, T.V.; Kramarov, V.M. A Strong Strand Displacement Activity of Thermostable DNA Polymerase Markedly Improves the Results of DNA Amplification. BioTechniques 2014, 57, 81–87. [Google Scholar] [CrossRef] [PubMed]
- Kamtekar, S.; Berman, A.J.; Wang, J.M.; Lazaro, J.M.; de Vega, M.; Blanco, L.; Salas, M.; Steitz, T.A. Insights into Strand Displacement and Processivity from the Crystal Structure of the Protein-Primed DNA Polymerase of Bacteriophage Phi. Mol. Cell 2004, 16, 1035–1036. [Google Scholar] [CrossRef]
- Merkens, L.S.; Bryan, S.K.; Moses, R.E. Inactivation of the 5′-3′ Exonuclease of Thermus Aquaticus DNA Polymerase. Biochim. Biophys. Acta 1995, 1264, 243–248. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Prosen, D.E.; Mei, L.; Sullivan, J.C.; Finney, M.; Vander Horn, P.B. A Novel Strategy to Engineer DNA Polymerases for Enhanced Processivity and Improved Performance in Vitro. Nucleic Acids Res 2004, 32, 1197–1207. [Google Scholar] [CrossRef]
- Belousova, E.A.; Rechkunova, N.I.; Lavrik, O.I. Thermostable DNA Polymerases Can Perform Translesion Synthesis Using 8-Oxoguanine and Tetrahydrofuran-Containing DNA Templates. Biochim. Biophys. Acta BBA-Proteins Proteom. 2006, 1764, 97–104. [Google Scholar] [CrossRef]
- Olejniczak, M.; Kozlowski, P.; Sobczak, K.; Krzyzosiak, W.J. Accurate and Sensitive Analysis of Triplet Repeat Expansions by Capillary Electrophoresis. Electrophoresis 2005, 26, 2198–2207. [Google Scholar] [CrossRef]
- Díaz Michelena, M.; Kilian, R.; Baeza, O.; Rios, F.; Rivero, M.Á.; Mesa, J.L.; González, V.; Ordoñez, A.A.; Langlais, B.; Rocca, M.C.L.; et al. The Formation of a Giant Collapse Caprock Sinkhole on the Barda Negra Plateau Basalts (Argentina): Magnetic, Mineralogical and Morphostructural Evidences. Geomorphology 2020, 367, 107297. [Google Scholar] [CrossRef]
- Dąbrowski, S.; Kur, J. Cloning and Expression in Escherichia coli of the Recombinant His-Tagged DNA Polymerases from Pyrococcus furiosus and Pyrococcus woesei. Protein Expr. Purif. 1998, 14, 131–138. [Google Scholar] [CrossRef]
- Pwo Super Yield DNA Polymerase Suitable for PCR. Available online: http://www.sigmaaldrich.com/ (accessed on 11 April 2023).
- Green, M.R.; Sambrook, J. E. Coli DNA Polymerase I and the Klenow Fragment. Cold Spring Harb. Protoc. 2020, 2020, 100743. [Google Scholar] [CrossRef]
- Walker, G.T.; Little, M.C.; Nadeau, J.G.; Shank, D.D. Isothermal in Vitro Amplification of DNA by a Restriction Enzyme/DNA Polymerase System. Proc. Natl. Acad. Sci. USA 1992, 89, 392–396. [Google Scholar] [CrossRef]
- Phang, S.-M.; Teo, C.-Y.; Lo, E.; Thi Wong, V.W. Cloning and Complete Sequence of the DNA Polymerase-Encoding Gene (BstpolI) and Characterisation of the Klenow-Like Fragment from Bacillus Stearothermophilus. Gene 1995, 163, 65–68. [Google Scholar] [CrossRef]
- Aliotta, J.M.; Pelletier, J.J.; Ware, J.L.; Moran, L.S.; Benner, J.S.; Kong, H. Thermostable Bst DNA Polymerase I Lacks a 3′ → 5′ Proofreading Exonuclease Activity. Genet. Anal.-Biomol. Eng. 1996, 12, 185–195. [Google Scholar] [CrossRef]
- Manosas, M.; Spiering, M.M.; Ding, F.; Bensimon, D.; Allemand, J.-F.; Benkovic, S.J.; Croquette, V. Mechanism of Strand Displacement Synthesis by DNA Replicative Polymerases. Nucleic Acids Res. 2012, 40, 6174–6186. [Google Scholar] [CrossRef]
- Rodriguez, I.; Lazaro, J.M.; Blanco, L.; Kamtekar, S.; Berman, A.J.; Wang, J.; Steitz, T.A.; Salas, M.; de Vega, M. A Specific Subdomain in 29 DNA Polymerase Confers Both Processivity and Strand-Displacement Capacity. Proc. Natl. Acad. Sci. USA 2005, 102, 6407–6412. [Google Scholar] [CrossRef]
- Rodriguez, I.; Lazaro, J.M.; Salas, M.; de Vega, M. Involvement of the TPR2 Subdomain Movement in the Activities of Phi29 DNA Polymerase. Nucleic Acids Res. 2009, 37, 193–203. [Google Scholar] [CrossRef]
- de Paz, A.M.; Cybulski, T.R.; Marblestone, A.H.; Zamft, B.M.; Church, G.M.; Boyden, E.S.; Kording, K.P.; Tyo, K.E.J. High-Resolution Mapping of DNA Polymerase Fidelity Using Nucleotide Imbalances and next-Generation Sequencing. Nucleic Acids Res 2018, 46, e78. [Google Scholar] [CrossRef]
- McInerney, P.; Adams, P.; Hadi, M.Z. Error Rate Comparison during Polymerase Chain Reaction by DNA Polymerase. Mol. Biol. Int. 2014, 2014, 287430. [Google Scholar] [CrossRef]
- McCulloch, S.D.; Kunkel, T.A. The Fidelity of DNA Synthesis by Eukaryotic Replicative and Translesion Synthesis Polymerases. Cell Res. 2008, 18, 148–161. [Google Scholar] [CrossRef]
- Shcherbakova, P.V.; Pavlov, Y.I.; Chilkova, O.; Rogozin, I.B.; Johansson, E.; Kunkel, T.A. Unique Error Signature of the Four-Subunit Yeast DNA Polymerase ϵ. J. Biol. Chem. 2003, 278, 43770–43780. [Google Scholar] [CrossRef]
- Bernad, A.; Blanco, L.; Lázaro, J.M.; Martín, G.; Salas, M. A Conserved 3′ → 5′ Exonuclease Active Site in Prokaryotic and Eukaryotic DNA Polymerases. Cell 1989, 59, 219–228. [Google Scholar] [CrossRef]
- Reha-Krantz, L.J. DNA Polymerase Proofreading: Multiple Roles Maintain Genome Stability. Biochim. Biophys. Acta-Proteins Proteom. 2010, 1804, 1049–1063. [Google Scholar] [CrossRef]
- Eckert, K.A.; Kunkel, T.A. High Fidelity DNA Synthesis by the Thermus Aquaticus DNA Polymerase. Nucleic Acids Res. 1990, 18, 3739–3744. [Google Scholar] [CrossRef]
- Hamilton, S.C.; Farchaus, J.W.; Davis, M.C. DNA Polymerases as Engines for Biotechnology. BioTechniques 2001, 31, 370–383. [Google Scholar] [CrossRef]
- Hong, G.F.; Feng, Z. DNA Polymerase with Proof-Reading 3′-5′ Exonuclease Activity Bacillus Stearothermophilus. U.S. Patent 5747298A, 5 May 1998. [Google Scholar]
- Mattila, P.; Korpela, J.; Tenkanen, T.; Pitkanen, K.; Pitkämem, K. Fidelity of DNA Synthesis by the Thermococcus Litoralis DNA Polymerase—An Exttemely Heat Stable Enzyme with Proofreading Activity. Nucleic Acids Res. 1991, 19, 4967–4973. [Google Scholar] [CrossRef]
- Esteban, J.A.; Salas, M.; Blanco, L. Fidelity of Φ29 DNA Polymerase. Comparison between Protein-Primed Initiation and DNA Polymerization. J. Biol. Chem. 1993, 268, 2719–2726. [Google Scholar] [CrossRef]
- Kunkel, T.A.; Bebenek, K. DNA Replication Fidelity. Annu. Rev. Biochem. 2000, 69, 497–529. [Google Scholar] [CrossRef]
- Kokoska, R.J.; Bebenek, K.; Boudsocq, F.; Woodgate, R.; Kunkel, T.A. Low Fidelity DNA Synthesis by a Y Family DNA Polymerase Due to Misalignment in the Active Site. J. Biol. Chem. 2002, 277, 19633–19638. [Google Scholar] [CrossRef]
- McDonald, J.P. Novel Thermostable Y-Family Polymerases: Applications for the PCR Amplification of Damaged or Ancient DNAs. Nucleic Acids Res. 2006, 34, 1102–1111. [Google Scholar] [CrossRef]
- Zhang, L.; Kang, M.; Xu, J.; Huang, Y. Archaeal DNA Polymerases in Biotechnology. Appl. Microbiol. Biotechnol. 2015, 99, 6585–6597. [Google Scholar] [CrossRef]
- Hartwig, A. Role of Magnesium in Genomic Stability. Mutat. Res. Mol. Mech. Mutagen. 2001, 475, 113–121. [Google Scholar] [CrossRef] [PubMed]
- McIntyre, J. Polymerase Iota—An Odd Sibling among Y Family Polymerases. DNA Repair 2020, 86, 102753. [Google Scholar] [CrossRef] [PubMed]
- Calvo, P.A.; Sastre-Moreno, G.; Perpiñá, C.; Guerra, S.; Martínez-Jiménez, M.I.; Blanco, L. The Invariant Glutamate of Human PrimPol DxE Motif Is Critical for Its Mn2+-Dependent Distinctive Activities. DNA Repair 2019, 77, 65–75. [Google Scholar] [CrossRef] [PubMed]
- Bock, C.W.; Katz, A.K.; Markham, G.D.; Glusker, J.P. Manganese as a Replacement for Magnesium and Zinc: Functional Comparison of the Divalent Ions. J. Am. Chem. Soc. 1999, 121, 7360–7372. [Google Scholar] [CrossRef]
- Pelletier, H.; Sawaya, M.R. Characterization of the Metal Ion Binding Helix−Hairpin−Helix Motifs in Human DNA Polymerase β by X-Ray Structural Analysis. Biochemistry 1996, 35, 12778–12787. [Google Scholar] [CrossRef]
- Villani, G. Effect of Manganese on In Vitro Replication of Damaged DNA Catalyzed by the Herpes Simplex Virus Type-1 DNA Polymerase. Nucleic Acids Res. 2002, 30, 3323–3332. [Google Scholar] [CrossRef]
- Kumar Vashishtha, A.; Konigsberg, W.H. The Effect of Different Divalent Cations on the Kinetics and Fidelity of Bacillus Stearothermophilus DNA Polymerase. AIMS Biophys. 2018, 5, 125–143. [Google Scholar] [CrossRef]
- Kittler, R.; Stoneking, M.; Kayser, M. A Whole Genome Amplification Method to Generate Long Fragments from Low Quantities of Genomic DNA. Anal. Biochem. 2002, 300, 237–244. [Google Scholar] [CrossRef]
- Baruch-Torres, N.; Brieba, L.G. Plant Organellar DNA Polymerases Are Replicative and Translesion DNA Synthesis Polymerases. Nucleic Acids Res. 2017, 45, 10751–10763. [Google Scholar] [CrossRef]
- Nevin, P.; Gabbai, C.C.; Marians, K.J. Replisome-Mediated Translesion Synthesis by a Cellular Replicase. J. Biol. Chem. 2017, 292, 13833–13842. [Google Scholar] [CrossRef]
- Pastor-Palacios, G.; Lopez-Ramirez, V.; Cardona-Felix, C.S.; Brieba, L.G. A Transposon-Derived DNA Polymerase from Entamoeba Histolytica Displays Intrinsic Strand Displacement, Processivity and Lesion Bypass. PLoS ONE 2012, 7, 12. [Google Scholar] [CrossRef] [PubMed]
- Sabouri, N.; Johansson, E. Translesion Synthesis of Abasic Sites by Yeast DNA Polymerase Epsilon. J. Biol. Chem. 2009, 284, 31555–31563. [Google Scholar] [CrossRef] [PubMed]
- Ordóñez, C.D.; Lechuga, A.; Salas, M.; Redrejo-Rodríguez, M. Engineered Viral DNA Polymerase with Enhanced DNA Amplification Capacity: A Proof-of-Concept of Isothermal Amplification of Damaged DNA. Sci. Rep. 2020, 10, 15046. [Google Scholar] [CrossRef] [PubMed]
- Ordóñez, C.D. Novel Activities and New Members of B-Family Polymerases with Applications in Biotechnology. Ph.D. Thesis, Universidad Autónoma de Madrid, Madrid, Spain, 2022. [Google Scholar]
- Chien, A.; Edgar, D.B.; Trela, J.M. Deoxyribonucleic Acid Polymerase from the Extreme Thermophile Thermus Aquaticus. J. Bacteriol. 1976, 127, 1550–1557. [Google Scholar] [CrossRef] [PubMed]
- Lasken, R.S.; Stockwell, T.B. Mechanism of Chimera Formation during the Multiple Displacement Amplification Reaction. BMC Biotechnol. 2007, 7, 19. [Google Scholar] [CrossRef]
- Povilaitis, T.; Alzbutas, G.; Sukackaite, R.; Siurkus, J.; Skirgaila, R. In Vitro Evolution of Phi29 DNA Polymerase Using Isothermal Compartmentalized Self Replication Technique. Protein Eng. Des. Sel. 2016, 29, 617–628. [Google Scholar] [CrossRef]
- Povilaitis, T.; Skirgaila, R. Phi29 DNA Polymerase Mutants Having Increased Thermostability and Processivity Comprising M8R, V51A, M97T, L123S, G197D, K209E, E221K, E239G, Q497P, K512E, E515A, and F526L 2014. Eur. Patent 281357 B1, 15 June 2016. [Google Scholar]
- Salas, M.; Lázaro, J.M.; de Vega, M.; Rodrígez, I.; Serrano, L.; Delgado, J. Bacteriophage Phi29 Dna Polymerase Variants Having Improved Thermoactivity. Worldwide Applications WO2017109262A1, 29 June 2017. [Google Scholar]
- Stepanauskas, R.; Fergusson, E.A.; Brown, J.; Poulton, N.J.; Tupper, B.; Labonté, J.M.; Becraft, E.D.; Brown, J.M.; Pachiadaki, M.G.; Povilaitis, T.; et al. Improved Genome Recovery and Integrated Cell-Size Analyses of Individual Uncultured Microbial Cells and Viral Particles. Nat. Commun. 2017, 8, 84. [Google Scholar] [CrossRef]
- Paik, I.; Ngo, P.H.T.; Shroff, R.; Diaz, D.J.; Maranhao, A.C.; Walker, D.J.F.; Bhadra, S.; Ellington, A.D. Improved Bst DNA Polymerase Variants Derived via a Machine Learning Approach. Biochemistry 2021, 62, 410–418. [Google Scholar] [CrossRef]
- Navin, N.E. Cancer Genomics: One Cell at a Time. Genome Biol. 2014, 15, 452. [Google Scholar] [CrossRef]
- Alsmadi, O.; Alkayal, F.; Monies, D.; Meyer, B.F. Specific and Complete Human Genome Amplification with Improved Yield Achieved by Phi29 DNA Polymerase and a Novel Primer at Elevated Temperature. BMC Res. Notes 2009, 2, 48. [Google Scholar] [CrossRef]
- Zyrina, N.V.; Antipova, V.N.; Zheleznaya, L.A. Ab Initio Synthesis by DNA Polymerases. FEMS Microbiol. Lett. 2014, 351, 1–6. [Google Scholar] [CrossRef]
- Zyrina, N.V.; Antipova, V.N. Nonspecific Synthesis in the Reactions of Isothermal Nucleic Acid Amplification. Biochem. Mosc. 2021, 86, 887–897. [Google Scholar] [CrossRef] [PubMed]
- Sobol, M.S.; Kaster, A.-K. Back to Basics: A Simplified Improvement to Multiple Displacement Amplification for Microbial Single-Cell Genomics. Int. J. Mol. Sci. 2023, 24, 4270. [Google Scholar] [CrossRef]
- Takahashi, H.; Yamazaki, H.; Akanuma, S.; Kanahara, H.; Saito, T.; Chimuro, T.; Kobayashi, T.; Ohtani, T.; Yamamoto, K.; Sugiyama, S.; et al. Preparation of Phi29 DNA Polymerase Free of Amplifiable DNA Using Ethidium Monoazide, an Ultraviolet-Free Light-Emitting Diode Lamp and Trehalose. PLoS ONE 2014, 9, e82624. [Google Scholar] [CrossRef]
- Woyke, T.; Sczyrba, A.; Lee, J.; Rinke, C.; Tighe, D.; Clingenpeel, S.; Malmstrom, R.; Stepanauskas, R.; Cheng, J.F. Decontamination of MDA Reagents for Single Cell Whole Genome Amplification. PLoS ONE 2011, 6, 5. [Google Scholar] [CrossRef]
- Antipova, V.N.; Zheleznaya, L.A.; Zyrina, N.V. Ab Initio DNA Synthesis by Bst Polymerase in the Presence of Nicking Endonucleases Nt.AlwI, Nb.BbvCI, and Nb.BsmI. FEMS Microbiol. Lett. 2014, 357, 144–150. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.; Jensen, K.; Frank-Kamenetskii, M.D. Very Efficient Template/Primer-Independent DNA Synthesis by Thermophilic DNA Polymerase in the Presence of a Thermophilic Restriction Endonuclease. Biochemistry 2004, 43, 13459–13466. [Google Scholar] [CrossRef] [PubMed]
- Ogata, N.; Miura, T. Creation of Genetic Information by DNA Polymerase of the Thermophilic Bacterium Thermus Thermophilus. Nucleic Acids Res. 1998, 26, 4657–4661. [Google Scholar] [CrossRef]
- Ogata, N.; Miura, T. Genetic Information “created” by Archaebacterial DNA Polymerase. Biochem. J. 1997, 324, 667–671. [Google Scholar] [CrossRef]
- Garafutdinov, R.R.; Gilvanov, A.R.; Sakhabutdinova, A.R. The Influence of Reaction Conditions on DNA Multimerization During Isothermal Amplification with Bst Exo− DNA Polymerase. Appl. Biochem. Biotechnol. 2020, 190, 758–771. [Google Scholar] [CrossRef]
- Liang, X.; Kato, T.; Asanuma, H. Unexpected Efficient Ab Initio DNA Synthesis at Low Temperature by Using Thermophilic DNA Polymerase. Nucleic Acids Symp. Ser. 2007, 51, 351–352. [Google Scholar] [CrossRef] [PubMed]
- Pinard, R.; de Winter, A.; Sarkis, G.J.; Gerstein, M.B.; Tartaro, K.R.; Plant, R.N.; Egholm, M.; Rothberg, J.M.; Leamon, J.H. Assessment of Whole Genome Amplification-Induced Bias through High-Throughput, Massively Parallel Whole Genome Sequencing. BMC Genom. 2006, 7, 216. [Google Scholar] [CrossRef] [PubMed]
- Biezuner, T.; Raz, O.; Amir, S.; Milo, L.; Adar, R.; Fried, Y.; Ainbinder, E.; Shapiro, E. Comparison of Seven Single Cell Whole Genome Amplification Commercial Kits Using Targeted Sequencing. Sci. Rep. 2021, 11, 17171. [Google Scholar] [CrossRef] [PubMed]
- Keown, R.A.; Dums, J.T.; Brumm, P.J.; MacDonald, J.; Mead, D.A.; Ferrell, B.D.; Moore, R.M.; Harrison, A.O.; Polson, S.W.; Wommack, K.E. Novel Viral DNA Polymerases From Metagenomes Suggest Genomic Sources of Strand-Displacing Biochemical Phenotypes. Front. Microbiol. 2022, 13, 858366. [Google Scholar] [CrossRef]
- Piotrowski, Y.; Gurung, M.K.; Larsen, A.N. Characterization and Engineering of a DNA Polymerase Reveals a Single Amino-Acid Substitution in the Fingers Subdomain to Increase Strand-Displacement Activity of A-Family Prokaryotic DNA Polymerases. BMC Mol. Cell Biol. 2019, 20, 31. [Google Scholar] [CrossRef]
- Zhu, B.; Wang, L.; Mitsunobu, H.; Lu, X.; Hernandez, A.J.; Yoshida-Takashima, Y.; Nunoura, T.; Tabor, S.; Richardson, C.C. Deep-Sea Vent Phage DNA Polymerase Specifically Initiates DNA Synthesis in the Absence of Primers. Proc. Natl. Acad. Sci. USA 2017, 114, E2310–E2318. [Google Scholar] [CrossRef]
- Coulther, T.A.; Stern, H.R.; Beuning, P.J. Engineering Polymerases for New Functions. Trends Biotechnol. 2019, 37, 1091–1103. [Google Scholar] [CrossRef]
- Davidson, J.F. Insertion of the T3 DNA Polymerase Thioredoxin Binding Domain Enhances the Processivity and Fidelity of Taq DNA Polymerase. Nucleic Acids Res. 2003, 31, 4702–4709. [Google Scholar] [CrossRef][Green Version]
- de Vega, M.; Lazaro, J.M.; Mencia, M.; Blanco, L.; Salas, M. Improvement of Φ29 DNA Polymerase Amplification Performance by Fusion of DNA Binding Motifs. Proc. Natl. Acad. Sci. USA 2010, 107, 16506–16511. [Google Scholar] [CrossRef]
- Wang, L.; Liang, C.; Wu, J.; Liu, L.; Tyo, K.E.J. Increased Processivity, Misincorporation, and Nucleotide Incorporation Efficiency in Sulfolobus Solfataricus Dpo4 Thumb Domain Mutants. Appl. Environ. Microbiol. 2017, 83, e01013-17. [Google Scholar] [CrossRef]
- Zhai, B.; Chow, J.; Cheng, Q. Two Approaches to Enhance the Processivity and Salt Tolerance of Staphylococcus Aureus DNA Polymerase. Protein J. 2019, 38, 190–198. [Google Scholar] [CrossRef]
- Cho, S.S.; Yu, M.; Kim, S.H.; Kwon, S.T. Enhanced PCR Efficiency of High-Fidelity DNA Polymerase from Thermococcus Waiotapuensis. Enzyme Microb. Technol. 2014, 63, 39–45. [Google Scholar] [CrossRef] [PubMed]
- Pavlov, A.R.; Belova, G.I.; Kozyavkin, S.A.; Slesarev, A.I. Helix-Hairpin-Helix Motifs Confer Salt Resistance and Processivity on Chimeric DNA Polymerases. Proc. Natl. Acad. Sci. USA 2002, 99, 13510–13515. [Google Scholar] [CrossRef] [PubMed]
- Baar, C.; D’Abbadie, M.; Vaisman, A.; Arana, M.E.; Hofreiter, M.; Woodgate, R.; Kunkel, T.A.; Holliger, P. Molecular Breeding of Polymerases for Resistance to Environmental Inhibitors. Nucleic Acids Res. 2011, 39, e51. [Google Scholar] [CrossRef][Green Version]
- Śpibida, M.; Krawczyk, B.; Zalewska-Piątek, B.; Piątek Rafałand Wysocka, M.; Olszewski, M. Fusion of DNA-Binding Domain of Pyrococcus furiosus Ligase with TaqStoffel DNA Polymerase as a Useful Tool in PCR with Difficult Targets. Appl. Microbiol. Biotechnol. 2018, 102, 713–721. [Google Scholar] [CrossRef] [PubMed]
- Yamagami, T.; Ishino, S.; Kawarabayasi, Y.; Ishino, Y. Mutant Taq DNA Polymerases with Improved Elongation Ability as a Useful Reagent for Genetic Engineering. Front. Microbiol. 2014, 5, 461. [Google Scholar] [CrossRef] [PubMed]
- Olszewski, M.; Śpibida, M.; Bilek, M.; Krawczyk, B. Fusion of Taq DNA Polymerase with Single-Stranded DNA Binding-like Protein of Nanoarchaeum Equitans—Expression and Characterization. PLoS ONE 2017, 12, e0184162. [Google Scholar] [CrossRef]
- Villbrandt, B.; Sobek, H.; Frey, B.; Schomburg, D. Domain Exchange: Chimeras of Thermus Aquaticus DNA Polymerase, Escherichia coli DNA Polymerase I and Thermotoga Neapolitana DNA Polymerase. Protein Eng. 2000, 13, 645–654. [Google Scholar] [CrossRef]
- Torres, L.L.; Pinheiro, V.B. Xenobiotic Nucleic Acid (XNA) Synthesis by Phi29 DNA Polymerase. Curr. Protoc. Chem. Biol. 2018, 10, e41. [Google Scholar] [CrossRef]
- Rai, D.K.; Diaz-San Segundo, F.; Campagnola, G.; Keith, A.; Schafer, E.A.; Kloc, A.; de los Santos, T.; Peersen, O.; Rieder, E. Attenuation of Foot-and-Mouth Disease Virus by Engineered Viral Polymerase Fidelity. J. Virol. 2017, 91, e00081-17. [Google Scholar] [CrossRef]
- Reha-Krantz, L.J.; Woodgate, S.; Goodman, M.F. Engineering Processive DNA Polymerases with Maximum Benefit at Minimum Cost. Front. Microbiol. 2014, 5, 380. [Google Scholar] [CrossRef] [PubMed]
- Elshawadfy, A.M.; Keith, B.J.; Ooi, E.H.; Kinsman, T.; Heslop, P.; Connolly, B.A. DNA Polymerase Hybrids Derived from the Family-B Enzymes of Pyrococcus furiosus and Thermococcus Kodakarensis: Improving Performance in the Polymerase Chain Reaction. Front. Microbiol. 2014, 5, 224. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; de Paz, A.; Zamft, B.M.; Marblestone, A.H.; Boyden, E.S.; Kording, K.P.; Tyo, K.E.J. DNA Binding Strength Increases the Processivity and Activity of a Y-Family DNA Polymerase. Sci. Rep. 2017, 7, 4756. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Romesberg, F.E. Directed Polymerase Evolution. FEBS Lett. 2014, 588, 219–229. [Google Scholar] [CrossRef]
- Agudo, R.; Calvo, P.A.; Martínez-Jiménez, M.; Blanco, L. Engineering Human PrimPol into an Efficient RNA-Dependent-DNA Primase/Polymerase. Nucleic Acids Res. 2017, 45, 9046–9058. [Google Scholar] [CrossRef]
- Sebestova, E.; Bendl, J.; Brezovsky, J.; Damborsky, J. Computational Tools for Designing Smart Libraries. Methods Mol. Biol. 2014, 1179, 291–314. [Google Scholar] [CrossRef]
- Zeymer, C.; Hilvert, D. Directed Evolution of Protein Catalysts. Annu. Rev. Biochem. 2018, 87, 131–157. [Google Scholar] [CrossRef]
- Forloni, M.; Liu, A.Y.; Wajapeyee, N. Random Mutagenesis Using Error-Prone DNA Polymerases. Cold Spring Harb. Protoc. 2018, 2018, 220–230. [Google Scholar] [CrossRef]
- Oren, A.; Garrity, G.M. Valid Publication of the Names of Forty-Two Phyla of Prokaryotes. Int. J. Syst. Evol. Microbiol. 2021, 71, 005056. [Google Scholar] [CrossRef]
- D’Abbadie, M.; Hofreiter, M.; Vaisman, A.; Loakes, D.; Gasparutto, D.; Cadet, J.; Woodgate, R.; Pääbo, S.; Holliger, P. Molecular Breeding of Polymerases for Amplification of Ancient DNA. Nat. Biotechnol. 2007, 25, 939–943. [Google Scholar] [CrossRef]
- Crameri, A.; Raillard, S.A.; Bermudez, E.; Stemmer, W.P.C. DNA Shuffling of a Family of Genes from Diverse Species Accelerates Directed Evolution. Nature 1998, 391, 288–291. [Google Scholar] [CrossRef] [PubMed]
- Milligan, J.N.; Shroff, R.; Garry, D.J.; Ellington, A.D. Evolution of a Thermophilic Strand-Displacing Polymerase Using High-Temperature Isothermal Compartmentalized Self-Replication. Biochemistry 2018, 57, 4607–4619. [Google Scholar] [CrossRef] [PubMed]
- Jestin, J.L.; Kristensen, P.; Winter, G. A Method for the Selection of Catalytic Activity Using Phage Display and Proximity Coupling. Angew. Chem. Int. Ed. 1999, 38, 1124–1127. [Google Scholar] [CrossRef]
- Leconte, A.M.; Chen, L.; Romesberg, F.E. Polymerase Evolution: Efforts toward Expansion of the Genetic Code. J. Am. Chem. Soc. 2005, 127, 12470–12471. [Google Scholar] [CrossRef]
- Larsen, A.C.; Dunn, M.R.; Hatch, A.; Sau, S.P.; Youngbull, C.; Chaput, J.C. A General Strategy for Expanding Polymerase Function by Droplet Microfluidics. Nat. Commun. 2016, 7, 11235. [Google Scholar] [CrossRef]
- Takahashi, M.; Takahashi, E.; Joudeh, L.I.; Marini, M.; Das, G.; Elshenawy, M.M.; Akal, A.; Sakashita, K.; Alam, I.; Tehseen, M.; et al. Dynamic Structure Mediates Halophilic Adaptation of a DNA Polymerase from the Deep-Sea Brines of the Red Sea. FASEB J. 2018, 32, 3346–3360. [Google Scholar] [CrossRef]
- Pavlov, A.R.; Pavlova, N.V.; Kozyavkin, S.A.; Slesarev, A.I. Cooperation between Catalytic and DNA Binding Domains Enhances Thermostability and Supports DNA Synthesis at Higher Temperatures by Thermostable DNA Polymerases. Biochemistry 2012, 51, 2032–2043. [Google Scholar] [CrossRef]
- Wang, Y. Nucleic Acid Modifying Enzymes. U.S. Patent 6627424B1, 30 September 2003. [Google Scholar]
- Wang, Y.; Horn, P.V.; Xi, L. Methods of Using Improved Polymerases. Eur. Patent 1456394B2, 27 April 2011. [Google Scholar]
- Wang, Y. Sso7-Polymerase Conjugate Proteins. U.S. Patent 20040081963A1, 23 October 2002. [Google Scholar]
- Oscorbin, I.P.; Belousova, E.A.; Boyarskikh, U.A.; Zakabunin, A.I.; Khrapov, E.A.; Filipenko, M.L. Derivatives of Bst-like Gss-Polymerase with Improved Processivity and Inhibitor Tolerance. Nucleic Acids Res. 2017, 45, 9595–9610. [Google Scholar] [CrossRef]
- Paik, I.; Bhadra, S.; Ellington, A.D. Charge Engineering Improves the Performance of Bst DNA Polymerase Fusions. ACS Synth. Biol. 2022, 11, 1488–1496. [Google Scholar] [CrossRef]
- Falgueras, M.S.; Jose, M.D.V.; Bolos, J.M.L.; Davila, L.B.; Caballero, M.M. Phage Phi29 DNA Polymerase Chimera. U.S. Patent 8404808B2, 26 March 2013. [Google Scholar]
- Salas, M.; Holguera, I.; Redrejo-Rodriguez, M.; de Vega, M. DNA-Binding Proteins Essential for Protein-Primed Bacteriophage Phi29 DNA Replication. Front. Mol. Biosci. 2016, 3, 37. [Google Scholar] [CrossRef]
| DNAP Family | Bacteria | Archaea | Eukarya | Virus |
|---|---|---|---|---|
| A | Pol I [Bst, Taq] | ND | Pol γ | T7 |
| B | Pol II | Pol B1, B2, B3 [Pwo] | Pol α, δ, ε, ζ | Φ29, Bam35, HSV, RB69 |
| C | Pol III | ND | ND | ND |
| D | ND | Pol D | ND | ND |
| X | Pol X | ND | Pol β, λ, μ | ASFV |
| Y | Pol IV (DinB), Pol V (UmuCD) | Dpo4, Dbh | Pol η, ι, κ, Rev1 | ND |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ordóñez, C.D.; Redrejo-Rodríguez, M. DNA Polymerases for Whole Genome Amplification: Considerations and Future Directions. Int. J. Mol. Sci. 2023, 24, 9331. https://doi.org/10.3390/ijms24119331
Ordóñez CD, Redrejo-Rodríguez M. DNA Polymerases for Whole Genome Amplification: Considerations and Future Directions. International Journal of Molecular Sciences. 2023; 24(11):9331. https://doi.org/10.3390/ijms24119331
Chicago/Turabian StyleOrdóñez, Carlos D., and Modesto Redrejo-Rodríguez. 2023. "DNA Polymerases for Whole Genome Amplification: Considerations and Future Directions" International Journal of Molecular Sciences 24, no. 11: 9331. https://doi.org/10.3390/ijms24119331
APA StyleOrdóñez, C. D., & Redrejo-Rodríguez, M. (2023). DNA Polymerases for Whole Genome Amplification: Considerations and Future Directions. International Journal of Molecular Sciences, 24(11), 9331. https://doi.org/10.3390/ijms24119331






