De Novo Transcriptome Profiling for the Generation and Validation of Microsatellite Markers, Transcription Factors, and Database Development for Andrographis paniculata
Abstract
1. Introduction
2. Results
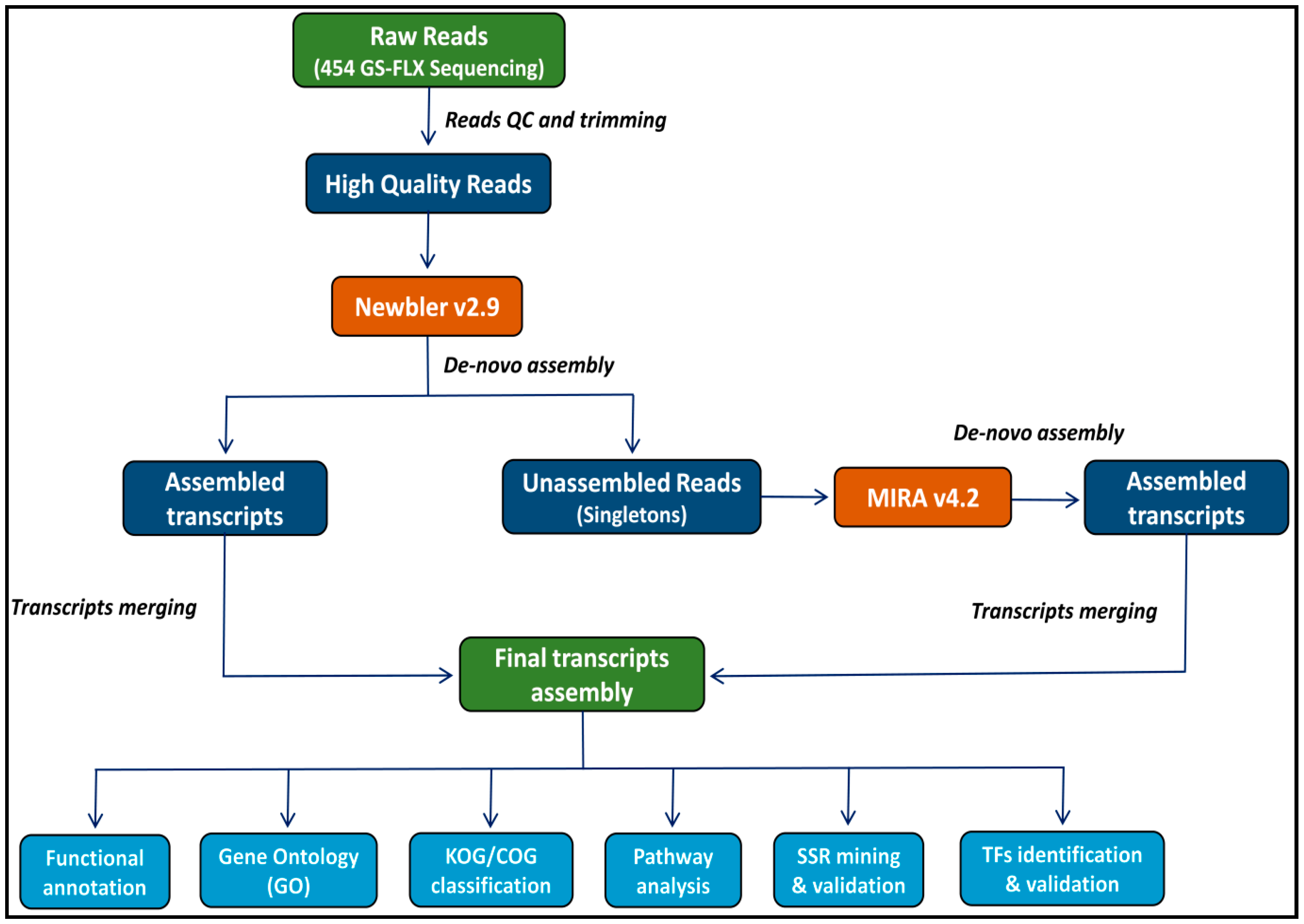
2.1. 454 GS-FLX Sequencing and De Novo Transcriptome Assembly
2.2. Functional Annotation and Classification of Transcripts
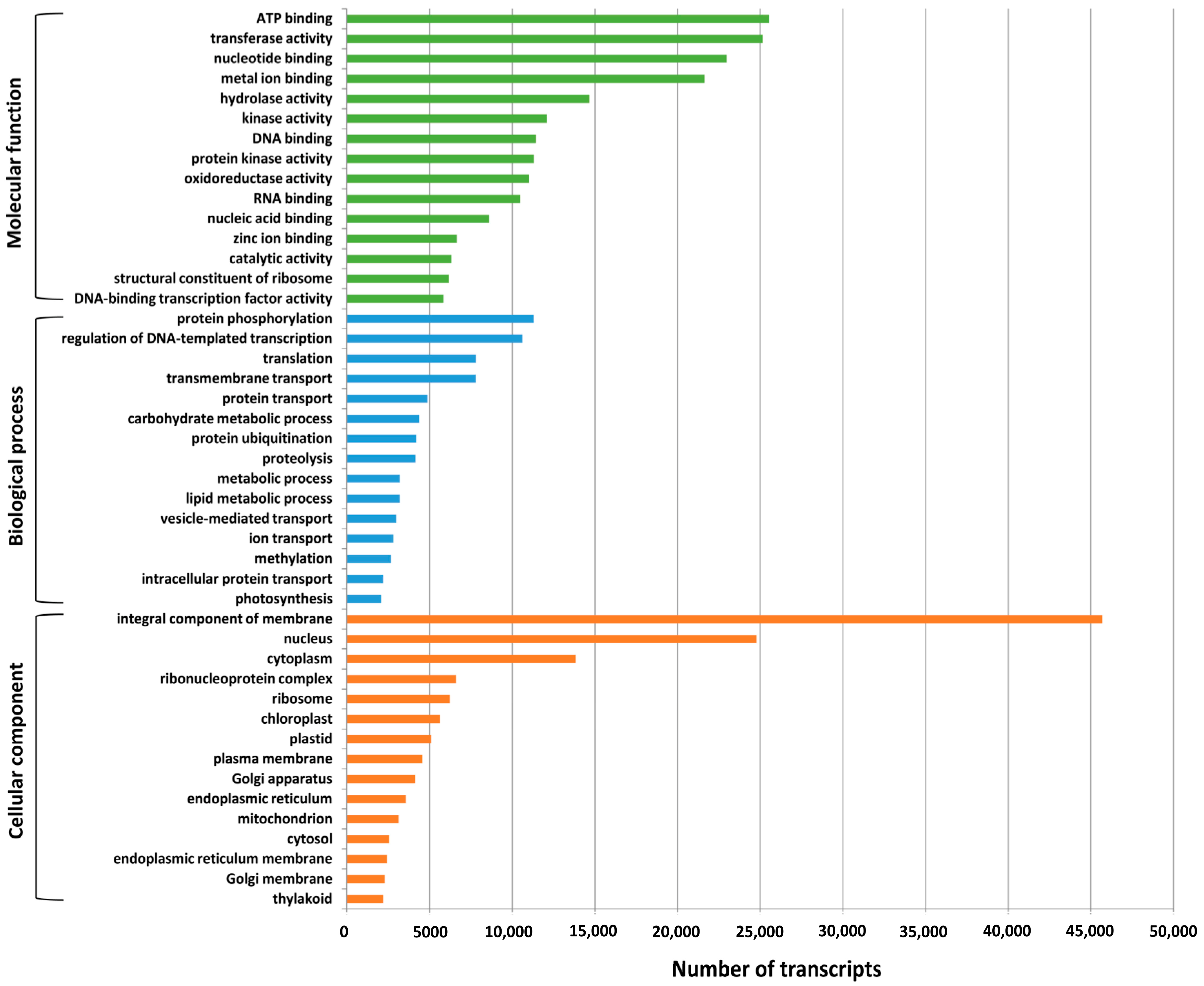
2.3. Gene Ontology (GO) Annotation
2.4. COG Analysis
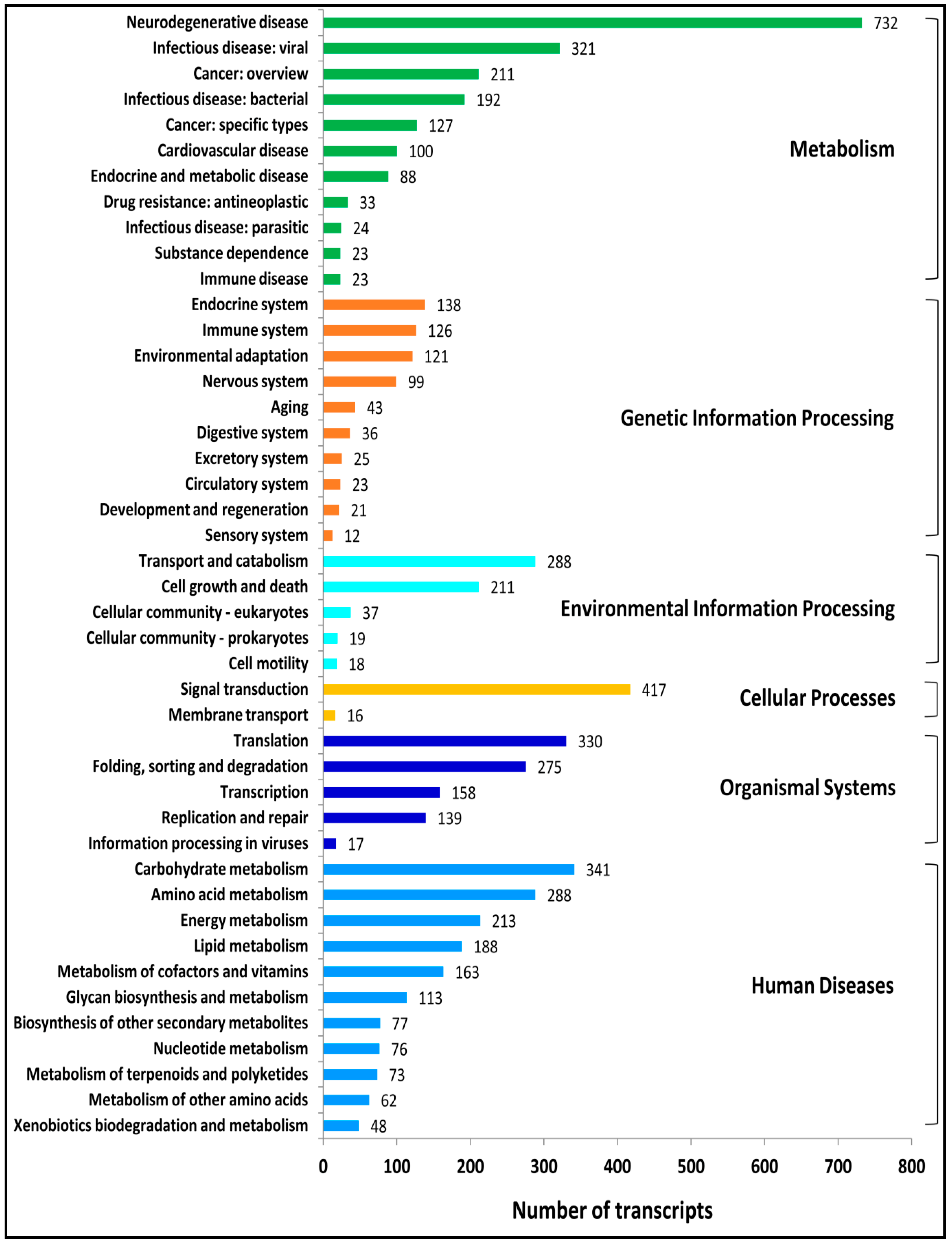
2.5. Metabolic Pathway Enrichment Analysis
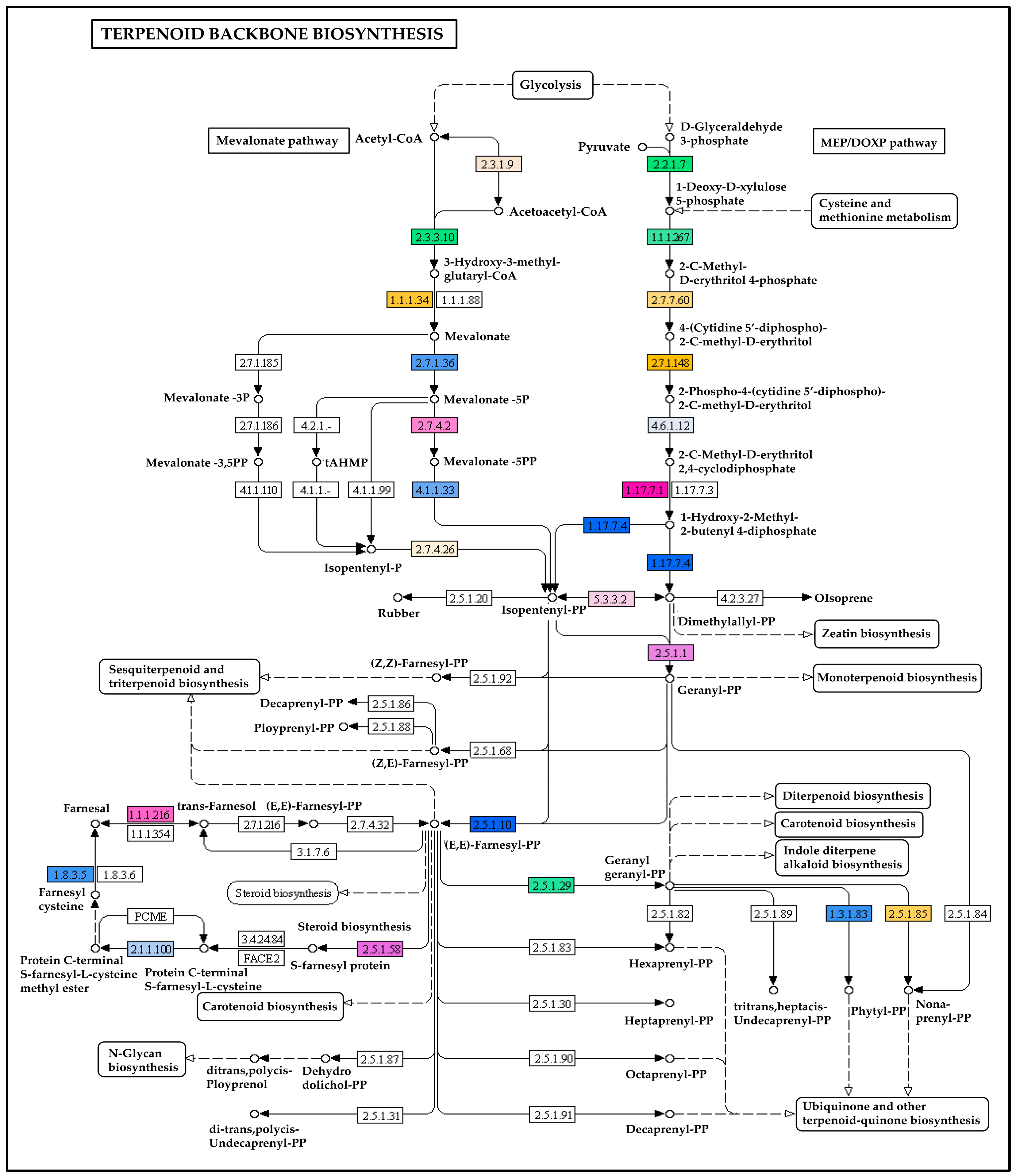
2.6. Transcripts Encoding for the Enzymes Involved in Terpenoids Biosynthesis
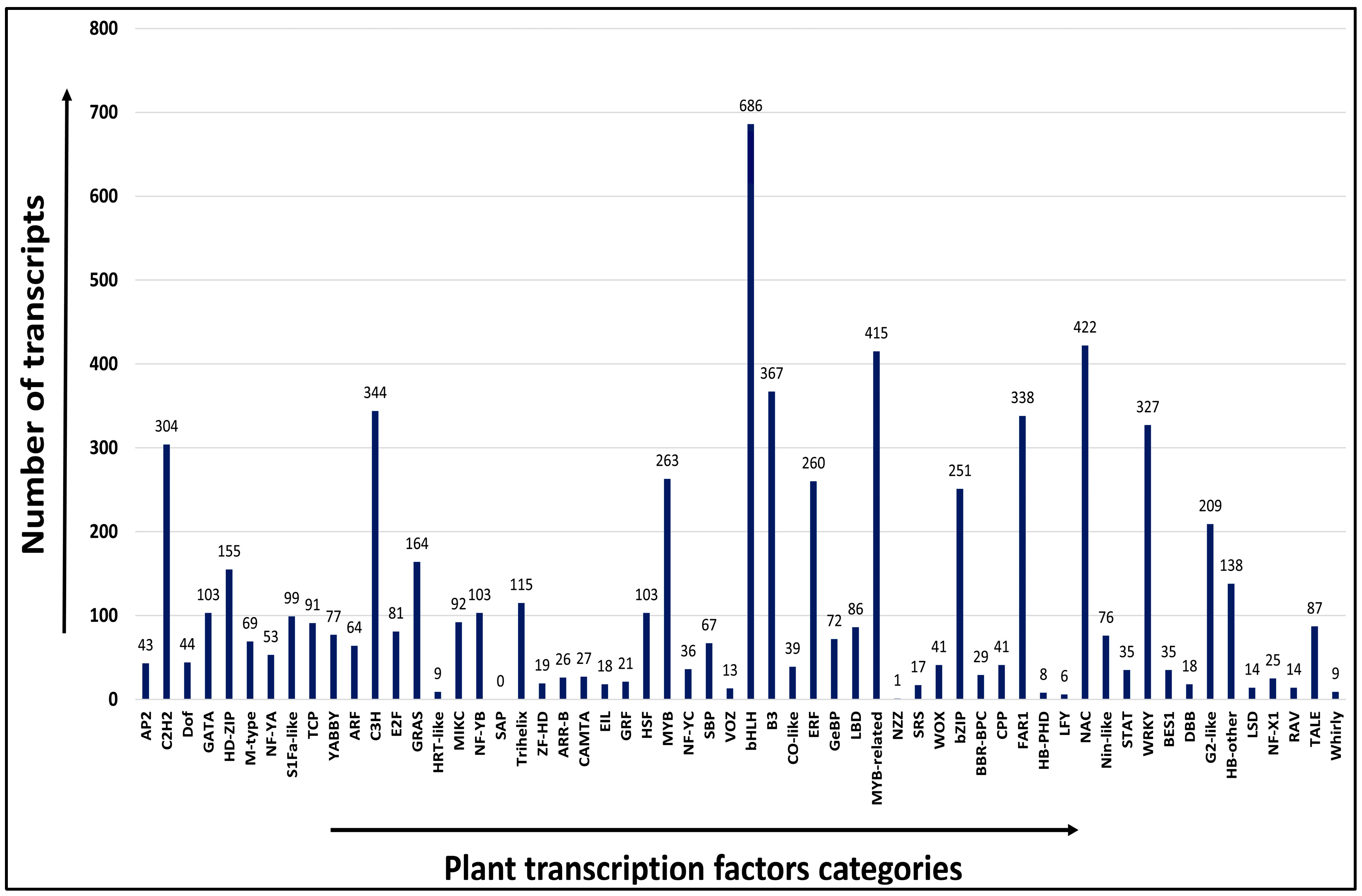
2.7. Transcription Factor (TF) Analysis
2.8. Validation of Transcription Factors
2.9. Frequency and Distribution of EST-SSRs
2.10. Assessment of Genetic Diversity in A. paniculata
2.11. Meta-Transcriptome Assembly and Data Analysis
2.12. Database Resource of A. paniculata
3. Discussion
4. Materials and Methods
4.1. Plant Materials and RNA Isolation
4.2. cDNA Library Preparation and Sequencing
4.3. Reads Quality Assessment and De Novo Transcriptome Assembly
4.4. Functional Annotation and Biological Pathways Assignment
4.5. Identification of Transcription Factors and Gene Families
4.6. Selection of TF Genes, Primer Designing, and PCR Amplification
4.7. Identification of EST-SSR Markers and Primer Designing
4.8. Assessment of EST-SSR Polymorphism
4.9. Genetic Diversity Data Analysis
4.10. Meta-Transcriptome Assembly for the Generation of a Database Resource
4.11. Meta-Transcriptome Analysis
4.12. Database Resource Development
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kumar, S.; Singh, B.; Bajpai, V. Andrographis paniculata (Burm.f.) Nees: Traditional uses, phytochemistry, pharmacological properties, and quality control/quality assurance. J. Ethnopharmacol. 2021, 275, 114054. [Google Scholar] [CrossRef] [PubMed]
- Chao, W.W.; Lin, B.F. Isolation and identification of bioactive compounds in Andrographis paniculata (Chuanxinlian). Chin. Med. 2010, 5, 17. [Google Scholar] [CrossRef] [PubMed]
- Gan, L.; Zheng, Y.; Deng, L.; Sun, P.; Ye, J.; Wei, X.; Liu, F.; Yu, L.; Ye, W.; Fan, C.; et al. Diterpenoid Lactones with Anti-Inflammatory Effects from the Aerial Parts of Andrographis paniculata. Molecules 2019, 24, 2726. [Google Scholar] [CrossRef] [PubMed]
- Zhou, K.L.; Chen, L.X.; Zhuang, Y.L.; Wang, N.L.; Yao, X.S.; Qiu, F. Two new ent-labdane diterpenoid glycosides from the aerial parts of Andrographis paniculata. J. Asian Nat. Prod. Res. 2008, 10, 939–943. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, X.L.; Lai, C.J.S.; Wang, R.S.; Kang, L.P.; Ma, T.; Zhao, Z.H.; Gao, W.; Huang, L.Q. Functional characterization of three flavonoid glycosyl transferases from Andrographis paniculata. R. Soc. Open Sci. 2019, 6, 190150. [Google Scholar] [CrossRef] [PubMed]
- Sivananthan, M.; Elamaran, M. Medicinal and Pharmacological properties of Andrographis paniculata. Int. J. Biomol. Biomed. 2013, 3, 1–12. [Google Scholar]
- Hossain, S.; Urbi, Z.; Karuniawati, H.; Mohiuddin, R.B.; Moh Qrimida, A.; Allzrag, A.M.; Ming, L.C.; Pagano, E.; Capasso, R. Andrographis paniculata (Burm. f.) wall. ex nees: An updated review of Phytochemistry, antimicrobial pharmacology, and clinical safety and efficacy. Life 2021, 11, 348. [Google Scholar] [CrossRef]
- Patel, A.A.; Shukla, Y.M.; Kumar, S.; Sakure, A.A.; Parekh, M.J.; Zala, H.N. Transcriptome analysis for molecular landscaping of genes controlling diterpene andrographolide biosynthesis in Andrographis paniculata (Burm. f.) nees. 3 Biotech 2020, 10, 512. [Google Scholar] [CrossRef]
- Patidar, S.; Gontia, A.S.; Upadhyay, A.; Nayak, P.S. Biochemical Constituents in Kalmegh (Andrographis paniculata Nees.) Under Various Row Spacing’s and Nitrogen Levels. World Appl. Sci. J. 2011, 15, 1095–1099. [Google Scholar]
- Yao, W.; An, T.; Xu, Z.; Zhang, L.; Gao, H.; Sun, W.; Liao, B.; Jiang, C.; Liu, Z.; Duan, L.; et al. Genomic-wide identification and expression analysis of AP2/ERF transcription factors related to andrographolide biosynthesis in Andrographis paniculata. Ind. Crops Prod. 2020, 157, 112878. [Google Scholar] [CrossRef]
- Vlk, D.; Řepková, J. Application of next-generation sequencing in plant breeding. Czech J. Genet. Plant Breed. 2017, 53, 89–96. [Google Scholar] [CrossRef]
- Singh, V.K.; Singh, A.K.; Singh, S.; Singh, B.D. Next-Generation Sequencing (NGS) Tools and Impact in Plant Breeding. In Advances in Plant Breeding Strategies: Breeding, Biotechnology and Molecular Tools; Springer: Berlin/Heideberg, Germany, 2015; pp. 563–612. [Google Scholar]
- Sun, W.; Leng, L.; Yin, Q.; Xu, M.; Huang, M.; Xu, Z.; Chen, S.; Jiang, C.; Xie, N.; Zheng, X.; et al. The genome of the medicinal plant Andrographis paniculata provides insight into the biosynthesis of the bioactive diterpenoid neoandrographolide. Plant J. 2019, 97, 841–857. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Chen, S.; Wei, K.; Yang, Z.; Duan, S.; Du, Y.; Qu, P.; Miao, J.; Chen, W.; Dong, Y. Chromosome Level Genome Assembly of Andrographis paniculata. Front. Genet. 2020, 11, 701. [Google Scholar] [CrossRef]
- Garg, A.; Agrawal, L.; Misra, R.C.; Sharma, S.; Ghosh, S. Andrographis paniculata transcriptome provides molecular insights into tissue-specific accumulation of medicinal diterpenes. BMC Genom. 2015, 16, 659. [Google Scholar] [CrossRef] [PubMed]
- Cherukupalli, N.; Divate, M.; Mittapelli, S.R.; Khareedu, V.R.; Vudem, D.R. De novo Assembly of Leaf Transcriptome in the Medicinal Plant Andrographis paniculata. Front. Plant Sci. 2016, 7, 1203. [Google Scholar] [CrossRef]
- Li, J.; Chen, X.; Zhan, R.; He, R. Transcriptome profiling reveals metabolic alteration in Andrographis paniculata in response to continuous cropping. Ind. Crops Prod. 2019, 137, 585–596. [Google Scholar] [CrossRef]
- Sharma, P.; Kumar, S.; Beriwal, S.; Sharma, P.; Bhairappanavar, S.B.; Verma, R.J.; Das, J. Comparative transcriptome profiling and co-expression network analysis reveals functionally coordinated genes associated with metabolic processes of Andrographis paniculata. Plant Gene 2020, 23, 100234. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, X.; Wang, Y.H.; Shen, S.K. De Novo Assembly of Transcriptome and Development of Novel EST-SSR Markers in Rhododendron rex Lévl. through Illumina Sequencing. Front. Plant Sci. 2017, 26, 1664. [Google Scholar] [CrossRef]
- Fang, X.; Zhang, Y.; Cui, J.; Yue, L.; Tao, J.; Wang, Y.; Zhang, R.; Liu, J.; Jiang, A.; Zhang, J. Genetic Map Construction, QTL Mapping, and Candidate Genes Screening of Grain Size Traits in Common Buckwheat (Fagopyrum esculentum M.). Agronomy 2022, 12, 2062. [Google Scholar] [CrossRef]
- Liu, J.J.; Sniezko, R.A.; Sissons, R.; Krakowski, J.; Alger, G.; Schoettle, A.W.; Williams, H.; Zamany, A.; Zitomer, R.A.; Kegley, A. Association Mapping and Development of Marker-Assisted Selection Tools for the Resistance to White Pine Blister Rust in the Alberta Limber Pine Populations. Front. Plant Sci. 2020, 11, 557672. [Google Scholar] [CrossRef]
- Padmesh, P.; Sabu, K.K.; Seeni, S.; Pushpangadan, P. The use of RAPD in assessing genetic variability in Andrographis paniculata Nees, a hepatoprotective drug. Curr. Sci. 1999, 76, 833–835. [Google Scholar]
- Sharma, S.N.; Sinha, R.K.; Sharma, D.K.; Jha, Z. Assessment of intra-specific variability at morphological, molecular and biochemical level of Andrographis paniculata (Kalmegh). Curr. Sci. 2009, 96, 3. [Google Scholar]
- Minz, P.L.; Singh, N.; Mishra, S.K.; Koche, V. Genetic variability among Andrographis paniculata in Chhattisgarh region assessed by RAPD markers. Afr. J. Biotechnol. 2013, 12, 5714–5722. [Google Scholar]
- Valdiani, A.; Talei, D.; Javanmard, A.; Tan, S.G.; Kadir, M.A.; Maziah, M. Morpho-molecular analysis as a prognostic model for repulsive feedback of the medicinal plant “Andrographis paniculata” to allogamy. Gene 2014, 542, 156–167. [Google Scholar] [CrossRef]
- Gangopadhyay, S.; Patel, M.A.; Mistry, J.G.; Kumar, S. RAPD based genetic variability assessment in Andrographis paniculata (burm. f.) wall. Ex Nees (kalmegh): An important hepatoprotective herb. Med. Plants Int. J. Phytomedicines Relat. Ind. 2014, 6, 47. [Google Scholar] [CrossRef]
- Sabu, K.K.; Padmesh, P.; Seeni, S. Intraspecific variation in active principle content and isozymes of Andrographis paniculata Nees (Kalmegh): A traditional hepatoprotective medicinal herb of India. J. Med. Aromat. Plant Sci. 2001, 23, 637–647. [Google Scholar]
- Maison, T.; Volkaert, H.A.; Boonprakob, U.; Paisooksantivatana, Y. Genetic Diversity of Andrographis paniculata Wall. ex Nees as Revealed by Morphological Characters and Molecular Markers. Kasetsart J. (Nat. Sci.) 2005, 39, 388–399. [Google Scholar]
- Wijarat, P.; Keeratinijakal, V.; Toojinda, T.; Vanavichit, A.; Tragoonrung, S. Genetic evaluation of Andrographis paniculata (Burm. f.) Nees revealed by SSR, AFLP and RAPD markers. J. Med. Plants Res. 2012, 6, 2777–2788. [Google Scholar] [CrossRef]
- Chen, R.; Wang, X.Y.; Song, Y.N.; Zhu, Y.F.; Wang, P.L.; Li, M.; Zhong, G.Y. Genetic diversity analysis of Andrographis paniculata in China based on SRAP and SNP. China J. Chin. Mater. Med. 2014, 39, 4559–4565. [Google Scholar]
- Tiwari, G.; Singh, R.; Singh, N.; Choudhury, D.R.; Paliwal, R.; Kumar, A.; Gupta, V. Study of arbitrarily amplified (RAPD and ISSR) and gene targeted (SCoT and CBDP) markers for genetic diversity and population structure in kalmegh [Andrographis paniculata (Burm. f.) nees]. Ind. Crops Prod. 2016, 86, 1–11. [Google Scholar] [CrossRef]
- Manjesh, G.N.; Kaipa, H.B.; Chinapolaiah, A.; Suryanarayana, M.A.; Halesh, G.K. Genetic diversity analysis using molecular marker in Kalmegh (Andrographis paniculata). Eco. Env. Cons. 2016, 22, S1–S6. [Google Scholar]
- Hiremath, C.; Greeshma, M.; Gupta, N.; Kuppusamy, B.; Shanker, K.; Sundaresan, V. Morphometric, chemotypic, and Molecular Diversity Studies in Andrographis paniculata. J. Herbs Spices Med. Plants 2020, 27, 109–122. [Google Scholar] [CrossRef]
- Arif, M.F.; Subositi, D.; Sari, A.N.; Aristya, G.R.; Lesmana, I.; Kasiamdari, S. Genetic diversity of green chireta (Andrographis paniculata (Burm.f.) Wall. Ex Nees.) from Indonesia based on ISSR and RAPD markers. Malays. Appl. Biol. 2020, 49, 61–68. [Google Scholar] [CrossRef]
- Kumar, R.; Kumar, C.; Paliwal, R.; Roy Choudhury, D.; Singh, I.; Kumar, A.; Kumari, A.; Singh, R. Development of novel genomic simple sequence repeat (g-SSR) markers and their validation for genetic diversity analyses in Kalmegh [Andrographis paniculata (Burm. f.) nees]. Plants 2020, 9, 1734. [Google Scholar] [CrossRef]
- Burke, R.; Schwarze, J.; Sherwood, O.L.; Jnaid, Y.; McCabe, P.F.; Kacprzyk, J. Stressed to Death: The Role of Transcription Factors in Plant Programmed Cell Death Induced by Abiotic and Biotic Stimuli. Front. Plant Sci. 2020, 11, 1235. [Google Scholar] [CrossRef]
- Shahzad, R.; Jamil, S.; Ahmad, S.; Nisar, A.; Amina, Z.; Saleem, S.; Iqbal, M.Z.; Atif, R.M.; Wang, X. Harnessing the potential of plant transcription factors in developing climate resilient crops to improve global food security: Current and future perspectives. Saudi J. Biol. Sci. 2021, 28, 2323–2341. [Google Scholar] [CrossRef] [PubMed]
- Biswal, B.; Jena, B.; Giri, A.K.; Acharya, L. De novo transcriptome and tissue specific expression analysis of genes associated with biosynthesis of secondary metabolites in Operculina turpethum (L.). Sci. Rep. 2021, 11, 22539. [Google Scholar] [CrossRef]
- Kalra, S.; Puniya, B.L.; Kulshreshtha, D.; Kumar, S.; Kaur, J.; Ramachandran, S.; Singh, K. De Novo Transcriptome Sequencing Reveals Important Molecular Networks and Metabolic Pathways of the Plant, Chlorophytum borivilianum. PLoS ONE 2013, 8, e83336. [Google Scholar] [CrossRef] [PubMed]
- Parasar, N.R.; Kumar, R.; Natarajan, P.; Parani, M. De novo assembly, annotation and molecular marker identification from the leaf transcriptome of Ocimum gratissimum L. Plant Genet. Resour. Charact. Util. 2021, 19, 469–476. [Google Scholar]
- Padmanabhan, P.; Srikakulam, N.; Velayudha, V.K.K.; Christdas, J.; Pandi, G. Comprehensive Leaf Transcriptome of a Non-model Plant, Abelmoschus esculentus for the Functional Genomics Studies. J. Genet. Genome Res. 2018, 5, 036. [Google Scholar] [CrossRef]
- Li, F.; Wu, C.; Gao, M.; Jiao, M.; Qu, C.; Gonzalez-Uriarte, A.; Luo, C. Transcriptome sequencing, molecular markers, and transcription factor discovery of Platanus acerifolia in the presence of Corythucha ciliata. Sci. Data 2019, 6, 128. [Google Scholar] [CrossRef] [PubMed]
- Chang, P.; Zhu, L.; Zhao, M.; Li, C.; Zhang, Y.; Li, L. The first transcriptome sequencing and analysis of the endangered plant species Picea neoveitchii Mast. and potential EST-SSR markers development. Biotechnol. Biotechnol. Equip. 2019, 33, 967–973. [Google Scholar] [CrossRef]
- Liu, W.; Jia, X.; Liu, Z.; Zhang, Z.; Wang, Y.; Liu, Z.; Xie, W. Development and Characterization of Transcription Factor Gene-Derived Microsatellite (TFGM) Markers in Medicago truncatula and Their Transferability in Leguminous and Non-Leguminous Species. Molecules 2015, 20, 8759–8771. [Google Scholar] [CrossRef] [PubMed]
- Biswas, M.K.; Nath, U.K.; Howlader, J.; Bagchi, M.; Natarajan, S.; Kayum, M.A.; Kim, H.T.; Park, J.I.; Kang, J.G.; Nou, I.S. Exploration and Exploitation of Novel SSR Markers for Candidate Transcription Factor Genes in Lilium Species. Genes 2018, 9, 97. [Google Scholar] [CrossRef]
- Parmar, R.; Seth, R.; Sharma, R.K. Genome-wide identification and characterization of functionally relevant microsatellite markers from transcription factor genes of Tea (Camellia sinensis (L.) O. Kuntze). Sci. Rep. 2022, 12, 201. [Google Scholar] [CrossRef] [PubMed]
- Backiyarani, S.; Chandrasekar, A.; Uma, S.; Saraswathi, M.S. MusatransSSRDB (a transcriptome derived SSR database)—An advanced tool for banana improvement. J. Biosci. 2019, 44, 4. [Google Scholar] [CrossRef] [PubMed]
- Jasrotia, R.S.; Iquebal, M.A.; Yadav, P.K.; Kumar, N.; Jaiswal, S.; Angadi, U.B.; Rai, A.; Kumar, D. Development of transcriptome based web genomic resources of yellow mosaic disease in Vigna mungo. Physiol. Mol. Biol. Plants 2017, 23, 767–777. [Google Scholar] [CrossRef]
- Gebeyehu, A.; Hammenhag, C.; Tesfaye, K.; Vetukuri, R.R.; Ortiz, R.; Geleta, M. RNA-Seq Provides Novel Genomic Resources for Noug (Guizotia abyssinica) and Reveals Microsatellite Frequency and Distribution in Its Transcriptome. Front. Plant Sci. 2022, 13, 882136. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; Kumar, R.; Mahato, A.K.; Paliwal, R.; Singh, A.K.; Kumar, S.; Marla, S.S.; Kumar, A.; Singh, N.K. De novo transcriptome sequencing facilitates genomic resource generation in Tinospora cordifolia. Funct. Integr. Genom. 2016, 16, 581–591. [Google Scholar] [CrossRef]
- Garg, R.; Patel, R.K.; Jhanwar, S.; Priya, P.; Bhattacharjee, A.; Yadav, G.; Bhatia, S.; Chattopadhyay, D.; Tyagi, A.K.; Jain, M. Gene Discovery and Tissue-Specific Transcriptome Analysis in Chickpea with Massively Parallel Pyrosequencing and Web Resource Development. Plant Physiol. 2011, 156, 1661–1678. [Google Scholar] [CrossRef]
- Venkata Bindu, B.B.; Srinath, M.; Shailaja, A.; Giri, C.C. Comparative protein profile studies and in silico structural/functional analysis of HMGR (ApHMGR) in Andrographis paniculata (Burmf.) Wall. ex Nees. Ann. Phytomed. 2017, 6, 30–44. [Google Scholar] [CrossRef]
- Movahedi, A.; Wei, H.; Pucker, B.; Ghaderi-Zefrehei, M.; Rasouli, F.; Kiani-Pouya, A.; Jiang, T.; Zhuge, Q.; Ynag, L.; Zhou, X. Isoprenoid biosynthesis regulation in poplars by methylerythritol phosphate and mevalonic acid pathways. Front. Plant Sci. 2022, 13, 968780. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Zhang, C.; Jiang, X.; Liu, Q.; Liu, Q.; Wang, K. De Novo Transcriptomic Analysis and Development of EST–SSRs for Styrax japonicus. Forests 2018, 9, 748. [Google Scholar] [CrossRef]
- Feng, S.; Zhao, L.; Liu, Z.; Liu, Y.; Yang, T.; Wei, A. De novo transcriptome assembly of Zanthoxylum bungeanum using Illumina sequencing for evolutionary analysis and simple sequence repeat marker development. Sci. Rep. 2017, 7, 16754. [Google Scholar] [CrossRef]
- Taheri, S.; Abdullah, T.L.; Rafii, M.Y.; Harikrishna, J.A.; Werbrouck, S.P.; Teo, C.H.; Sahebi, M.; Azizi, P. De novo assembly of transcriptomes, mining, and development of novel EST-SSR markers in Curcuma alismatifolia (Zingiberaceae family) through Illumina sequencing. Sci. Rep. 2019, 9, 3047. [Google Scholar] [CrossRef]
- Sangwan, R.S.; Tripathi, S.; Singh, J.; Narnoliya, L.K.; Sangwan, N.S. De novo sequencing and assembly of Centella asiatica leaf transcriptome for mapping of structural, functional and regulatory genes with special reference to secondary metabolism. Gene 2013, 525, 58–76. [Google Scholar] [CrossRef]
- Yue, H.; Yue, H.; Wang, L.; Liu, H.; Yue, W.; Du, X.; Song, W.; Nie, X. De novo assembly and characterization of the transcriptome of broomcorn millet (Panicum miliaceum L.) for gene discovery and marker development. Front. Plant Sci. 2016, 7, 1083. [Google Scholar] [CrossRef]
- Hina, F.; Yisilam, G.; Wang, S.; Li, P.; Fu, C. De novo Transcriptome Assembly, Gene Annotation and SSR Marker Development in the Moon Seed Genus Menispermum (Menispermaceae). Front Genet. 2020, 8, 380. [Google Scholar] [CrossRef]
- Qian, Y.; Zhang, T.; Yu, Y.; Gou, L.; Yang, J.; Xu, J.; Pi, E. Regulatory Mechanisms of bHLH Transcription Factors in Plant Adaptive Responses to Various Abiotic Stresses. Front. Plant Sci. 2021, 12, 677611. [Google Scholar] [CrossRef]
- Wani, S.H.; Anand, S.; Singh, B.; Bohra, A.; Joshi, R. WRKY transcription factors and plant defense responses: Latest discoveries and future prospects. Plant Cell Rep. 2021, 40, 1071–1085. [Google Scholar] [CrossRef]
- Nuruzzaman, M.; Sharoni, A.M.; Kikuchi, S. Roles of NAC transcription factors in the regulation of biotic and abiotic stress responses in plants. Front. Microbiol. 2013, 4, 248. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Kumar, C.; Jain, R.; Maurya, A.; Kumar, A.; Kumari, A.; Singh, R. Molecular cloning and In-silico characterization of NAC86 of Kalmegh (Andrographis paniculata). Indian J. Hortic. 2022, 79, 9–14. [Google Scholar] [CrossRef]
- Singh, R.; Mahato, A.K.; Singh, A.; Kumar, R.; Singh, A.K.; Kumar, S.; Marla, S.S.; Kumar, A.; Singh, N.K. TinoTranscriptDB: A Database of Transcripts and Microsatellite Markers of Tinospora cordifolia, an Important Medicinal Plant. Genes 2022, 13, 1433. [Google Scholar] [CrossRef] [PubMed]
- Yazaki, K. ABC transporters involved in the transport of plant secondary metabolites. FEBS Lett. 2005, 580, 1183–1191. [Google Scholar] [CrossRef]
- Dutta, S.; Kumawat, G.; Singh, B.P.; Gupta, D.K.; Singh, S.; Dogra, V.; Gaikwad, K.; Sharma, T.R.; Raje, R.S.; Bandhopadhya, T.K.; et al. Development of genic-SSR markers by deep transcriptome sequencing in pigeonpea [Cajanus cajan (L.) Millspaugh]. BMC Plant Biol. 2011, 11, 17. [Google Scholar] [CrossRef] [PubMed]
- Metzgar, D.; Bytof, J.; Wills, C. Selection against frameshift mutations limits microsatellite expansion in coding DNA. Genome Res. 2000, 10, 72–80. [Google Scholar]
- Zeng, S.; Xiao, G.; Guo, J.; Fei, Z.; Xu, Y.; Roe, B.A.; Wang, Y. Development of a EST dataset and characterization of EST-SSRs in a traditional Chinese medicinal plant, Epimedium sagittatum (Sieb. Et zucc.) Maxim. BMC Genom. 2010, 11, 94. [Google Scholar] [CrossRef]
- Zhou, Q.; Luo, D.; Ma, L.; Xie, W.; Wang, Y.; Wang, Y.; Liu, Z. Development and cross-species transferability of EST-SSR markers in Siberian wild rye (Elymus sibiricus L.) using Illumina sequencing. Sci. Rep. 2016, 6, 20549. [Google Scholar] [CrossRef]
- Hu, Z.; Zhang, T.; Gao, X.X.; Wang, Y.; Zhang, Q.; Zhou, H.J. De novo assembly and characterization of the leaf, bud, and fruit transcriptome from the vulnerable tree Juglans mandshurica for the development of 20 new microsatellite markers using Illumina sequencing. Mol. Genet. Genom. 2016, 291, 849–862. [Google Scholar] [CrossRef]
- Wu, J.; Cai, C.; Cheng, F.; Cui, H.; Zhou, H. Characterisation and development of EST-SSR markers in tree peony using transcriptome sequences. Mol. Breed. 2014, 34, 1853–1866. [Google Scholar] [CrossRef]
- Varshney, R.K.; Graner, A.; Sorrells, M.E. Genic microsatellite markers in plants: Features and applications. Trends Biotechnol. 2005, 23, 48–55. [Google Scholar] [CrossRef] [PubMed]
- Shen, W.; Le, S.; Li, Y.; Hu, F. SeqKit: A Cross-Platform and Ultrafast Toolkit for FASTA/Q File Manipulation. PLoS ONE 2016, 5, e0163962. [Google Scholar] [CrossRef] [PubMed]
- Conesa, A.; Gotz, S.; Garcia-Gomez, J.M.; Terol, J.; Talon, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef] [PubMed]
- Moriya, Y.; Itoh, M.; Okuda, S.; Yoshizawa, A.C.; Kanehisa, M. KAAS: An automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 2007, 35, W182–W185. [Google Scholar] [CrossRef]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3—New capabilities and interfaces. Nucleic Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef]
- Doyle, J.J.T.; Doyle, J.L. Isolation of plant DNA from fresh tissue. Focus 1990, 12, 13–15. [Google Scholar]
- Yeh, F.C.; Yang, R.; Boyle, T.; Ye, Z.; Mao, J.; Yang, R.C.; Boyle, T.; Ye, Z. PopGene, the User-Friendly Shareware for Population Genetic Analysis; Molecular Biology and Biotechnology Centre, University of Alberta: Edmonton, AB, Canada, 1997. [Google Scholar]
- Liu, K.; Muse, S.V. PowerMarker: An integrated analysis environment for genetic marker analysis. Bioinformatics 2005, 1, 2128–2129. [Google Scholar] [CrossRef]
- Rohlf, F.J. NTSYS-pc: Numerical Taxonomy and Multivariate Analysis System, Version 2.1. Exeter Software; Applied Biostatistics Inc.: New York, NY, USA, 2000. [Google Scholar]
- Du, L.; Zhang, C.; Liu, Q.; Zhang, X.; Yue, B. Krait: An ultrafast tool for genome-wide survey of microsatellites and primer design. Bioinformatics 2018, 34, 681–683. [Google Scholar] [CrossRef]



| S. No. | Raw Data | Statistics |
|---|---|---|
| 1. | No. of raw reads | 748,409 |
| 2. | Length of raw reads (bp) | 305,186,073 |
| 3. | No. of trimmed reads | 730,670 |
| 4. | Length of trimmed reads (bp) | 301,674,417 |
| 5. | The average length of trimmed reads (bp) | 500 |
| Newbler Assembly | Statistics | |
| 6. | No. of transcripts | 16,149 |
| 7. | Size of assembly (bp) | 15,635,774 |
| 8. | N50 (bp) | 1184 |
| 9. | Average transcript length (bp) | 968.2 |
| 10. | Longest transcript length (bp) | 5975 |
| 11. | Smallest transcript length (bp) | 200 |
| 12. | Total no. of singletons (by Newbler) | 65,004 |
| MIRA Assembly | Statistics | |
| 13. | No. of transcripts | 6253 |
| 14. | Size of assembly (bp) | 4,186,517 |
| 15. | N50 (bp) | 650 |
| 16. | Average transcript length (bp) | 669.5 |
| 17. | Longest transcript length (bp) | 2067 |
| 18. | Smallest transcript length (bp) | 297 |
| Final Assembly | Statistics | |
| 19. | No. of transcripts in final assembly | 22,402 |
| 20. | Size of final assembly (bp) | 19,822,291 |
| 21. | N50 (bp) | 1007 |
| 22. | Average transcript length (bp) | 884.8 |
| 23. | Longest transcript length (bp) | 5975 |
| 24. | Smallest transcript length (bp) | 200 |
| 25. | GC% | 45.69 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Singh, R.; Singh, A.; Mahato, A.K.; Paliwal, R.; Tiwari, G.; Kumar, A. De Novo Transcriptome Profiling for the Generation and Validation of Microsatellite Markers, Transcription Factors, and Database Development for Andrographis paniculata. Int. J. Mol. Sci. 2023, 24, 9212. https://doi.org/10.3390/ijms24119212
Singh R, Singh A, Mahato AK, Paliwal R, Tiwari G, Kumar A. De Novo Transcriptome Profiling for the Generation and Validation of Microsatellite Markers, Transcription Factors, and Database Development for Andrographis paniculata. International Journal of Molecular Sciences. 2023; 24(11):9212. https://doi.org/10.3390/ijms24119212
Chicago/Turabian StyleSingh, Rakesh, Akshay Singh, Ajay Kumar Mahato, Ritu Paliwal, Gunjan Tiwari, and Ashok Kumar. 2023. "De Novo Transcriptome Profiling for the Generation and Validation of Microsatellite Markers, Transcription Factors, and Database Development for Andrographis paniculata" International Journal of Molecular Sciences 24, no. 11: 9212. https://doi.org/10.3390/ijms24119212
APA StyleSingh, R., Singh, A., Mahato, A. K., Paliwal, R., Tiwari, G., & Kumar, A. (2023). De Novo Transcriptome Profiling for the Generation and Validation of Microsatellite Markers, Transcription Factors, and Database Development for Andrographis paniculata. International Journal of Molecular Sciences, 24(11), 9212. https://doi.org/10.3390/ijms24119212








