Emerging Role of DREAM in Healthy Brain and Neurological Diseases
Abstract
1. Introduction
1.1. Control of Gene Expression by DREAM in the Nucleus
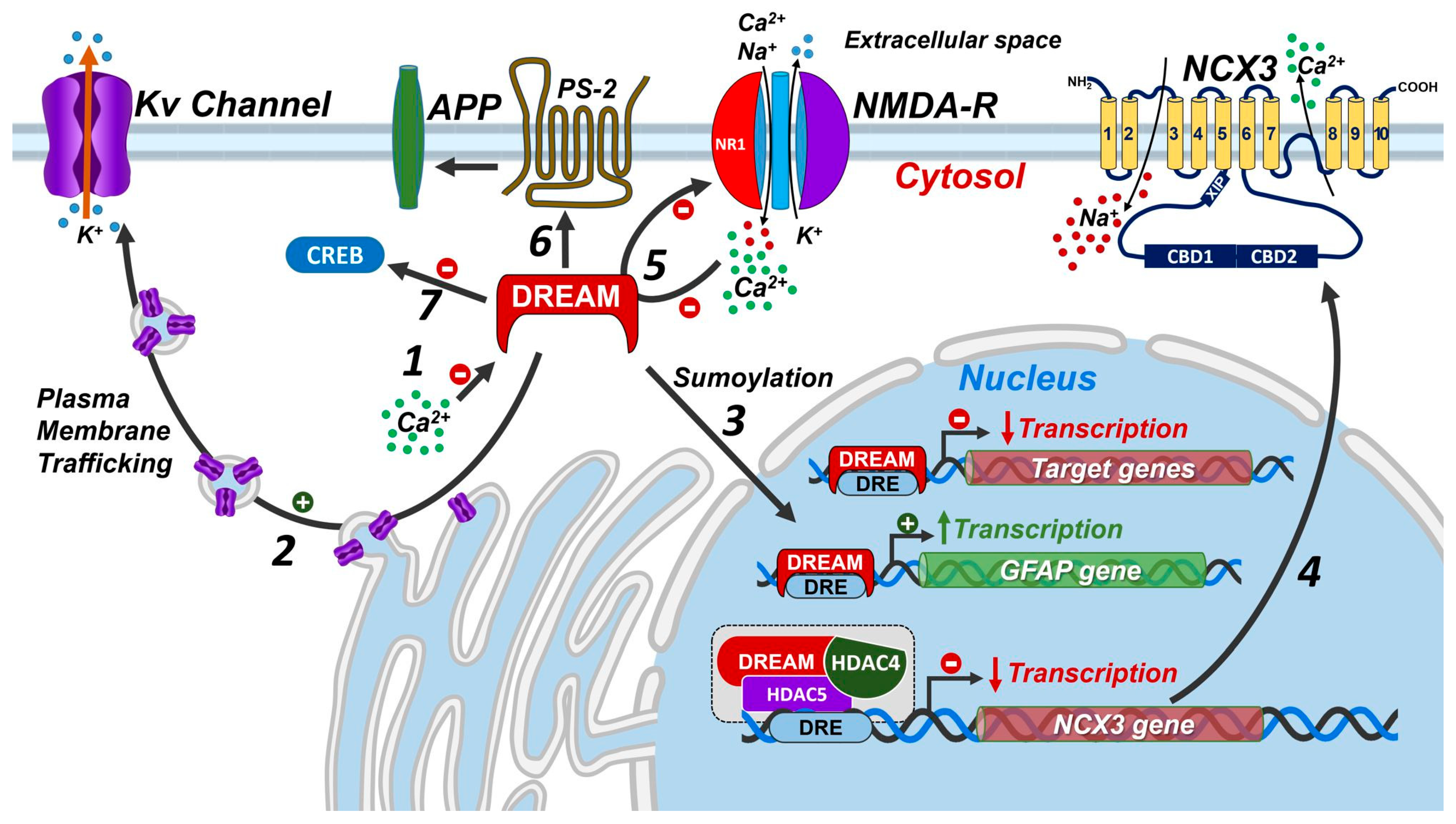
1.2. Control of Protein Activity by DREAM Outside the Nucleus
2. DREAM in CNS
2.1. Anatomical/Cellular Distribution
2.2. Physiological Roles of DREAM in CNS
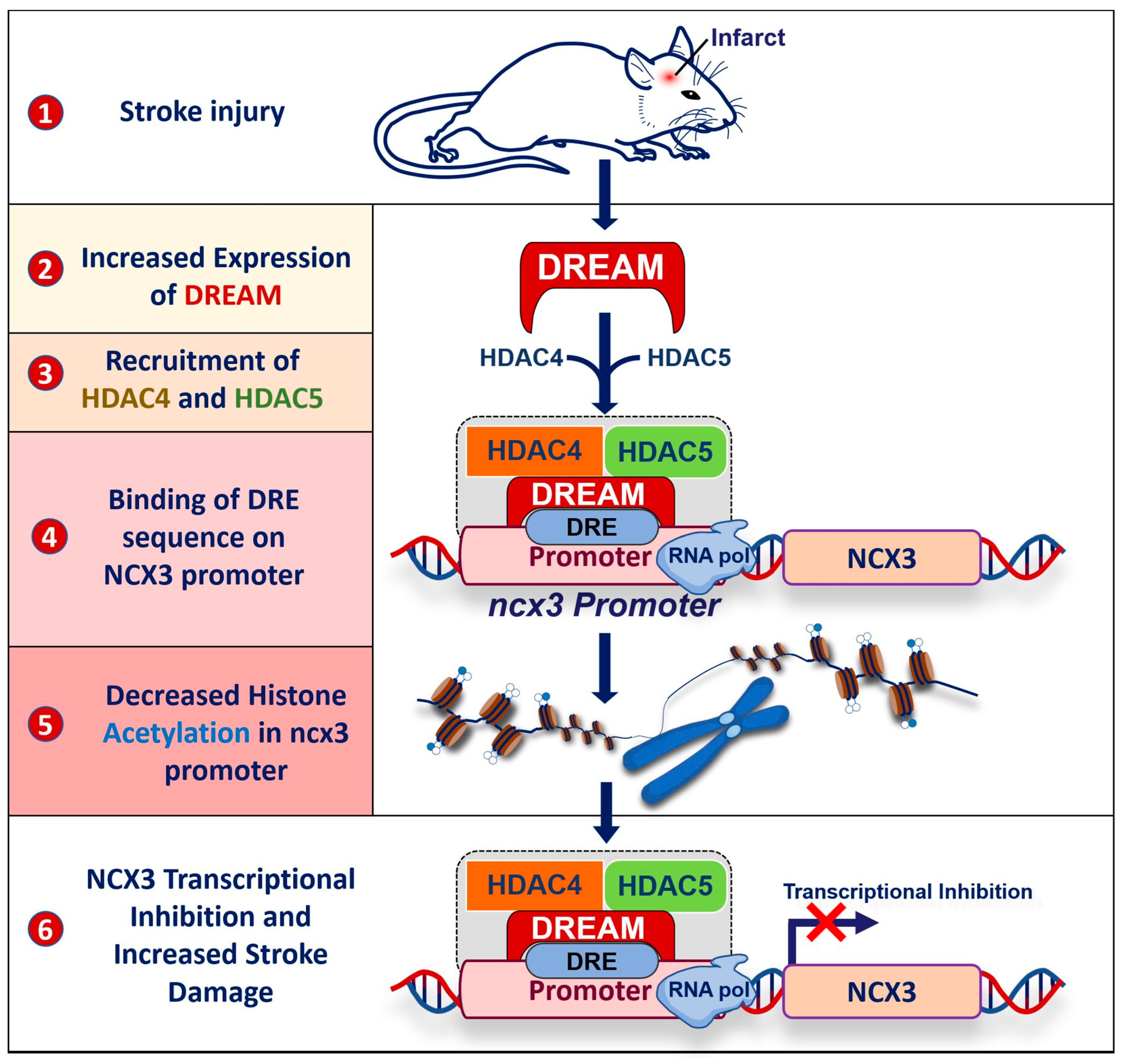
3. DREAM and Stroke
4. DREAM in Neurodegenerative Diseases
4.1. Amyotrophic Lateral Sclerosis (ALS)
4.2. Alzheimer’s Disease
4.3. DREAM and Huntington’s Disease (HD)
4.4. DREAM and Neuropathic Pain
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Carrion, A.M.; Link, W.A.; Ledo, F.; Mellstrom, B.; Naranjo, J.R. DREAM is a Ca2+-regulated transcriptional repressor. Nature 1999, 398, 80–84. [Google Scholar] [CrossRef] [PubMed]
- Ledo, F.; Carrion, A.M.; Link, W.A.; Mellstrom, B.; Naranjo, J.R. DREAM-alphaCREM interaction via leucine-charged domains derepresses downstream regulatory element-dependent transcription. Mol. Cell. Biol. 2000, 20, 9120–9126. [Google Scholar] [CrossRef] [PubMed]
- An, W.F.; Bowlby, M.R.; Betty, M.; Cao, J.; Ling, H.P.; Mendoza, G.; Hinson, J.W.; Mattsson, K.I.; Strassle, B.W.; Trimmer, J.S.; et al. Modulation of A-type potassium channels by a family of calcium sensors. Nature 2000, 403, 553–556. [Google Scholar] [CrossRef] [PubMed]
- Palczewska, M.; Casafont, I.; Ghimire, K.; Rojas, A.M.; Valencia, A.; Lafarga, M.; Mellstrom, B.; Naranjo, J.R. Sumoylation regulates nuclear localization of repressor DREAM. Biochim. Biophys. Acta 2011, 1813, 1050–1058. [Google Scholar] [CrossRef]
- Carrion, A.M.; Mellstrom, B.; Naranjo, J.R. Protein kinase A-dependent derepression of the human prodynorphin gene via differential binding to an intragenic silencer element. Mol. Cell. Biol. 1998, 18, 6921–6929. [Google Scholar] [CrossRef]
- Ledo, F.; Kremer, L.; Mellstrom, B.; Naranjo, J.R. Ca2+-dependent block of CREB-CBP transcription by repressor DREAM. EMBO J. 2002, 21, 4583–4592. [Google Scholar] [CrossRef]
- Rivas, M.; Mellstrom, B.; Naranjo, J.R.; Santisteban, P. Transcriptional repressor DREAM interacts with thyroid transcription factor-1 and regulates thyroglobulin gene expression. J. Biol. Chem. 2004, 279, 33114–33122. [Google Scholar] [CrossRef]
- Scsucova, S.; Palacios, D.; Savignac, M.; Mellstrom, B.; Naranjo, J.R.; Aranda, A. The repressor DREAM acts as a transcriptional activator on Vitamin D and retinoic acid response elements. Nucleic Acids Res. 2005, 33, 2269–2279. [Google Scholar] [CrossRef]
- Zaidi, N.F.; Kuplast, K.G.; Washicosky, K.J.; Kajiwara, Y.; Buxbaum, J.D.; Wasco, W. Calsenilin interacts with transcriptional co-repressor C-terminal binding protein(s). J. Neurochem. 2006, 98, 1290–1301. [Google Scholar] [CrossRef]
- Gomez-Villafuertes, R.; Torres, B.; Barrio, J.; Savignac, M.; Gabellini, N.; Rizzato, F.; Pintado, B.; Gutierrez-Adan, A.; Mellstrom, B.; Carafoli, E.; et al. Downstream regulatory element antagonist modulator regulates Ca2+ homeostasis and viability in cerebellar neurons. J. Neurosci. 2005, 25, 10822–10830. [Google Scholar] [CrossRef]
- Mellstrom, B.; Torres, B.; Link, W.A.; Naranjo, J.R. The BDNF gene: Exemplifying complexity in Ca2+ -dependent gene expression. Crit. Rev. Neurobiol. 2004, 16, 43–49. [Google Scholar] [CrossRef]
- Rivera-Arconada, I.; Benedet, T.; Roza, C.; Torres, B.; Barrio, J.; Krzyzanowska, A.; Avendano, C.; Mellstrom, B.; Lopez-Garcia, J.A.; Naranjo, J.R. DREAM regulates BDNF-dependent spinal sensitization. Mol. Pain 2010, 6, 95. [Google Scholar] [CrossRef]
- Benedet, T.; Gonzalez, P.; Oliveros, J.C.; Dopazo, J.M.; Ghimire, K.; Palczewska, M.; Mellstrom, B.; Naranjo, J.R. Transcriptional repressor DREAM regulates trigeminal noxious perception. J. Neurochem. 2017, 141, 544–552. [Google Scholar] [CrossRef]
- Cheng, H.Y.; Pitcher, G.M.; Laviolette, S.R.; Whishaw, I.Q.; Tong, K.I.; Kockeritz, L.K.; Wada, T.; Joza, N.A.; Crackower, M.; Goncalves, J.; et al. DREAM is a critical transcriptional repressor for pain modulation. Cell 2002, 108, 31–43. [Google Scholar] [CrossRef]
- Abraham, W.C.; Dragunow, M.; Tate, W.P. The role of immediate early genes in the stabilization of long-term potentiation. Mol. Neurobiol. 1991, 5, 297–314. [Google Scholar] [CrossRef]
- Morgan, J.I.; Curran, T. Stimulus-transcription coupling in the nervous system: Involvement of the inducible proto-oncogenes fos and jun. Annu. Rev. Neurosci. 1991, 14, 421–451. [Google Scholar] [CrossRef]
- Dragunow, M.; Beilharz, E.; Sirimanne, E.; Lawlor, P.; Williams, C.; Bravo, R.; Gluckman, P. Immediate-early gene protein expression in neurons undergoing delayed death, but not necrosis, following hypoxic-ischaemic injury to the young rat brain. Brain Res. Mol. Brain Res. 1994, 25, 19–33. [Google Scholar] [CrossRef]
- Baczyk, M.; Alami, N.O.; Delestree, N.; Martinot, C.; Tang, L.; Commisso, B.; Bayer, D.; Doisne, N.; Frankel, W.; Manuel, M.; et al. Synaptic restoration by cAMP/PKA drives activity-dependent neuroprotection to motoneurons in ALS. J. Exp. Med. 2020, 217, e20191734. [Google Scholar] [CrossRef]
- Rawat, V.; Goux, W.; Piechaczyk, M.; SR, D.M. c-Fos Protects Neurons through a Noncanonical Mechanism Involving HDAC3 Interaction: Identification of a 21-Amino Acid Fragment with Neuroprotective Activity. Mol. Neurobiol. 2016, 53, 1165–1180. [Google Scholar] [CrossRef]
- Harris, J.A. Using c-fos as a neural marker of pain. Brain Res. Bull. 1998, 45, 1–8. [Google Scholar] [CrossRef]
- Lee, J.; Kim, I.; Oh, S.R.; Ko, S.J.; Lim, M.K.; Kim, D.G.; Kim, C.H. Regulation of DREAM Expression by Group I mGluR. Korean J. Physiol. Pharmacol. 2011, 15, 95–100. [Google Scholar] [CrossRef] [PubMed]
- Edling, Y.; Ingelman-Sundberg, M.; Simi, A. Glutamate activates c-fos in glial cells via a novel mechanism involving the glutamate receptor subtype mGlu5 and the transcriptional repressor DREAM. Glia 2007, 55, 328–340. [Google Scholar] [CrossRef] [PubMed]
- Formisano, L.; Laudati, G.; Guida, N.; Mascolo, L.; Serani, A.; Cuomo, O.; Cantile, M.; Boscia, F.; Molinaro, P.; Anzilotti, S.; et al. HDAC4 and HDAC5 form a complex with DREAM that epigenetically down-regulates NCX3 gene and its pharmacological inhibition reduces neuronal stroke damage. J. Cereb. Blood Flow Metab. 2020, 40, 2081–2097. [Google Scholar] [CrossRef] [PubMed]
- Molinaro, P.; Cuomo, O.; Pignataro, G.; Boscia, F.; Sirabella, R.; Pannaccione, A.; Secondo, A.; Scorziello, A.; Adornetto, A.; Gala, R.; et al. Targeted disruption of Na+/Ca2+ exchanger 3 (NCX3) gene leads to a worsening of ischemic brain damage. J. Neurosci. 2008, 28, 1179–1184. [Google Scholar] [CrossRef] [PubMed]
- Molinaro, P.; Natale, S.; Serani, A.; Calabrese, L.; Secondo, A.; Tedeschi, V.; Valsecchi, V.; Pannaccione, A.; Scorziello, A.; Annunziato, L. Genetically modified mice to unravel physiological and pathophysiological roles played by NCX isoforms. Cell Calcium 2020, 87, 102189. [Google Scholar] [CrossRef]
- Sisalli, M.J.; Secondo, A.; Esposito, A.; Valsecchi, V.; Savoia, C.; Di Renzo, G.F.; Annunziato, L.; Scorziello, A. Endoplasmic reticulum refilling and mitochondrial calcium extrusion promoted in neurons by NCX1 and NCX3 in ischemic preconditioning are determinant for neuroprotection. Cell Death Differ. 2014, 21, 1142–1149. [Google Scholar] [CrossRef]
- Mellstrom, B.; Sahun, I.; Ruiz-Nuno, A.; Murtra, P.; Gomez-Villafuertes, R.; Savignac, M.; Oliveros, J.C.; Gonzalez, P.; Kastanauskaite, A.; Knafo, S.; et al. DREAM controls the on/off switch of specific activity-dependent transcription pathways. Mol. Cell. Biol. 2014, 34, 877–887. [Google Scholar] [CrossRef]
- Mellstrom, B.; Kastanauskaite, A.; Knafo, S.; Gonzalez, P.; Dopazo, X.M.; Ruiz-Nuno, A.; Jefferys, J.G.; Zhuo, M.; Bliss, T.V.; Naranjo, J.R.; et al. Specific cytoarchitectureal changes in hippocampal subareas in daDREAM mice. Mol. Brain 2016, 9, 22. [Google Scholar] [CrossRef]
- Buxbaum, J.D.; Choi, E.K.; Luo, Y.; Lilliehook, C.; Crowley, A.C.; Merriam, D.E.; Wasco, W. Calsenilin: A calcium-binding protein that interacts with the presenilins and regulates the levels of a presenilin fragment. Nat. Med. 1998, 4, 1177–1181. [Google Scholar] [CrossRef]
- Ruiz-Gomez, A.; Mellstrom, B.; Tornero, D.; Morato, E.; Savignac, M.; Holguin, H.; Aurrekoetxea, K.; Gonzalez, P.; Gonzalez-Garcia, C.; Cena, V.; et al. G protein-coupled receptor kinase 2-mediated phosphorylation of downstream regulatory element antagonist modulator regulates membrane trafficking of Kv4.2 potassium channel. J. Biol. Chem. 2007, 282, 1205–1215. [Google Scholar] [CrossRef]
- Zhang, Y.; Su, P.; Liang, P.; Liu, T.; Liu, X.; Liu, X.Y.; Zhang, B.; Han, T.; Zhu, Y.B.; Yin, D.M.; et al. The DREAM protein negatively regulates the NMDA receptor through interaction with the NR1 subunit. J. Neurosci. 2010, 30, 7575–7586. [Google Scholar] [CrossRef]
- Wu, L.J.; Mellstrom, B.; Wang, H.; Ren, M.; Domingo, S.; Kim, S.S.; Li, X.Y.; Chen, T.; Naranjo, J.R.; Zhuo, M. DREAM (downstream regulatory element antagonist modulator) contributes to synaptic depression and contextual fear memory. Mol. Brain 2010, 3, 3. [Google Scholar] [CrossRef]
- Anderson, D.; Mehaffey, W.H.; Iftinca, M.; Rehak, R.; Engbers, J.D.; Hameed, S.; Zamponi, G.W.; Turner, R.W. Regulation of neuronal activity by Cav3-Kv4 channel signaling complexes. Nat. Neurosci. 2010, 13, 333–337. [Google Scholar] [CrossRef]
- Spreafico, F.; Barski, J.J.; Farina, C.; Meyer, M. Mouse DREAM/calsenilin/KChIP3: Gene structure, coding potential, and expression. Mol. Cell Neurosci. 2001, 17, 1–16. [Google Scholar] [CrossRef]
- Cebolla, B.; Fernandez-Perez, A.; Perea, G.; Araque, A.; Vallejo, M. DREAM mediates cAMP-dependent, Ca2+-induced stimulation of GFAP gene expression and regulates cortical astrogliogenesis. J. Neurosci. 2008, 28, 6703–6713. [Google Scholar] [CrossRef]
- Zaidi, N.F.; Berezovska, O.; Choi, E.K.; Miller, J.S.; Chan, H.; Lilliehook, C.; Hyman, B.T.; Buxbaum, J.D.; Wasco, W. Biochemical and immunocytochemical characterization of calsenilin in mouse brain. Neuroscience 2002, 114, 247–263. [Google Scholar] [CrossRef]
- Tunur, T.; Stelly, C.E.; Schrader, L.A. DREAM/calsenilin/KChIP3 modulates strategy selection and estradiol-dependent learning and memory. Learn. Mem. 2013, 20, 686–694. [Google Scholar] [CrossRef]
- Lilliehook, C.; Bozdagi, O.; Yao, J.; Gomez-Ramirez, M.; Zaidi, N.F.; Wasco, W.; Gandy, S.; Santucci, A.C.; Haroutunian, V.; Huntley, G.W.; et al. Altered Abeta formation and long-term potentiation in a calsenilin knock-out. J. Neurosci. 2003, 23, 9097–9106. [Google Scholar] [CrossRef]
- Alexander, J.C.; McDermott, C.M.; Tunur, T.; Rands, V.; Stelly, C.; Karhson, D.; Bowlby, M.R.; An, W.F.; Sweatt, J.D.; Schrader, L.A. The role of calsenilin/DREAM/KChIP3 in contextual fear conditioning. Learn. Mem. 2009, 16, 167–177. [Google Scholar] [CrossRef]
- Fontan-Lozano, A.; Romero-Granados, R.; del-Pozo-Martin, Y.; Suarez-Pereira, I.; Delgado-Garcia, J.M.; Penninger, J.M.; Carrion, A.M. Lack of DREAM protein enhances learning and memory and slows brain aging. Curr. Biol. 2009, 19, 54–60. [Google Scholar] [CrossRef]
- Sirabella, R.; Secondo, A.; Pannaccione, A.; Molinaro, P.; Formisano, L.; Guida, N.; Di Renzo, G.; Annunziato, L.; Cataldi, M. ERK1/2, p38, and JNK regulate the expression and the activity of the three isoforms of the Na+/Ca2+ exchanger, NCX1, NCX2, and NCX3, in neuronal PC12 cells. J. Neurochem. 2012, 122, 911–922. [Google Scholar] [CrossRef] [PubMed]
- Boscia, F.; D’Avanzo, C.; Pannaccione, A.; Secondo, A.; Casamassa, A.; Formisano, L.; Guida, N.; Scorziello, A.; Di Renzo, G.; Annunziato, L. New roles of NCX in glial cells: Activation of microglia in ischemia and differentiation of oligodendrocytes. Adv. Exp. Med. Biol. 2013, 961, 307–316. [Google Scholar] [CrossRef] [PubMed]
- Molinaro, P.; Viggiano, D.; Nistico, R.; Sirabella, R.; Secondo, A.; Boscia, F.; Pannaccione, A.; Scorziello, A.; Mehdawy, B.; Sokolow, S.; et al. Na+-Ca2+ exchanger (NCX3) knock-out mice display an impairment in hippocampal long-term potentiation and spatial learning and memory. J. Neurosci. 2011, 31, 7312–7321. [Google Scholar] [CrossRef] [PubMed]
- Park, J.S.; Manzanero, S.; Chang, J.W.; Choi, Y.; Baik, S.H.; Cheng, Y.L.; Li, Y.I.; Gwon, A.R.; Woo, H.N.; Jang, J.; et al. Calsenilin contributes to neuronal cell death in ischemic stroke. Brain Pathol. 2013, 23, 402–412. [Google Scholar] [CrossRef] [PubMed]
- He, M.; Zhang, B.; Wei, X.; Wang, Z.; Fan, B.; Du, P.; Zhang, Y.; Jian, W.; Chen, L.; Wang, L.; et al. HDAC4/5-HMGB1 signalling mediated by NADPH oxidase activity contributes to cerebral ischaemia/reperfusion injury. J. Cell Mol. Med. 2013, 17, 531–542. [Google Scholar] [CrossRef]
- Annunziato, L.; Pignataro, G.; Di Renzo, G.F. Pharmacology of brain Na+/Ca2+ exchanger: From molecular biology to therapeutic perspectives. Pharmacol. Rev. 2004, 56, 633–654. [Google Scholar] [CrossRef]
- Formisano, L.; Guida, N.; Mascolo, L.; Serani, A.; Laudati, G.; Pizzorusso, V.; Annunziato, L. Transcriptional and epigenetic regulation of ncx1 and ncx3 in the brain. Cell Calcium 2020, 87, 102194. [Google Scholar] [CrossRef]
- Kiernan, M.C.; Vucic, S.; Cheah, B.C.; Turner, M.R.; Eisen, A.; Hardiman, O.; Burrell, J.R.; Zoing, M.C. Amyotrophic lateral sclerosis. Lancet 2011, 377, 942–955. [Google Scholar] [CrossRef]
- Hardiman, O.; van den Berg, L.H.; Kiernan, M.C. Clinical diagnosis and management of amyotrophic lateral sclerosis. Nat. Rev. Neurol. 2011, 7, 639–649. [Google Scholar] [CrossRef]
- Westeneng, H.J.; Debray, T.P.A.; Visser, A.E.; van Eijk, R.P.A.; Rooney, J.P.K.; Calvo, A.; Martin, S.; McDermott, C.J.; Thompson, A.G.; Pinto, S.; et al. Prognosis for patients with amyotrophic lateral sclerosis: Development and validation of a personalised prediction model. Lancet Neurol. 2018, 17, 423–433. [Google Scholar] [CrossRef]
- Guida, N.; Laudati, G.; Serani, A.; Mascolo, L.; Molinaro, P.; Montuori, P.; Di Renzo, G.; Canzoniero, L.M.T.; Formisano, L. The neurotoxicant PCB-95 by increasing the neuronal transcriptional repressor REST down-regulates caspase-8 and increases Ripk1, Ripk3 and MLKL expression determining necroptotic neuronal death. Biochem. Pharmacol. 2017, 142, 229–241. [Google Scholar] [CrossRef]
- Guida, N.; Laudati, G.; Anzilotti, S.; Sirabella, R.; Cuomo, O.; Brancaccio, P.; Santopaolo, M.; Galgani, M.; Montuori, P.; Di Renzo, G.; et al. Methylmercury upregulates RE-1 silencing transcription factor (REST) in SH-SY5Y cells and mouse cerebellum. Neurotoxicology 2016, 52, 89–97. [Google Scholar] [CrossRef]
- Formisano, L.; Guida, N.; Laudati, G.; Mascolo, L.; Di Renzo, G.; Canzoniero, L.M. MS-275 Inhibits Aroclor 1254-Induced SH-SY5Y Neuronal Cell Toxicity by Preventing the Formation of the HDAC3/REST Complex on the Synapsin-1 Promoter. J. Pharmacol. Exp. Ther. 2015, 352, 236–243. [Google Scholar] [CrossRef]
- Guida, N.; Laudati, G.; Mascolo, L.; Valsecchi, V.; Sirabella, R.; Selleri, C.; Di Renzo, G.; Canzoniero, L.M.; Formisano, L. p38/Sp1/Sp4/HDAC4/BDNF Axis Is a Novel Molecular Pathway of the Neurotoxic Effect of the Methylmercury. Front. Neurosci. 2017, 11, 8. [Google Scholar] [CrossRef]
- Larrode, P.; Calvo, A.C.; Moreno-Martinez, L.; de la Torre, M.; Moreno-Garcia, L.; Molina, N.; Castiella, T.; Iniguez, C.; Pascual, L.F.; Mena, F.J.M.; et al. DREAM-Dependent Activation of Astrocytes in Amyotrophic Lateral Sclerosis. Mol. Neurobiol. 2018, 55, 1–12. [Google Scholar] [CrossRef]
- Anzilotti, S.; Brancaccio, P.; Simeone, G.; Valsecchi, V.; Vinciguerra, A.; Secondo, A.; Petrozziello, T.; Guida, N.; Sirabella, R.; Cuomo, O.; et al. Preconditioning, induced by sub-toxic dose of the neurotoxin L-BMAA, delays ALS progression in mice and prevents Na+/Ca2+ exchanger 3 downregulation. Cell Death Dis. 2018, 9, 206. [Google Scholar] [CrossRef]
- Laudati, G.; Mascolo, L.; Guida, N.; Sirabella, R.; Pizzorusso, V.; Bruzzaniti, S.; Serani, A.; Di Renzo, G.; Canzoniero, L.M.T.; Formisano, L. Resveratrol treatment reduces the vulnerability of SH-SY5Y cells and cortical neurons overexpressing SOD1-G93A to Thimerosal toxicity through SIRT1/DREAM/PDYN pathway. Neurotoxicology 2019, 71, 6–15. [Google Scholar] [CrossRef]
- Vasilopoulou, C.; McDaid-McCloskey, S.L.; McCluskey, G.; Duguez, S.; Morris, A.P.; Duddy, W. Genome-Wide Gene-Set Analysis Identifies Molecular Mechanisms Associated with ALS. Int. J. Mol. Sci. 2023, 24, 4021. [Google Scholar] [CrossRef]
- Hardy, J.A.; Higgins, G.A. Alzheimer’s disease: The amyloid cascade hypothesis. Science 1992, 256, 184–185. [Google Scholar] [CrossRef]
- Ferretti, G.; Romano, A.; Sirabella, R.; Serafini, S.; Maier, T.J.; Matrone, C. An increase in Semaphorin 3A biases the axonal direction and induces an aberrant dendritic arborization in an in vitro model of human neural progenitor differentiation. Cell Biosci. 2022, 12, 182. [Google Scholar] [CrossRef]
- Pannaccione, A.; Secondo, A.; Molinaro, P.; D’Avanzo, C.; Cantile, M.; Esposito, A.; Boscia, F.; Scorziello, A.; Sirabella, R.; Sokolow, S.; et al. A new concept: Aβ1-42 generates a hyperfunctional proteolytic NCX3 fragment that delays caspase-12 activation and neuronal death. J. Neurosci. 2012, 32, 10609–10617. [Google Scholar] [CrossRef]
- Pannaccione, A.; Piccialli, I.; Secondo, A.; Ciccone, R.; Molinaro, P.; Boscia, F.; Annunziato, L. The Na(+)/Ca(2+) exchanger in Alzheimer’s disease. Cell calcium 2020, 87, 102190. [Google Scholar] [CrossRef] [PubMed]
- Jo, D.G.; Lee, J.Y.; Hong, Y.M.; Song, S.; Mook-Jung, I.; Koh, J.Y.; Jung, Y.K. Induction of pro-apoptotic calsenilin/DREAM/KChIP3 in Alzheimer’s disease and cultured neurons after amyloid-beta exposure. J. Neurochem. 2004, 88, 604–611. [Google Scholar] [CrossRef] [PubMed]
- Jo, D.G.; Kim, M.J.; Choi, Y.H.; Kim, I.K.; Song, Y.H.; Woo, H.N.; Chung, C.W.; Jung, Y.K. Pro-apoptotic function of calsenilin/DREAM/KChIP3. FASEB J. 2001, 15, 589–591. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Hurtado, A.; Burgos, D.F.; Gonzalez, P.; Dopazo, X.M.; Gonzalez, V.; Rabano, A.; Mellstrom, B.; Naranjo, J.R. Inhibition of DREAM-ATF6 interaction delays onset of cognition deficit in a mouse model of Huntington’s disease. Mol. Brain 2018, 11, 13. [Google Scholar] [CrossRef]
- Lopez-Hurtado, A.; Peraza, D.A.; Cercos, P.; Lagartera, L.; Gonzalez, P.; Dopazo, X.M.; Herranz, R.; Gonzalez, T.; Martin-Martinez, M.; Mellstrom, B.; et al. Targeting the neuronal calcium sensor DREAM with small-molecules for Huntington’s disease treatment. Sci. Rep. 2019, 9, 7260. [Google Scholar] [CrossRef]
- Naranjo, J.R.; Zhang, H.; Villar, D.; Gonzalez, P.; Dopazo, X.M.; Moron-Oset, J.; Higueras, E.; Oliveros, J.C.; Arrabal, M.D.; Prieto, A.; et al. Activating transcription factor 6 derepression mediates neuroprotection in Huntington disease. J. Clin. Investig. 2016, 126, 627–638. [Google Scholar] [CrossRef]
- Randic, M.; Cheng, G.; Kojic, L. Kappa-opioid receptor agonists modulate excitatory transmission in substantia gelatinosa neurons of the rat spinal cord. J. Neurosci. 1995, 15, 6809–6826. [Google Scholar] [CrossRef]
- Long, I.; Suppian, R.; Ismail, Z. The Effects of Pre-emptive Administration of Ketamine and norBNI on Pain Behavior, c-Fos, and Prodynorphin Protein Expression in the Rat Spinal Cord after Formalin-induced Pain Is Modulated by the DREAM Protein. Korean J. Pain 2013, 26, 255–264. [Google Scholar] [CrossRef]
- Tian, N.X.; Xu, Y.; Yang, J.Y.; Li, L.; Sun, X.H.; Wang, Y.; Zhang, Y. KChIP3 N-Terminal 31-50 Fragment Mediates Its Association with TRPV1 and Alleviates Inflammatory Hyperalgesia in Rats. J. Neurosci. 2018, 38, 1756–1773. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Molinaro, P.; Sanguigno, L.; Casamassa, A.; Valsecchi, V.; Sirabella, R.; Pignataro, G.; Annunziato, L.; Formisano, L. Emerging Role of DREAM in Healthy Brain and Neurological Diseases. Int. J. Mol. Sci. 2023, 24, 9177. https://doi.org/10.3390/ijms24119177
Molinaro P, Sanguigno L, Casamassa A, Valsecchi V, Sirabella R, Pignataro G, Annunziato L, Formisano L. Emerging Role of DREAM in Healthy Brain and Neurological Diseases. International Journal of Molecular Sciences. 2023; 24(11):9177. https://doi.org/10.3390/ijms24119177
Chicago/Turabian StyleMolinaro, Pasquale, Luca Sanguigno, Antonella Casamassa, Valeria Valsecchi, Rossana Sirabella, Giuseppe Pignataro, Lucio Annunziato, and Luigi Formisano. 2023. "Emerging Role of DREAM in Healthy Brain and Neurological Diseases" International Journal of Molecular Sciences 24, no. 11: 9177. https://doi.org/10.3390/ijms24119177
APA StyleMolinaro, P., Sanguigno, L., Casamassa, A., Valsecchi, V., Sirabella, R., Pignataro, G., Annunziato, L., & Formisano, L. (2023). Emerging Role of DREAM in Healthy Brain and Neurological Diseases. International Journal of Molecular Sciences, 24(11), 9177. https://doi.org/10.3390/ijms24119177






