Recent Advances in Computer-Aided Structure-Based Drug Design on Ion Channels
Abstract
1. Introduction
1.1. Classification of Ion Channels
- Voltage-gated sodium (NaV) channels, which are responsible for the onset and propagation of the action potential in neurons [13].
- Voltage-gated potassium (KV) channels, which among other functions, determine the shape and duration of the action potential, control the membrane resting potential, and modulate hormone secretion [14].
- Voltage-gated calcium (CaV) channels, which take part in various processes, including cardiac action potential propagation, muscle contraction, and calcium-dependent gene expression [15].
- Voltage-gated chloride channels (CICs), which contribute to regulating neuronal excitability and cell volume [16].
1.2. Mechanism of Action of Ion Channels

2. Ion Channels as Drug Targets
2.1. Voltage-Gated Sodium (NaV) Channels
2.2. Voltage-Gated Potassium (KV) Channels
2.3. Voltage-Gated Calcium (CaV) Channels
2.4. Ligand-Gated Ion Channels
2.5. Transient Receptor Potential (TRP) Channels
2.6. Ion Channel-Drug Interactions
3. Computer-Aided Drug Design Approaches
3.1. Structure-Based Drug Design
3.2. Ligand-Based Drug Design
3.3. Virtual Ligand Libraries for Ion Channel Drug Discovery
4. Computational Drug Design on Ion Channels: Recent Discoveries
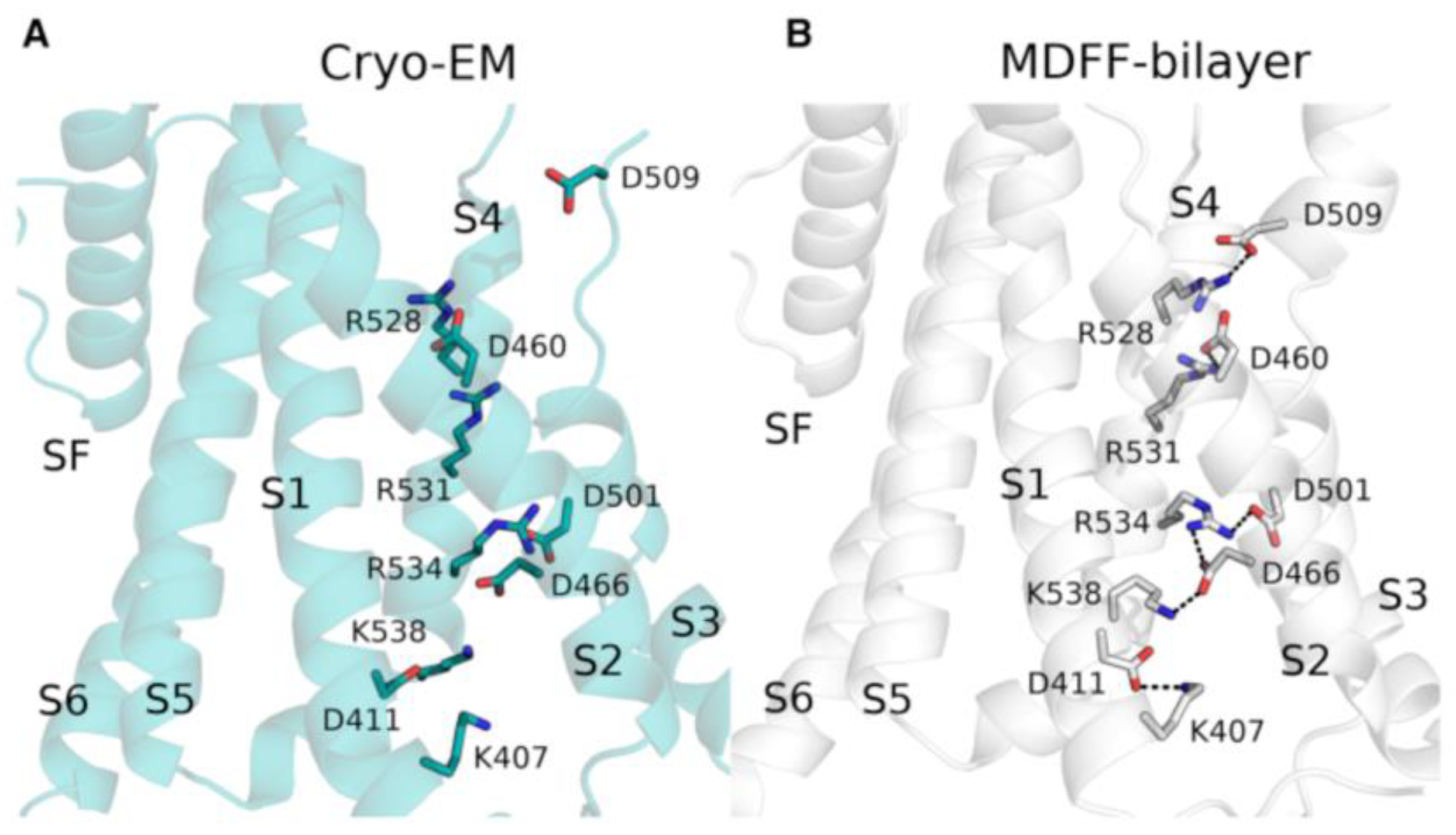
4.1. Computational Refinement of Cryo-EM Structures
4.2. Characterization of Drug-Ion Channel Interactions by Molecular Dynamics Simulations
4.3. Identification of Ion Channel Ligands by Virtual Docking Techniques
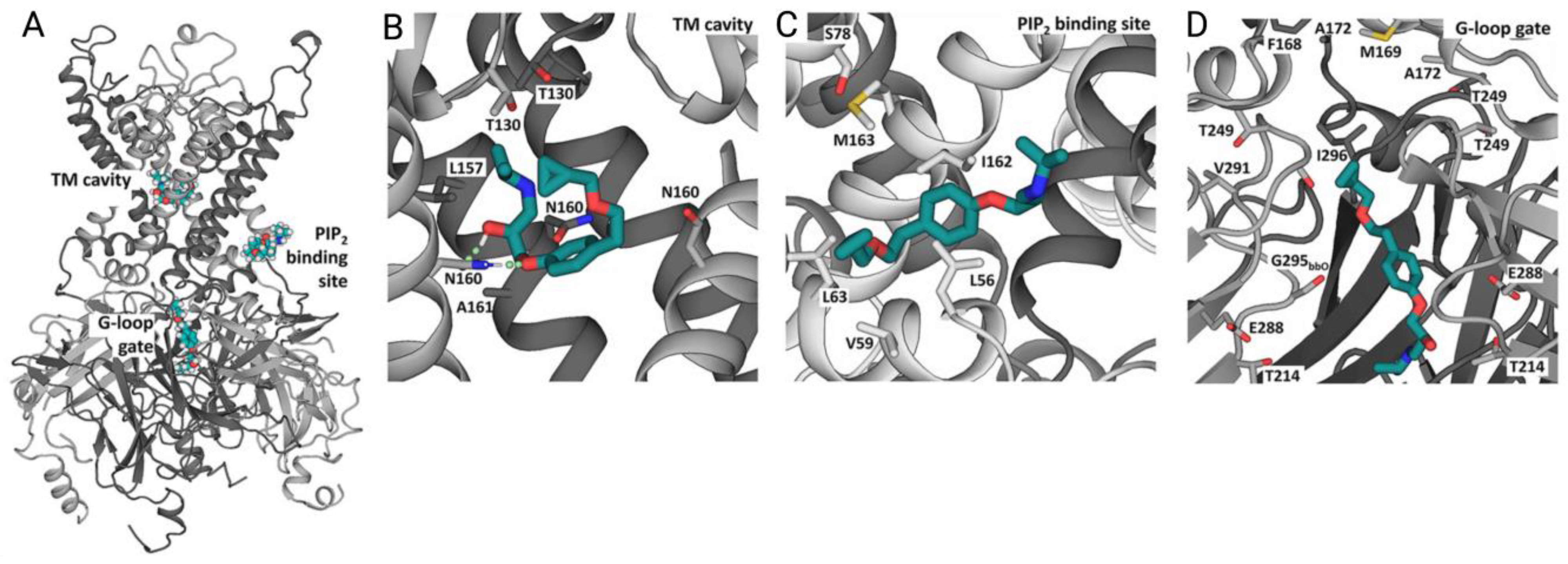
4.4. Prediction of Ion Channel Ligands with Structure-Based Virtual Screening
5. Conclusions and Further Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tanner, M.R.; Beeton, C. Differences in Ion Channel Phenotype and Function between Humans and Animal Models. Front. Biosci. Landmark 2018, 23, 43–64. [Google Scholar] [CrossRef]
- Zaydman, M.A.; Silva, J.R.; Cui, J. Ion Channel Associated Diseases: Overview of Molecular Mechanisms. Chem. Rev. 2012, 112, 6319–6333. [Google Scholar] [CrossRef]
- Cox, B. Ion Channel Drug Discovery: A Historical Perspective; University of Sussex: Brighton, UK, 2015; Volume 2015, ISBN 9781849735087. [Google Scholar]
- Yan, P.; Ke, B.; Fang, X. Ion Channels as a Therapeutic Target for Renal Fibrosis. Front. Physiol. 2022, 13, 1019028. [Google Scholar] [CrossRef] [PubMed]
- Boyle, Y.; Johns, T.G.; Fletcher, E. V Potassium Ion Channels in Malignant Central Nervous System Cancers. Cancers 2022, 14, 4767. [Google Scholar] [CrossRef]
- Fakih, D.; Migeon, T.; Moreau, N.; Baudouin, C.; Réaux-Le Goazigo, A.; Mélik Parsadaniantz, S. Transient Receptor Potential Channels: Important Players in Ocular Pain and Dry Eye Disease. Pharmaceutics 2022, 14, 1859. [Google Scholar] [CrossRef] [PubMed]
- Santos, R.; Ursu, O.; Gaulton, A.; Bento, A.P.; Donadi, R.S.; Bologa, C.G.; Karlsson, A.; Al-lazikani, B.; Hersey, A.; Oprea, T.I.; et al. A Comprehensive Map of Molecular Drug Targets. Nat. Rev. Drug Discov. 2016, 16, 19–34. [Google Scholar] [CrossRef]
- Doyle, D.A.; Cabral, M.; Pfuetzner, R.A.; Kuo, A.; Gulbis, J.M.; Cohen, S.L.; Chait, B.T.; Mackinnon, R. The Structure of the Potassium Channel: Molecular Basis of K+ Conduction and Selectivity. Science 1998, 280, 69–77. [Google Scholar] [CrossRef]
- Membrane Proteins of Known Structure. Available online: https://blanco.biomol.uci.edu/mpstruc/ (accessed on 10 March 2023).
- (OPM) Database—University of Michigan. Available online: https://opm.phar.umich.edu (accessed on 20 March 2023).
- SCOP: Structural Classification of Proteins. Available online: http://scop.mrc-lmb.cam.ac.uk/ (accessed on 20 March 2022).
- TCDB HOME. Available online: https://tcdb.org/ (accessed on 20 March 2022).
- De Lera Ruiz, M.; Kraus, R.L. Voltage-Gated Sodium Channels: Structure, Function, Pharmacology, and Clinical Indications. J. Med. Chem. 2015, 58, 7093–7118. [Google Scholar] [CrossRef]
- Kuang, Q.; Purhonen, P.; Hebert, H. Structure of Potassium Channels. Cell. Mol. Life Sci. 2015, 72, 3677–3693. [Google Scholar] [CrossRef]
- Qu, Y.; Sun, Y.; Yang, Z.; Ding, C. Calcium Ions Signaling: Targets for Attack and Utilization by Viruses. Front. Microbiol. 2022, 13, 889374. [Google Scholar] [CrossRef]
- Jentsch, T.J.; Stein, V.; Weinreich, F.; Zdebik, A.A. Molecular Structure and Physiological Function of Chloride Channels. Physiol. Rev. 2002, 82, 503–568. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Ritzel, R.M.; Wu, J. Functions and Mechanisms of the Voltage-Gated Proton Channel Hv1 in Brain and Spinal Cord Injury. Front. Cell. Neurosci. 2021, 15, 662971. [Google Scholar] [CrossRef] [PubMed]
- Alexander, S.P.H.; Mathie, A.; Peters, J.A. Guide to Receptors and Channels (GRAC), 5th edition. Br. J. Pharmacol. 2011, 164 (Suppl. S1), S1–S324. [Google Scholar] [CrossRef] [PubMed]
- Ligand-Gated Ion Channels. IUPHAR/BPS Guide to PHARMACOLOGY. Available online: https://www.guidetopharmacology.org/GRAC/FamilyDisplayForward?familyId=697 (accessed on 17 February 2023).
- Pan, Z.; Yang, H.; Reinach, P.S. Transient Receptor Potential (TRP) Gene Superfamily Encoding Cation Channels. Hum. Genom. 2011, 5, 108–116. [Google Scholar] [CrossRef]
- Khalil, M.; Alliger, K.; Weidinger, C.; Yerinde, C.; Wirtz, S.; Becker, C.; Engel, M.A. Functional Role of Transient Receptor Potential Channels in Immune Cells and Epithelia. Front. Immunol. 2018, 9, 174. [Google Scholar] [CrossRef]
- Ciardo, M.G.; Ferrer-Montiel, A. Lipids as Central Modulators of Sensory TRP Channels. Biochim. Biophys. Acta Biomembr. 2017, 1859, 1615–1628. [Google Scholar] [CrossRef] [PubMed]
- Natale, A.M.; Deal, P.E.; Minor, D.L. Structural Insights into the Mechanisms and Pharmacology of K2P Potassium Channels. J. Mol. Biol. 2021, 433, 166995. [Google Scholar] [CrossRef] [PubMed]
- Bezanilla, F. Voltage-Gated Ion Channels. IEEE Trans. NanoBioscience 2005, 4, 34–48. [Google Scholar] [CrossRef]
- Catterall, W.A. Structure and Function of Voltage-Gated Ion Channels. Annu. Rev. Biochem. 1995, 64, 493–531. [Google Scholar] [CrossRef]
- Jiang, Y.; Ruta, V.; Chen, J.; Lee, A.; Mackinnon, R. The Principle of Gating Charge Movement in a Voltage-Dependent K+ Channel. Nature 2003, 423, 42–48. [Google Scholar] [CrossRef]
- Michalakis, S.; Becirovic, E.; Biel, M. Retinal Cyclic Nucleotide-Gated Channels: From Pathophysiology to Therapy. Int. J. Mol. Sci. 2018, 19, 749. [Google Scholar] [CrossRef] [PubMed]
- Thompson, A.J.; Lester, H.A.; Lummis, S.C.R. The Structural Basis of Function in Cys-Loop Receptors. Q. Rev. Biophys. 2010, 43, 449–499. [Google Scholar] [CrossRef] [PubMed]
- Vithlani, M.; Terunuma, M.; Moss, S.J. The Dynamic Modulation of GABAa Receptor Trafficking and Its Role in Regulating the Plasticity of Inhibitory Synapses. Physiol. Rev. 2011, 91, 1009–1022. [Google Scholar] [CrossRef]
- Tretter, V.; Jacob, T.C.; Mukherjee, J.; Fritschy, J.M.; Pangalos, M.N.; Moss, S.J. The Clustering of GABAA Receptor Subtypes at Inhibitory Synapses Is Facilitated via the Direct Binding of Receptor A2 Subunits to Gephyrin. J. Neurosci. 2008, 28, 1356–1365. [Google Scholar] [CrossRef]
- Mayer, M.L. Structure and Mechanism of Glutamate Receptor Ion Channel Assembly, Activation and Modulation. Curr. Opin. Neurobiol. 2011, 21, 283–290. [Google Scholar] [CrossRef][Green Version]
- Traynelis, S.F.; Wollmuth, L.P.; McBain, C.J.; Menniti, F.S.; Vance, K.M.; Ogden, K.K.; Hansen, K.B.; Yuan, H.; Myers, S.J.; Dingledine, R. Glutamate Receptor Ion Channels: Structure, Regulation, and Function. Pharmacol. Rev. 2010, 62, 405–496. [Google Scholar] [CrossRef]
- Twomey, E.C.; Sobolevsky, A.I. Structural Mechanisms of Gating in Ionotropic Glutamate Receptors. Biochemistry 2018, 57, 267–276. [Google Scholar] [CrossRef]
- Ruan, N.; Tribble, J.; Peterson, A.M.; Jiang, Q.; Wang, J.Q.; Chu, X.P. Acid-Sensing Ion Channels and Mechanosensation. Int. J. Mol. Sci. 2021, 22, 4810. [Google Scholar] [CrossRef]
- Sherwood, T.W.; Frey, E.N.; Askwith, C.C. Structure and Activity of the Acid-Sensing Ion Channels. Am. J. Physiol. Cell Physiol. 2012, 303, C699–C710. [Google Scholar] [CrossRef]
- Liu, Y.; Ma, J.; DesJarlais, R.L.; Hagan, R.; Rech, J.; Lin, D.; Liu, C.; Miller, R.; Schoellerman, J.; Luo, J.; et al. Molecular Mechanism and Structural Basis of Small-Molecule Modulation of the Gating of Acid-Sensing Ion Channel 1. Commun. Biol. 2021, 4, 174. [Google Scholar] [CrossRef]
- Ma, D.; Zhong, L.; Yan, Z.; Yao, J.; Zhang, Y.; Ye, F.; Huang, Y.; Lai, D.; Yang, W.; Hou, P.; et al. Structural Mechanisms for the Activation of Human Cardiac KCNQ1 Channel by Electro-Mechanical Coupling Enhancers. Proc. Natl. Acad. Sci. USA 2022, 119, e2207067119. [Google Scholar] [CrossRef] [PubMed]
- Noviello, C.M.; Gharpure, A.; Mukhtasimova, N.; Cabuco, R.; Baxter, L.; Borek, D.; Sine, S.M.; Hibbs, R.E. Structure and Gating Mechanism of the A7 Nicotinic Acetylcholine Receptor. Cell 2021, 184, 2121–2134.e13. [Google Scholar] [CrossRef] [PubMed]
- Sobolevsky, A.I.; Rosconi, M.P.; Gouaux, E. X-ray Structure, Symmetry and Mechanism of an AMPA-Subtype Glutamate Receptor. Nature 2009, 462, 745–756. [Google Scholar] [CrossRef] [PubMed]
- Yoder, N.; Yoshioka, C.; Gouaux, E. Gating Mechanisms of Acid-Sensing Ion Channels. Nature 2018, 555, 397–401. [Google Scholar] [CrossRef]
- Imbrici, P.; Liantonio, A.; Camerino, G.M.; De Bellis, M.; Camerino, C.; Mele, A.; Giustino, A.; Pierno, S.; De Luca, A.; Tricarico, D.; et al. Therapeutic Approaches to Genetic Ion Channelopathies and Perspectives in Drug Discovery. Front. Pharmacol. 2016, 7, 121. [Google Scholar] [CrossRef]
- Bagal, S.K.; Brown, A.D.; Cox, P.J.; Omoto, K.; Owen, R.M.; Pryde, D.C.; Sidders, B.; Skerratt, S.E.; Stevens, E.B.; Storer, R.I.; et al. Ion Channels as Therapeutic Targets: A Drug Discovery Perspective. J. Med. Chem. 2013, 56, 593–624. [Google Scholar] [CrossRef]
- Bagal, S.K.; Marron, B.E.; Owen, R.M.; Storer, R.I.; Swain, N.A. Voltage Gated Sodium Channels as Drug Discovery Targets. Channels 2015, 9, 360–366. [Google Scholar] [CrossRef]
- Bachmann, M.; Li, W.; Edwards, M.J.; Ahmad, S.A.; Patel, S.; Szabo, I.; Gulbins, E. Voltage-Gated Potassium Channels as Regulators of Cell Death. Front. Cell Dev. Biol. 2020, 8, 611853. [Google Scholar] [CrossRef]
- Waszkielewicz, A.M.; Gunia, A.; Szkaradek, N.; Sloczynska, K.; Krupinska, S.; Marona, H. Ion Channels as Drug Targets in Central Nervous System Disorders. Curr. Med. Chem. 2013, 999, 29–35. [Google Scholar] [CrossRef][Green Version]
- Conte Camerino, D.; Tricarico, D.; Desaphy, J.F. Ion Channel Pharmacology. Neurotherapeutics 2007, 4, 184–198. [Google Scholar] [CrossRef]
- Wulff, H.; Castle, N.A.; Pardo, L.A. Voltage-Gated Potassium Channels as Therapeutic Targets. Nat. Rev. Drug Discov. 2009, 8, 982–1001. [Google Scholar] [CrossRef] [PubMed]
- Catterall, W.A. From Ionic Currents to Molecular Mechanisms: The Structure and Function of Voltage-Gated Sodium Channels. Neuron 2000, 26, 13–25. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Wong, A.H.C.; Liu, F. Ligand-Gated Ion Channel Interacting Proteins and Their Role in Neuroprotection. Front. Cell. Neurosci. 2014, 8, 3–7. [Google Scholar] [CrossRef]
- Krasowski, M.D.; Harrison, N.L. General Anaesthetic Actions on Ligand-Gated Ion Channels. Cell. Mol. Life Sci. 1999, 55, 1278–1303. [Google Scholar] [CrossRef] [PubMed]
- Mount, C.; Downton, C. Alzheimer Disease: Progress or Profit? Nat. Med. 2006, 12, 780–784. [Google Scholar] [CrossRef] [PubMed]
- Eren-Koçak, E.; Dalkara, T. Ion Channel Dysfunction and Neuroinflammation in Migraine and Depression. Front. Pharmacol. 2021, 12, 777607. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhang, Q.; Guo, P.; Fu, J.; Mei, L.; Lv, D.; Wang, J.; Lai, D.; Ye, S.; Yang, H.; et al. Molecular Basis for Ligand Activation of the Human KCNQ2 Channel. Cell Res. 2021, 31, 52–61. [Google Scholar] [CrossRef]
- Iorio, M.T.; Vogel, F.D.; Koniuszewski, F.; Scholze, P.; Rehman, S.; Simeone, X.; Schnürch, M.; Mihovilovic, M.D.; Ernst, M. GABAa Receptor Ligands Often Interact with Binding Sites in the Transmembrane Domain and in the Extracellular Domain—Can the Promiscuity Code Be Cracked? Int. J. Mol. Sci. 2020, 21, 334. [Google Scholar] [CrossRef]
- Elgarf, A.A.; Siebert, D.C.B.; Steudle, F.; Draxler, A.; Li, G.; Huang, S.; Cook, J.M.; Ernst, M.; Scholze, P. Different Benzodiazepines Bind with Distinct Binding Modes to GABA A Receptors. ACS Chem. Biol. 2018, 13, 2033–2039. [Google Scholar] [CrossRef]
- Knox, H.J.; Rego Campello, H.; Lester, H.A.; Gallagher, T.; Dougherty, D.A. Characterization of Binding Site Interactions and Selectivity Principles in the A3β4 Nicotinic Acetylcholine Receptor. J. Am. Chem. Soc. 2022, 144, 16101–16117. [Google Scholar] [CrossRef]
- Gharpure, A.; Teng, J.; Zhuang, Y.; Noviello, C.M.; Walsh, R.M.; Cabuco, R.; Howard, R.J.; Zaveri, N.T.; Lindahl, E.; Hibbs, R.E. Agonist Selectivity and Ion Permeation in the A3β4 Ganglionic Nicotinic Receptor. Neuron 2019, 104, 501–511.e6. [Google Scholar] [CrossRef]
- Gore, M.; Jagtap, U. Computational Drug Discovery and Design; Baron, R., Ed.; Springer: Berlin/Heidelberg, Germany, 2012; Volume 819, ISBN 978-1-4939-7755-0. [Google Scholar]
- Sliwoski, G.; Kothiwale, S.; Meiler, J.; Lowe, E.W. Computational Methods in Drug Discovery. Pharmacol. Rev. 2014, 66, 334–395. [Google Scholar] [CrossRef]
- Wilson, G.L.; Lill, M.A. Integrating Structure-Based and Ligand-Based Approaches for Computational Drug Design. Future Med. Chem. 2011, 3, 735–750. [Google Scholar] [CrossRef]
- Jhoti, H.; Leach, A.R. Structure-Based Drug Discovery Computer-Aided Drug Discovery; Springer: Dordrecht, The Netherlands, 2007; ISBN 9789048171231. [Google Scholar]
- Huxford, T. X-Ray Crystallography. In Brenner’s Encyclopedia of Genetics; Academic Press: Cambridge, MA, USA, 2013; pp. 366–368. [Google Scholar] [CrossRef]
- Hu, Y.; Cheng, K.; He, L.; Zhang, X.; Jiang, B.; Jiang, L.; Li, C.; Wang, G.; Yang, Y.; Liu, M. NMR-Based Methods for Protein Analysis. Anal. Chem. 2021, 93, 1866–1879. [Google Scholar] [CrossRef]
- Carroni, M.; Saibil, H.R. Cryo Electron Microscopy to Determine the Structure of Macromolecular Complexes. Methods 2016, 95, 78–85. [Google Scholar] [CrossRef]
- Muhammed, M.T.; Aki-Yalcin, E. Homology Modeling in Drug Discovery: Overview, Current Applications, and Future Perspectives. Chem. Biol. Drug Des. 2019, 93, 12–20. [Google Scholar] [CrossRef]
- AlphaFold. Available online: https://www.deepmind.com/research/highlighted-research/alphafold (accessed on 20 February 2023).
- Halperin, I.; Ma, B.; Wolfson, H.; Nussinov, R. Principles of Docking: An Overview of Search Algorithms and a Guide to Scoring Functions. Proteins Struct. Funct. Genet. 2002, 47, 409–443. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Wang, R. Classification of Current Scoring Functions. J. Chem. Inf. Model. 2015, 55, 475–482. [Google Scholar] [CrossRef]
- Li, J.; Fu, A.; Zhang, L. An Overview of Scoring Functions Used for Protein–Ligand Interactions in Molecular Docking. Interdiscip. Sci. Comput. Life Sci. 2019, 11, 320–328. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.Y.; Grinter, S.Z.; Zou, X. Scoring Functions and Their Evaluation Methods for Protein-Ligand Docking: Recent Advances and Future Directions. Phys. Chem. Chem. Phys. 2010, 12, 12899–12908. [Google Scholar] [CrossRef] [PubMed]
- Shen, Q.; Xiong, B.; Zheng, M.; Luo, X.; Luo, C.; Liu, X.; Du, Y.; Li, J.; Zhu, W.; Shen, J.; et al. Knowledge-Based Scoring Functions in Drug Design: 2. Can the Knowledge Base Be Enriched? J. Chem. Inf. Model. 2011, 51, 386–397. [Google Scholar] [CrossRef] [PubMed]
- Pason, L.P.; Sotriffer, C.A. Empirical Scoring Functions for Affinity Prediction of Protein-Ligand Complexes. Mol. Inform. 2016, 35, 541–548. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Sze, K.H.; Lu, G.; Ballester, P.J. Machine-Learning Scoring Functions for Structure-Based Virtual Screening. Wiley Interdiscip. Rev. Comput. Mol. Sci. 2021, 11, e1478. [Google Scholar] [CrossRef]
- Yang, C.; Chen, E.A.; Zhang, Y. Protein–Ligand Docking in the Machine-Learning Era. Molecules 2022, 27, 4568. [Google Scholar] [CrossRef]
- Li, Y.; Zhou, D.; Zheng, G.; Li, X.; Wu, D.; Yuan, Y. DyScore: A Boosting Scoring Method with Dynamic Properties for Identifying True Binders and Nonbinders in Structure-Based Drug Discovery. J. Chem. Inf. Model. 2022, 62, 5550–5567. [Google Scholar] [CrossRef] [PubMed]
- Schaller, D.; Šribar, D.; Noonan, T.; Deng, L.; Nguyen, T.N.; Pach, S.; Machalz, D.; Bermudez, M.; Wolber, G. Next Generation 3D Pharmacophore Modeling. Wiley Interdiscip. Rev. Comput. Mol. Sci. 2020, 10, e1468. [Google Scholar] [CrossRef]
- Hu, B.; Lill, M.A. Exploring the Potential of Protein-Based Pharmacophore Models in Ligand Pose Prediction and Ranking. J. Chem. Inf. Model. 2013, 53, 1179–1190. [Google Scholar] [CrossRef]
- Hu, B.; Lill, M.A. PharmDock: A Pharmacophore-Based Docking Program. J. Cheminform. 2014, 6, 14. [Google Scholar] [CrossRef]
- Gentile, F.; Agrawal, V.; Hsing, M.; Ton, A.T.; Ban, F.; Norinder, U.; Gleave, M.E.; Cherkasov, A. Deep Docking: A Deep Learning Platform for Augmentation of Structure Based Drug Discovery. ACS Cent. Sci. 2020, 6, 939–949. [Google Scholar] [CrossRef]
- Gentile, F.; Yaacoub, J.C.; Gleave, J.; Fernandez, M.; Ton, A.T.; Ban, F.; Stern, A.; Cherkasov, A. Artificial Intelligence–Enabled Virtual Screening of Ultra-Large Chemical Libraries with Deep Docking. Nat. Protoc. 2022, 17, 672–697. [Google Scholar] [CrossRef]
- Hollingsworth, S.A.; Dror, R.O. Molecular Dynamics Simulation for All. Neuron 2018, 99, 1129–1143. [Google Scholar] [CrossRef]
- Torrie, G.M.; Valleau, J.P. Monte Carlo Free Energy Estimates Using Non-Boltzmann Sampling: Application to the Sub-Critical Lennard-Jones Fluid. Chem. Phys. Lett. 1974, 28, 578–581. [Google Scholar] [CrossRef]
- Torrie, J.M.; Valleau, J.P. Nonphysical Sampling Distributions in Monte Carlo Free-Energy Estimation: Umbrella Sampling. J. Comput. Phys. 1977, 23, 187–199. [Google Scholar] [CrossRef]
- You, W.; Tang, Z.; Chang, C.E.A. Potential Mean Force from Umbrella Sampling Simulations: What Can We Learn and What Is Missed? J. Chem. Theory Comput. 2019, 15, 2433–2443. [Google Scholar] [CrossRef]
- Lüdemann, S.K.; Lounnas, V.; Wade, R.C. How Do Substrates Enter and Products Exit the Buried Active Site of Cytochrome P450cam? 1. Random Expulsion Molecular Dynamics Investigation of Ligand Access Channels and Mechanisms. J. Mol. Biol. 2000, 303, 797–811. [Google Scholar] [CrossRef] [PubMed]
- Kokh, D.B.; Amaral, M.; Bomke, J.; Grädler, U.; Musil, D.; Buchstaller, H.P.; Dreyer, M.K.; Frech, M.; Lowinski, M.; Vallee, F.; et al. Estimation of Drug-Target Residence Times by τ-Random Acceleration Molecular Dynamics Simulations. J. Chem. Theory Comput. 2018, 14, 3859–3869. [Google Scholar] [CrossRef]
- Furini, S.; Domene, C. Computational Studies of Transport in Ion Channels Using Metadynamics. Biochim. Biophys. Acta Biomembr. 2016, 1858, 1733–1740. [Google Scholar] [CrossRef] [PubMed]
- Bhakat, S. Collective Variable Discovery in the Age of Machine Learning: Reality, Hype and Everything in Between. RSC Adv. 2022, 12, 25010–25024. [Google Scholar] [CrossRef]
- Xu, J.; Hagler, A. Chemoinformatics and Drug Discovery. Molecules 2002, 7, 566–600. [Google Scholar] [CrossRef]
- Priya, S.; Tripathi, G.; Singh, D.B.; Jain, P.; Kumar, A. Machine Learning Approaches and Their Applications in Drug Discovery and Design. Chem. Biol. Drug Des. 2022, 100, 136–153. [Google Scholar] [CrossRef] [PubMed]
- Giordano, D.; Biancaniello, C.; Argenio, M.A.; Facchiano, A. Drug Design by Pharmacophore and Virtual Screening Approach. Pharmaceuticals 2022, 15, 646. [Google Scholar] [CrossRef]
- Staszak, M.; Staszak, K.; Wieszczycka, K.; Bajek, A.; Roszkowski, K.; Tylkowski, B. Machine Learning in Drug Design: Use of Artificial Intelligence to Explore the Chemical Structure–Biological Activity Relationship. Wiley Interdiscip. Rev. Comput. Mol. Sci. 2022, 12, e1568. [Google Scholar] [CrossRef]
- Menke, J.; Maskri, S.; Koch, O. Computational Ion Channel Research: From the Application of Artificial Intelligence to Molecular Dynamics Simulations. Cell. Physiol. Biochem. 2021, 55, 14–45. [Google Scholar] [CrossRef]
- Rodolpho, C.B.; Vinícius, M.A.; Meryck, F.B.S.; Eugene, M.; Denis, F.; Alexander, T.; Carolina, H.A. Tuning HERG out: Antitarget QSAR Models for Drug Development Rodolpho. Curr. Top. Med. Chem. 2014, 14, 1399–1415. [Google Scholar] [CrossRef]
- Braga, R.C.; Alves, V.M.; Silva, M.F.B.; Muratov, E.; Fourches, D.; Lião, L.M.; Tropsha, A.; Andrade, C.H. Pred-HERG: A Novel Web-Accessible Computational Tool for Predicting Cardiac Toxicity. Mol. Inform. 2015, 34, 698–701. [Google Scholar] [CrossRef]
- Pred-HERG 4.2—LabMol. Available online: http://predherg.labmol.com.br/ (accessed on 8 May 2023).
- Schyman, P.; Liu, R.; Desai, V.; Wallqvist, A. VNN Web Server for ADMET Predictions. Front. Pharmacol. 2017, 8, 889. [Google Scholar] [CrossRef] [PubMed]
- VNN-ADMET—BHSAI. Available online: https://vnnadmet.bhsai.org/vnnadmet/login.xhtml (accessed on 8 May 2023).
- Perron, Q.; Mirguet, O.; Tajmouati, H.; Skiredj, A.; Rojas, A.; Gohier, A.; Ducrot, P.; Bourguignon, M.P.; Sansilvestri-Morel, P.; Do Huu, N.; et al. Deep Generative Models for Ligand-Based de Novo Design Applied to Multi-Parametric Optimization. J. Comput. Chem. 2022, 43, 692–703. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Chen, H. De Novo Molecule Design Using Molecular Generative Models Constrained by Ligand-Protein Interactions. J. Chem. Inf. Model. 2022, 62, 3291–3306. [Google Scholar] [CrossRef]
- ChEMBL Database. Available online: https://www.ebi.ac.uk/chembl/ (accessed on 20 February 2023).
- Gaulton, A.; Bellis, L.J.; Bento, A.P.; Chambers, J.; Davies, M.; Hersey, A.; Light, Y.; McGlinchey, S.; Michalovich, D.; Al-Lazikani, B.; et al. ChEMBL: A Large-Scale Bioactivity Database for Drug Discovery. Nucleic Acids Res. 2012, 40, 1100–1107. [Google Scholar] [CrossRef] [PubMed]
- Gaulton, A.; Hersey, A.; Nowotka, M.L.; Bento, A.P.; Chambers, J.; Mendez, D.; Mutowo, P.; Atkinson, F.; Bellis, L.J.; Cibrian-Uhalte, E.; et al. The ChEMBL Database in 2017. Nucleic Acids Res. 2017, 45, D945–D954. [Google Scholar] [CrossRef]
- Andrei, C.; Mihai, D.P.; Zanfirescu, A.; Nitulescu, G.M.; Negres, S. In Silico Drug Repurposing Framework Predicts Repaglinide, Agomelatine and Protokylol as TRPV1 Modulators with Analgesic Activity. Pharmaceutics 2022, 14, 2563. [Google Scholar] [CrossRef] [PubMed]
- Kong, W.; Huang, W.; Peng, C.; Zhang, B.; Duan, G.; Ma, W.; Huang, Z. Multiple Machine Learning Methods Aided Virtual Screening of NaV1.5 Inhibitors. J. Cell. Mol. Med. 2023, 27, 266–276. [Google Scholar] [CrossRef]
- Heikamp, K.; Bajorath, J. Large-Scale Similarity Search Profiling of ChEMBL Compound Data Sets. J. Chem. Inf. Model. 2011, 51, 1831–1839. [Google Scholar] [CrossRef]
- Liang, L.; Ma, C.; Du, T.; Zhao, Y.; Zhao, X.; Liu, M.; Wang, Z.; Lin, J. Bioactivity-Explorer: A Web Application for Interactive Visualization and Exploration of Bioactivity Data. J. Cheminform. 2019, 11, 47. [Google Scholar] [CrossRef]
- Gadiya, Y.; Zaliani, A.; Gribbon, P.; Hofmann-Apitius, M. PEMT: A Patent Enrichment Tool for Drug Discovery. Bioinformatics 2023, 39, btac716. [Google Scholar] [CrossRef]
- Mok, N.Y.; Brenk, R. Mining the ChEMBL Database: An Efficient Chemoinformatics Workflow for Assembling an Ion Channel-Focused Screening Library. J. Chem. Inf. Model. 2011, 51, 2449–2454. [Google Scholar] [CrossRef] [PubMed]
- DrugBank Database. Available online: https://go.drugbank.com (accessed on 20 February 2023).
- Wishart, D.S.; Knox, C.; Guo, A.C.; Shrivastava, S.; Hassanali, M.; Stothard, P.; Chang, Z.; Woolsey, J. DrugBank: A Comprehensive Resource for in Silico Drug Discovery and Exploration. Nucleic Acids Res. 2006, 34, 668–672. [Google Scholar] [CrossRef]
- Wishart, D.S.; Feunang, Y.D.; Guo, A.C.; Lo, E.J.; Marcu, A.; Grant, J.R.; Sajed, T.; Johnson, D.; Li, C.; Sayeeda, Z.; et al. DrugBank 5.0: A Major Update to the DrugBank Database for 2018. Nucleic Acids Res. 2018, 46, D1074–D1082. [Google Scholar] [CrossRef]
- Feng, H.; Wei, G. Virtual Screening of DrugBank Database for HERG Blockers Using Topological Laplacian-Assisted AI Models. Comput. Biol. Med. 2023, 153, 106491. [Google Scholar] [CrossRef]
- Wu, X.; Zeng, W.; Lin, F.; Zhou, X. NeuRank: Learning to Rank with Neural Networks for Drug–Target Interaction Prediction. BMC Bioinform. 2021, 22, 567. [Google Scholar] [CrossRef]
- Kanehisa, M. A Database for Post-Genome Analysis. Trends Genet. 1997, 13, 375–376. [Google Scholar] [CrossRef]
- Mysinger, M.M.; Carchia, M.; Irwin, J.J.; Shoichet, B.K. Directory of Useful Decoys, Enhanced (DUD-E): Better Ligands and Decoys for Better Benchmarking. J. Med. Chem. 2012, 55, 6582–6594. [Google Scholar] [CrossRef]
- Enamine. Available online: https://enamine.net (accessed on 20 February 2023).
- ZINC Database. Available online: https://zinc.docking.org (accessed on 20 February 2023).
- ChemBridge. Available online: https://chembridge.com (accessed on 20 February 2023).
- Life Chemicals. Available online: https://lifechemicals.com (accessed on 20 February 2023).
- Schwede, T.; Kopp, J.; Guex, N.; Peitsch, M.C. SWISS-MODEL: An Automated Protein Homology-Modeling Server. Nucleic Acids Res. 2003, 31, 3381–3385. [Google Scholar] [CrossRef]
- Song, Y.; Dimaio, F.; Wang, R.Y.R.; Kim, D.; Miles, C.; Brunette, T.; Thompson, J.; Baker, D. High-Resolution Comparative Modeling with RosettaCM. Structure 2013, 21, 1735–1742. [Google Scholar] [CrossRef]
- Modeller. Available online: https://salilab.org/modeller/ (accessed on 20 February 2023).
- Baek, M.; DiMaio, F.; Anishchenko, I.; Dauparas, J.; Ovchinnikov, S.; Lee, G.R.; Wang, J.; Cong, Q.; Kinch, L.N.; Schaeffer, R.D.; et al. Accurate Prediction of Protein Structures and Interactions Using a Three-Track Neural Network. Science 2021, 373, 871–876. [Google Scholar] [CrossRef] [PubMed]
- Wu, R.; Ding, F.; Wang, R.; Shen, R.; Zhang, X.; Luo, S.; Su, C.; Wu, Z.; Xie, Q.; Bergerc, B.; et al. High-Resolution de Novo Structure Prediction from Primary Sequence. bioRxiv 2022. [Google Scholar] [CrossRef]
- Terashi, G.; Wang, X.; Kihara, D. Protein Model Refinement for Cryo-EM Maps Using AlphaFold 2 and the DAQ Score Research Papers. Acta Crystallogr. Sect. D Struct. Biol. 2023, 79, 10–21. [Google Scholar] [CrossRef] [PubMed]
- Khan, H.M.; Guo, J.; Duff, H.J.; Tieleman, D.P.; Noskov, S.Y. Refinement of a Cryo-EM Structure of HERG: Bridging Structure and Function. Biophys. J. 2021, 120, 738–748. [Google Scholar] [CrossRef]
- Vandenberg, J.I.; Perry, M.D.; Perrin, M.J.; Mann, S.A.; Ke, Y.; Hill, A.P. HERG K(+) Channels: Structure, Function, and Clinical Significance. Physiol. Rev. 2012, 92, 1393–1478. [Google Scholar] [CrossRef]
- Wang, W.; MacKinnon, R. Cryo-EM Structure of the Open Human Ether-à-Go-Go-Related K+ Channel HERG. Cell 2017, 169, 422–430.e10. [Google Scholar] [CrossRef]
- Trabuco, L.G.; Villa, E.; Schreiner, E.; Harrison, C.B.; Schulten, K. Molecular Dynamics Flexible Fitting: A Practical Guide to Combine Cryo-Electron Microscopy and X-Ray Crystallography. Methods 2009, 49, 174–180. [Google Scholar] [CrossRef] [PubMed]
- Dämgen, M.A.; Biggin, P.C. A Refined Open State of the Glycine Receptor Obtained via Molecular Dynamics Simulations. Structure 2020, 28, 130–139.e2. [Google Scholar] [CrossRef] [PubMed]
- San Martín, V.P.; Sazo, A.; Utreras, E.; Moraga-Cid, G.; Yévenes, G.E. Glycine Receptor Subtypes and Their Roles in Nociception and Chronic Pain. Front. Mol. Neurosci. 2022, 15, 848642. [Google Scholar] [CrossRef]
- Houtman, M.J.C.; Friesacher, T.; Chen, X.; Zangerl-Plessl, E.M.; van der Heyden, M.A.G.; Stary-Weinzinger, A. Development of IKATP Ion Channel Blockers Targeting Sulfonylurea Resistant Mutant KIR6.2 Based Channels for Treating DEND Syndrome. Front. Pharmacol. 2022, 12, 4051. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Garon, A.; Wieder, M.; Houtman, M.J.C.; Zangerl-Plessl, E.M.; Langer, T.; Van Der Heyden, M.A.G.; Stary-Weinzinger, A. Computational Identification of Novel Kir6 Channel Inhibitors. Front. Pharmacol. 2019, 10, 549. [Google Scholar] [CrossRef]
- Ashcroft, F.M.; Puljung, M.C.; Vedovato, N. Neonatal Diabetes and the KATP Channel: From Mutation to Therapy. Trends Endocrinol. Metab. 2017, 28, 377–387. [Google Scholar] [CrossRef]
- De Jong, D.H.; Singh, G.; Bennett, W.F.D.; Arnarez, C.; Wassenaar, T.A.; Schäfer, L.V.; Periole, X.; Tieleman, D.P.; Marrink, S.J. Improved Parameters for the Martini Coarse-Grained Protein Force Field. J. Chem. Theory Comput. 2013, 9, 687–697. [Google Scholar] [CrossRef]
- Yelshanskaya, M.V.; Singh, A.K.; Narangoda, C.; Williams, R.S.B.; Kurnikova, M.G.; Sobolevsky, A.I. Structural Basis of AMPA Receptor Inhibition by Trans-4-Butylcyclohexane Carboxylic Acid. Br. J. Pharmacol. 2022, 179, 3628–3644. [Google Scholar] [CrossRef]
- Hanada, T. Ionotropic Glutamate Receptors in Epilepsy: A Review Focusing on Ampa and Nmda Receptors. Biomolecules 2020, 10, 464. [Google Scholar] [CrossRef]
- Rugg-Gunn, F. Adverse Effects and Safety Profile of Perampanel: A Review of Pooled Data. Epilepsia 2014, 55, 13–15. [Google Scholar] [CrossRef]
- Shi, S.; Ma, B.; Sun, F.; Qu, C.; Li, G.; Shi, D.; Liu, W.; Zhang, H.; Hailong, A. Zafirlukast Inhibits the Growth of Lung Adenocarcinoma via Inhibiting TMEM16A Channel Activity. J. Biol. Chem. 2022, 298, 101731. [Google Scholar] [CrossRef] [PubMed]
- Ji, Q.; Guo, S.; Wang, X.; Pang, C.; Zhan, Y.; Chen, Y.; An, H. Recent Advances in TMEM16A: Structure, Function, and Disease. J. Cell. Physiol. 2019, 234, 7856–7873. [Google Scholar] [CrossRef]
- Zimova, L.; Ptakova, A.; Mitro, M.; Krusek, J.; Vlachova, V. Activity Dependent Inhibition of TRPC1/4/5 Channels by Duloxetine Involves Voltage Sensor-like Domain. Biomed. Pharmacother. 2022, 152, 113262. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues-Amorim, D.; Olivares, J.M.; Spuch, C.; Rivera-Baltanás, T. A Systematic Review of Efficacy, Safety, and Tolerability of Duloxetine. Front. Psychiatry 2020, 11, 554899. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, K.; Lennerz, J.K.; Hein, A.; Link, A.S.; Stefan Kaczmarek, J.; Delling, M.; Uysal, S.; Pfeifer, J.D.; Riccio, A.; Clapham, D.E. Transient Receptor Potential Cation Channel, Subfamily C, Member 5 (TRPC5) Is a Cold-Transducer in the Peripheral Nervous System. Proc. Natl. Acad. Sci. USA 2011, 108, 18114–18119. [Google Scholar] [CrossRef]
- Pushpakom, S.; Iorio, F.; Eyers, P.A.; Escott, K.J.; Hopper, S.; Wells, A.; Doig, A.; Guilliams, T.; Latimer, J.; McNamee, C.; et al. Drug Repurposing: Progress, Challenges and Recommendations. Nat. Rev. Drug Discov. 2018, 18, 41–58. [Google Scholar] [CrossRef]
- Toft-Bertelsen, T.L.; Jeppesen, M.G.; Tzortzini, E.; Xue, K.; Giller, K.; Becker, S.; Mujezinovic, A.; Bentzen, B.H.; Andreas, L.B.; Kolocouris, A.; et al. Amantadine Has Potential for the Treatment of COVID-19 Because It Inhibits Known and Novel Ion Channels Encoded by SARS-CoV-2. Commun. Biol. 2021, 4, 1347. [Google Scholar] [CrossRef]
- Jefferson, T.; Demicheli, V.; Di Pietrantonj, C.; Rivetti, D. Amantadine and Rimantadine for Influenza A in Adults. Cochrane Database Syst. Rev. 2006, 2006, CD001169. [Google Scholar] [CrossRef]
- Meng, X.-Y.; Zhang, H.-X.; Mezei, M.; Cui, M. Molecular Docking: A Powerful Approach for Structure-Based Drug Discovery. Curr. Comput. Aided Drug Des. 2012, 7, 146–157. [Google Scholar] [CrossRef]
- Abdelsayed, M.; Page, D.; Ruben, P.C. ARumenamides: A Novel Class of Potential Antiarrhythmic Compounds. Front. Pharmacol. 2022, 13, 976903. [Google Scholar] [CrossRef]
- Han, D.; Tan, H.; Sun, C.; Li, G. Dysfunctional Nav1.5 Channels Due to SCN5A Mutations. Exp. Biol. Med. 2018, 243, 852–863. [Google Scholar] [CrossRef] [PubMed]
- Balserfi, J.R.; Nuss, H.B.; Romashko, D.N.; Marban, E. Functional Consequences of Lidocaine Binding to Slow-Inactivated Sodium Channels. J. Gen. Physiol. 1996, 107, 643–658. [Google Scholar] [CrossRef]
- Sokolov, S.; Peters, C.H.; Rajamani, S.; Ruben, P.C. Proton-Dependent Inhibition of the Cardiac Sodium Channel Nav1.5 by Ranolazine. Front. Pharmacol. 2013, 4, 78. [Google Scholar] [CrossRef]
- Potet, F.; Egecioglu, D.E.; Burridge, P.W.; George, A.L. GS-967 and Eleclazine Block Sodium Channels in Human Induced Pluripotent Stem Cell-Derived Cardiomyocytes. Mol. Pharmacol. 2020, 98, 540–547. [Google Scholar] [CrossRef] [PubMed]
- Urdaneta, R.K.F.; Mcarthur, J.R.; Ohm, M.P.G.; Gaudet, R.; Tikhonov, D.B.; Zhorov, B.S.; French, R.J. Batrachotoxin Acts as a Stent to Hold Open Homotetrameric Prokaryotic Voltage-Gated Sodium Channels. J. Gen. Physiol. 2019, 151, 186–199. [Google Scholar] [CrossRef] [PubMed]
- Jensen, A.A.; Frølund, B.; Liljefors, T.; Krogsgaard-Larsen, P. Neuronal Nicotinic Acetylcholine Receptors: Structural Revelations, Target Identifications, and Therapeutic Inspirations. J. Med. Chem. 2005, 48, 4705–4745. [Google Scholar] [CrossRef] [PubMed]
- Lasala, M.; Fabiani, C.; Corradi, J.; Antollini, S.; Bouzat, C. Molecular Modulation of Human α 7 Nicotinic Receptor by Amyloid- β Peptides. Front. Cell Neurosci. 2019, 13, 37. [Google Scholar] [CrossRef]
- Laikowski, M.M.; Reisdorfer, F.; Moura, S. NAChR A4β2 Subtype and Their Relation with Nicotine Addiction, Cognition, Depression and Hyperactivity Disorder. Curr. Med. Chem. 2018, 26, 3792–3811. [Google Scholar] [CrossRef]
- Batista, V.S.; Gonçalves, A.M. Pharmacophore Mapping Combined with DbCICA Reveal New Structural Features for the Development of Novel Ligands Targeting A4β2 and A7 Nicotinic Acetylcholine Receptors. Molecules 2022, 27, 8236. [Google Scholar] [CrossRef]
- Doñate-Macian, P.; Duarte, Y.; Rubio-Moscardo, F.; Pérez-Vilaró, G.; Canan, J.; Díez, J.; González-Nilo, F.; Valverde, M.A. Structural Determinants of TRPV4 Inhibition and Identification of New Antagonists with Antiviral Activity. Br. J. Pharmacol. 2022, 179, 3576–3591. [Google Scholar] [CrossRef]
- Rajan, S.; Schremmer, C.; Weber, J.; Alt, P.; Geiger, F.; Dietrich, A. Ca2+ Signaling by TRPV4 Channels in Respiratory Function and Disease. Cells 2021, 10, 822. [Google Scholar] [CrossRef] [PubMed]
- NCI Open Database Compounds. Available online: https://cactus.nci.nih.gov/download/nci/ (accessed on 20 February 2023).
- Valverde, M.A.; Diez, J. The TRPV4 Channel Links Calcium in Fl Ux to DDX3X Activity and Viral Infectivity. Nat. Commun. 2018, 9, 2307. [Google Scholar] [CrossRef]
- Dallas, M. Patch Clamp Physiology; Molecular Devices, LLC.: San Jose, CA, USA, 2021; ISBN 9781071608173. [Google Scholar]
- Walters, W.P.; Stahl, M.T.; Murcko, M.A. Virtual Screening—An Overview. Drug Discov. Today 1998, 3, 160–178. [Google Scholar] [CrossRef]
- Liu, H.; Gao, Z.B.; Yao, Z.; Zheng, S.; Li, Y.; Zhu, W.; Tan, X.; Luo, X.; Shen, J.; Chen, K.; et al. Discovering Potassium Channel Blockers from Synthetic Compound Database by Using Structure-Based Virtual Screening in Conjunction with Electrophysiological Assay. J. Med. Chem. 2007, 50, 83–93. [Google Scholar] [CrossRef] [PubMed]
- MDL®. Available Chemicals Directory. Available online: http://www.mdli.com/acd/ (accessed on 13 March 2023).
- Pegoraro, S.; Lang, M.; Dreker, T.; Kraus, J.; Hamm, S.; Meere, C.; Feurle, J.; Tasler, S.; Prütting, S.; Kuras, Z.; et al. Inhibitors of Potassium Channels KV1.3 and IK-1 as Immunosuppressants. Bioorganic Med. Chem. Lett. 2009, 19, 2299–2304. [Google Scholar] [CrossRef]
- Teisseyre, A.; Palko-Labuz, A.; Sroda-Pomianek, K.; Michalak, K. Voltage-Gated Potassium Channel Kv1.3 as a Target in Therapy of Cancer. Front. Oncol. 2019, 9, 933. [Google Scholar] [CrossRef]
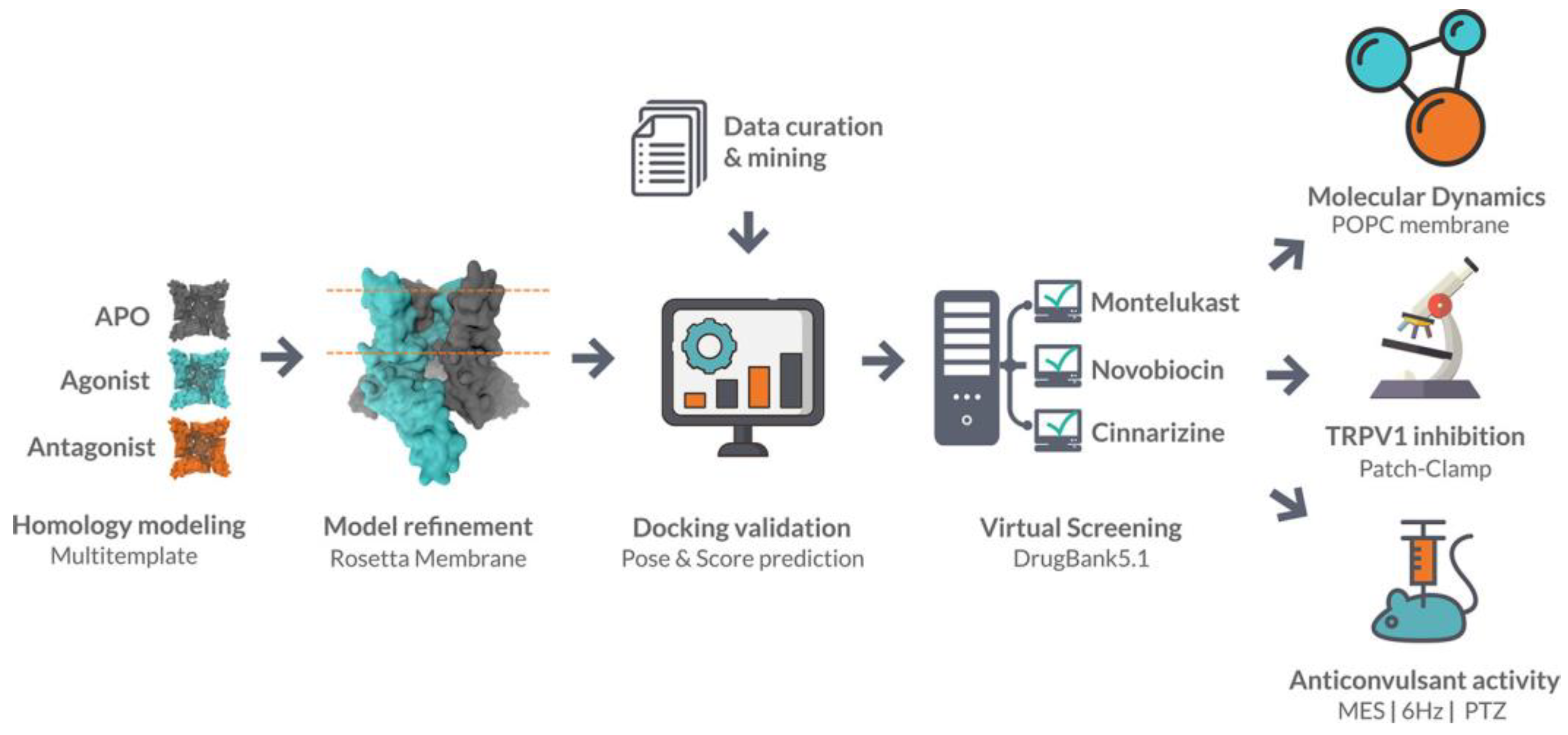
- Llanos, M.A.; Enrique, N.; Sbaraglini, M.L.; Garofalo, F.M.; Talevi, A.; Gavernet, L.; Mart, P. Structure-Based Virtual Screening Identifies Novobiocin, Montelukast, and Cinnarizine as TRPV1 Modulators with Anticonvulsant Activity In Vivo. J. Chem. Inf. Model. 2022, 62, 3008–3022. [Google Scholar] [CrossRef]
- Montell, C.; Caterina, M.J. Thermoregulation: Channels That Are Cool to the Core. Curr. Biol. 2007, 17, 885–887. [Google Scholar] [CrossRef]
- Cho, S.J.; Vaca, M.A.; Miranda, C.J.; Gouemo, P.N. Inhibition of Transient Potential Receptor Vanilloid Type 1 Suppresses Seizure Susceptibility in the Genetically Epilepsy-Prone Rat. CNS Neurosci. Ther. 2018, 24, 18–28. [Google Scholar] [CrossRef]
- Garami, A.; Pakai, E.; McDonald, H.A.; Reilly, R.M.; Gomtsyan, A.; Corrigan, J.J.; Pinter, E.; Zhu, D.X.D.; Lehto, S.G.; Gavva, N.R.; et al. TRPV1 Antagonists That Cause Hypothermia, Instead of Hyperthermia, in Rodents: Compounds’ Pharmacological Profiles, in Vivo Targets, Thermoeffectors Recruited and Implications for Drug Development. Acta Physiol. 2018, 223, e13038. [Google Scholar] [CrossRef]
- Pasqualetto, G.; Zuanon, M.; Brancale, A.; Young, M.T. Identification of a Novel P2X7 Antagonist Using Structure-Based Virtual Screening. Front. Pharmacol. 2023, 13, 1094607. [Google Scholar] [CrossRef] [PubMed]
- Di Virgilio, F.; Dal Ben, D.; Sarti, A.C.; Giuliani, A.L.; Falzoni, S. Review the P2X7 Receptor in Infection and Inflammation. Immunity 2017, 47, 15–31. [Google Scholar] [CrossRef]
- Specs. Available online: https://www.specs.net (accessed on 20 February 2023).
- Kimber, T.B.; Chen, Y.; Volkamer, A. Deep Learning in Virtual Screening: Recent Applications and Developments. Int. J. Mol. Sci. 2021, 22, 4435. [Google Scholar] [CrossRef] [PubMed]
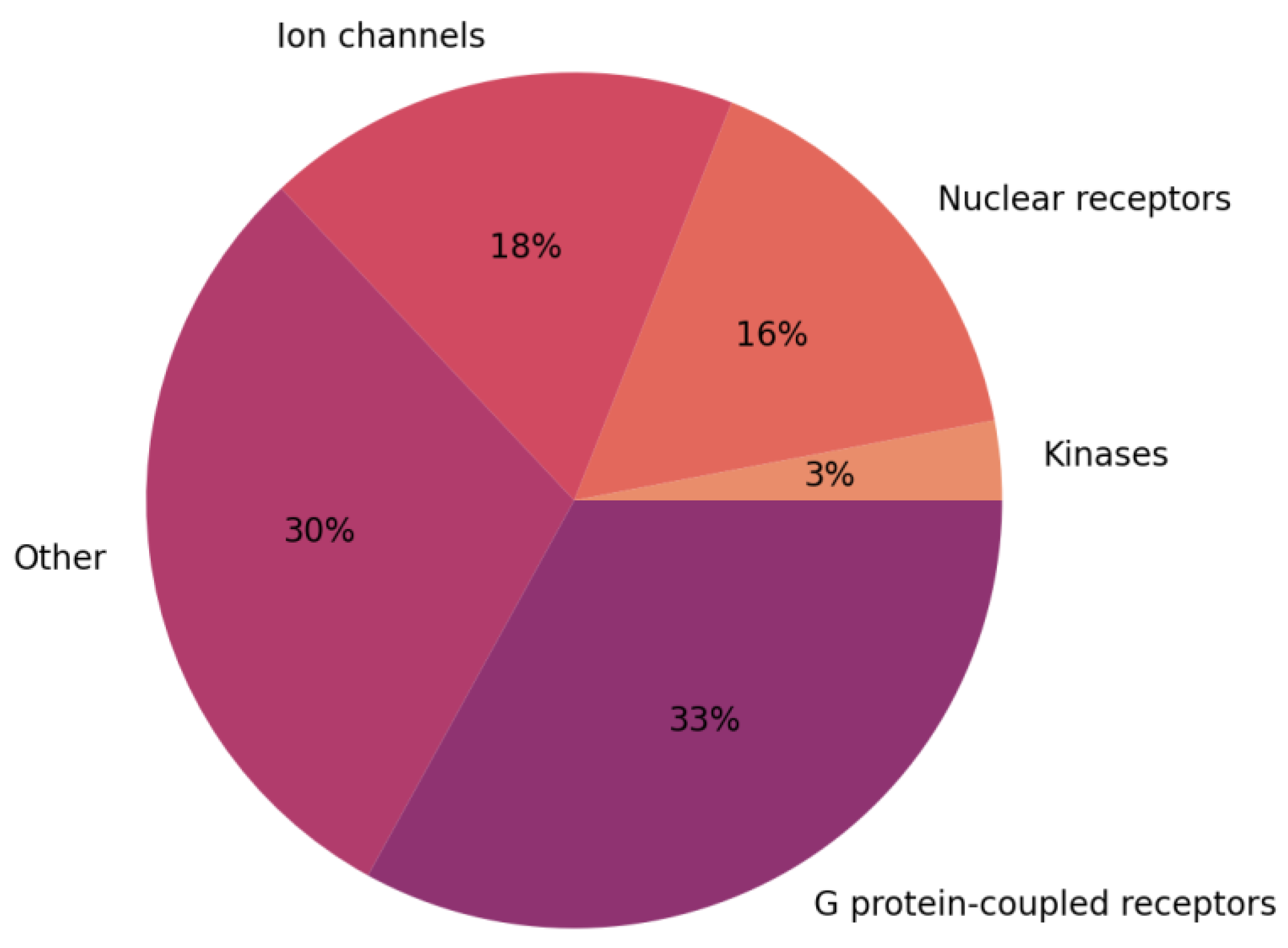
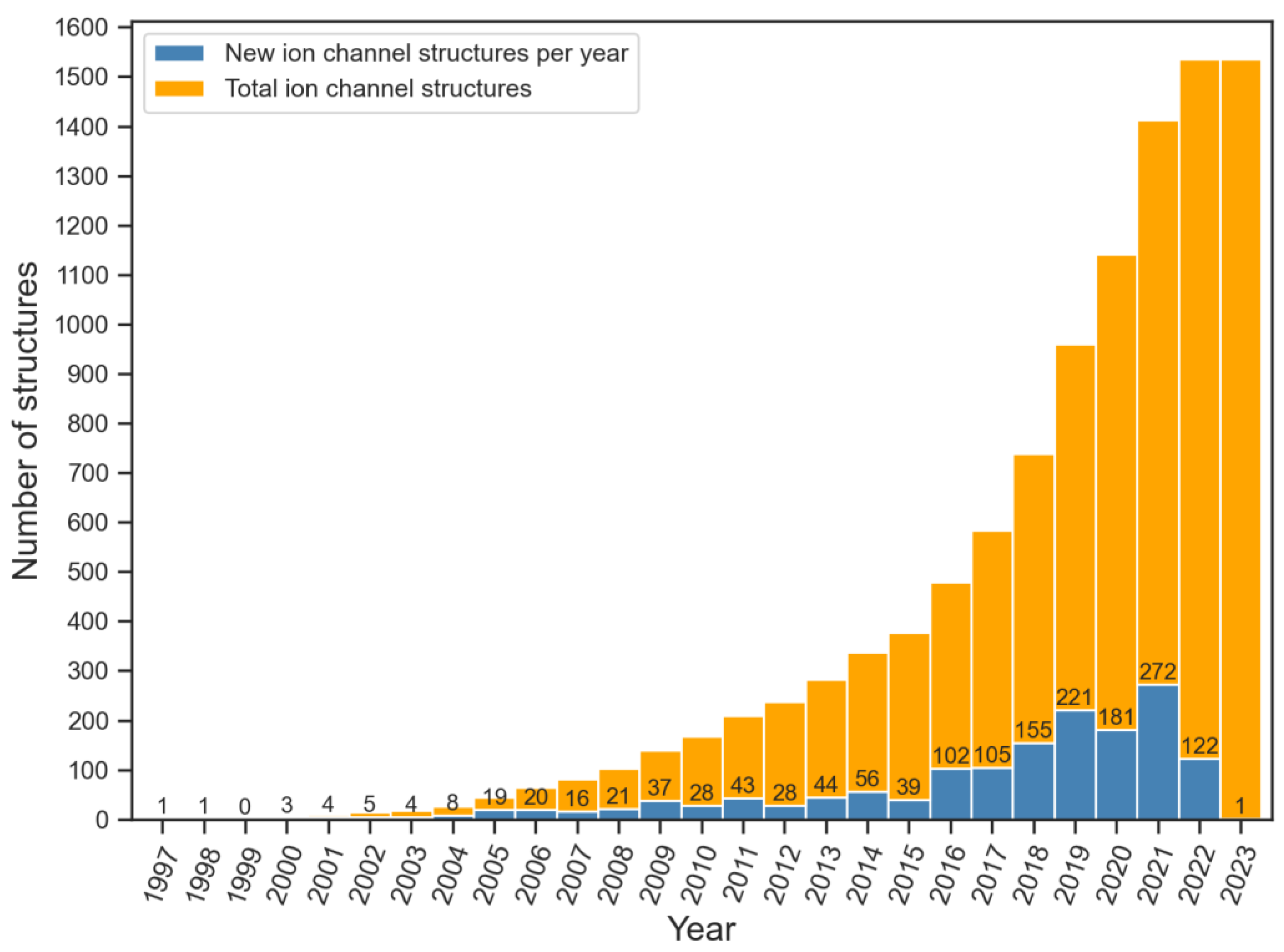
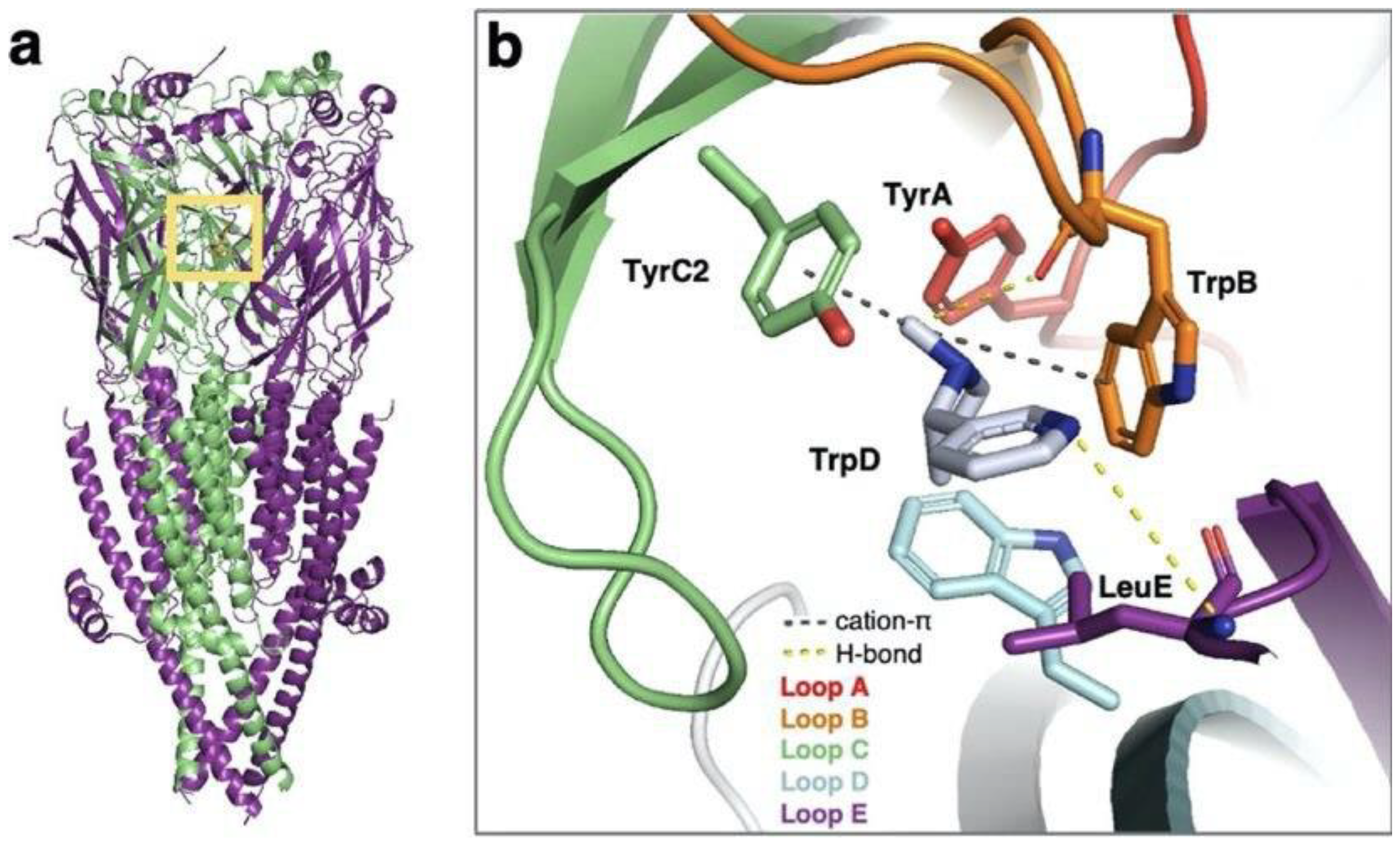
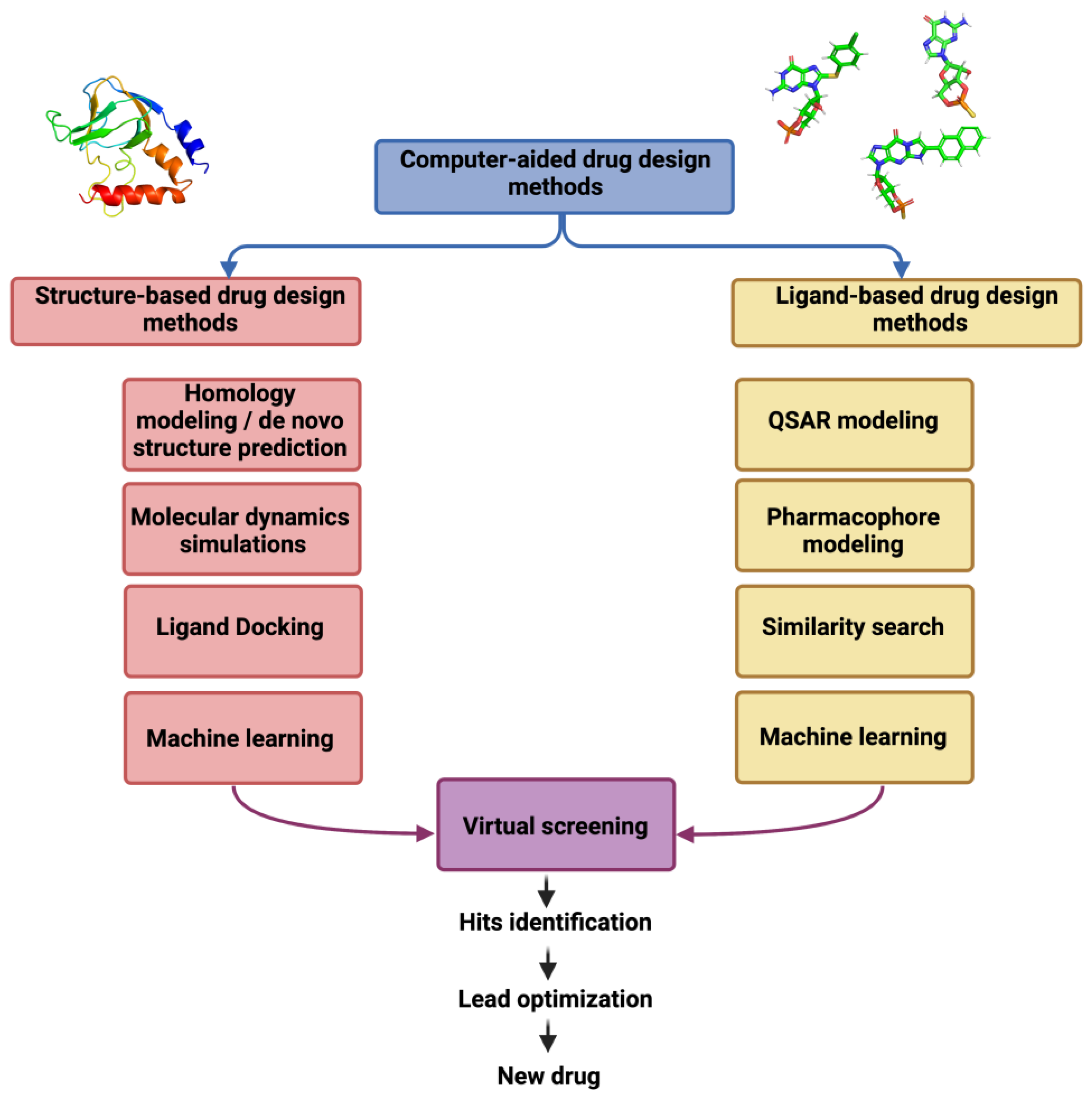
| Ion Channel Family | Number of Structures | Number of Structures in Complex with Ligands |
|---|---|---|
| Transient receptor potential channels | 222 | 165 |
| KcsA voltage-gated K+ channels | 61 | 51 |
| Cyclic nucleotide-gated ion channels | 53 | 45 |
| Inward rectifier potassium channels | 53 | 51 |
| Two pore Na+ channels | 46 | 45 |
| NaK channels | 36 | 22 |
| Ryanodine receptors | 26 | 25 |
| K+ channels MthK | 24 | 16 |
| Two pore Ca2+ channels | 23 | 18 |
| Voltage-sensitive calcium channel | 22 | 22 |
| Polycystin cation channels | 21 | 12 |
| KCNQ voltage-gated potassium channels | 19 | 17 |
| ATP-sensitive K+ channels | 16 | 15 |
| Slo potassium channels | 14 | 9 |
| Voltage-sensing proton channel | 6 | 3 |
| Calcium-activated SK potassium channels | 4 | 4 |
| Plant inwardly rectifying potassium channels | 4 | 4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pliushcheuskaya, P.; Künze, G. Recent Advances in Computer-Aided Structure-Based Drug Design on Ion Channels. Int. J. Mol. Sci. 2023, 24, 9226. https://doi.org/10.3390/ijms24119226
Pliushcheuskaya P, Künze G. Recent Advances in Computer-Aided Structure-Based Drug Design on Ion Channels. International Journal of Molecular Sciences. 2023; 24(11):9226. https://doi.org/10.3390/ijms24119226
Chicago/Turabian StylePliushcheuskaya, Palina, and Georg Künze. 2023. "Recent Advances in Computer-Aided Structure-Based Drug Design on Ion Channels" International Journal of Molecular Sciences 24, no. 11: 9226. https://doi.org/10.3390/ijms24119226
APA StylePliushcheuskaya, P., & Künze, G. (2023). Recent Advances in Computer-Aided Structure-Based Drug Design on Ion Channels. International Journal of Molecular Sciences, 24(11), 9226. https://doi.org/10.3390/ijms24119226





