Characterization of Arbuscular Mycorrhizal Effector Proteins
Abstract
1. Introduction
2. Fungal Effector Protein Functions: Suppression of Defense and Niche Occupation
3. Toward the Establishment of a Functional AM Symbiosis by Suppression of the Host Immunity
4. AMF Genomes Encode a Wide Effector Repertoire
5. Protein Domains Identified in the R. irregularis Effector Repertoire
6. Current Insights into AMF Effectors
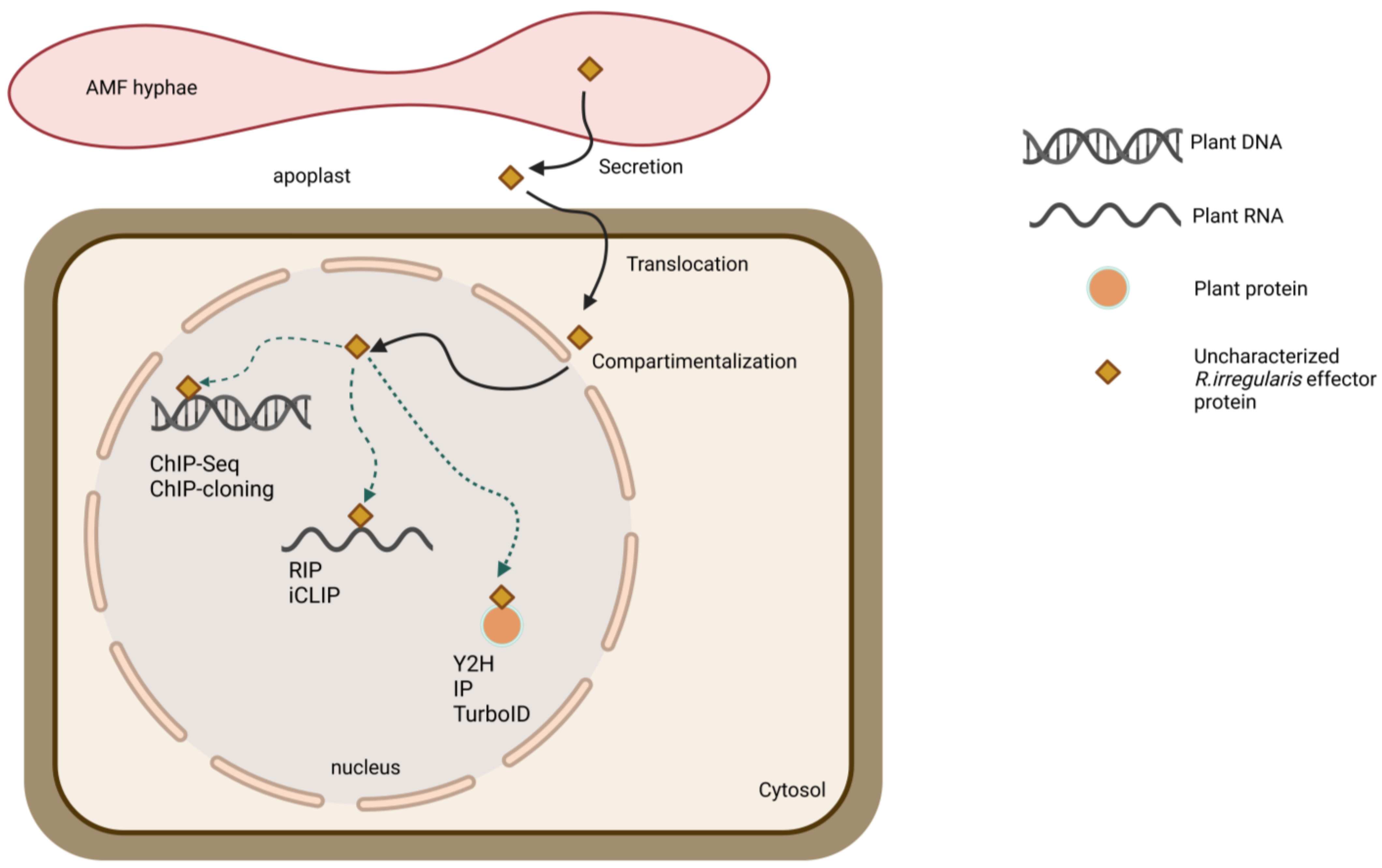
7. Approach to Tackle the Unknown
7.1. FIRST STEP: Bioinformatic Assessment of Effector Features
7.2. SECOND STEP: Functional Validation of Effector Proteins
7.3. THIRD STEP: Seeking the Hidden Interacting Plant Partners
8. Overview and Future Prospectives
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AMF | Arbuscular mycorrhizal fungi |
| AM | Arbuscular mycorrhiza |
| BiFC | Bimolecular fluorescence complementation |
| Ca2+ | Calcium |
| ChIP | Chromatin IP |
| ChIP-PCR | Chromatin IP coupled to PCR amplification |
| ChIP-seq | Chromatin IP coupled to NGS |
| Co-IP | Co-immunoprecipitation |
| CRN | Crinkler |
| CRN | Crinkler |
| CrY2H-seq | Cre reporter-mediated Y2H NGS |
| DAMPs | Damage-associated molecular patterns |
| DMI3 | Does not make the infection |
| EMSA | Electrophoresis mobility shift assay |
| ENOD | Early nodulin |
| ER | endoplasmic reticulum |
| ERF19 | Ethylene response transcription factor 19 |
| ERM | Extraradical mycelium |
| FAST | Fluorescence-Activating and absorption-Shifting Tag |
| FSD | Fungal Secretome Database |
| H2B | Histone 2b |
| H2O2 | Hydrogen peroxide |
| HIGS | Host-induced gene silencing |
| HMM | Hidden Markov Model |
| iCLIP | Cross-Linking RNA IP |
| iCLIP-NGS | Cross-Linking RNA IP coupled to NGS |
| IDRs | Intrinsic disorder regions |
| IP | Immunoprecipitation |
| IRM | Intraradical mycelium |
| LCM | Laser capture microdissection |
| LC-MS/MS | Liquid chromatography–tandem mass spectrometry |
| LRR | Leucine-rich repeat |
| MAMPs | Microbe-associated molecular patterns |
| ML | MD-2-related lipid |
| MtBCP1 | M. truncatula blue copper protein 1 |
| NGS | Next generation sequencing |
| NIS1 | Necrosis-inducing secreted protein 1 |
| NLSs | Nuclear localization signals |
| NO | Nitric oxide |
| PHYTOGB1 | Phytoglobin 1 |
| PPI | Protein-protein interaction |
| PRRs | Pattern recognition receptors |
| PT4 | Phosphate transporter 4 |
| PTI | Pattern-triggered immunity |
| rBiFC | Ratiometric BiFC |
| RiEF1α | R. irregularis elongation factor 1α |
| RiNLE1 | Nuclear-localized effector 1 |
| RIP | RNA-IP |
| RISCs | RNA-induced silencing complexes |
| RiSLM | Secreted LysM-containing effector |
| RNAi | Interference RNA |
| ROS | Reactive oxygen species |
| SIS1 | Strigolactone-induced secreted protein 1 |
| SP7 | Secreted protein 7 |
| SSPs | Small secreted proteins |
| STR | Stunted arbuscule |
| SUC2 | SUCROSE INVERTASE 2 |
| SWEET | SUGARS WILL EVENTUALLY BE EXPORTED TRANSPORTER |
| TF | Transcription factor |
| TOR | Target Of Rapamycin |
| WGA | Wheat Germ Agglutinin |
| Y1H | Yeast one-hybrid |
| Y2H | Yeast two-hybrid |
| Y2H-seq | Y2H sequencing |
References
- Parniske, M. Arbuscular mycorrhiza: The mother of plant root endosymbioses. Nat. Rev. Microbiol. 2008, 6, 763–775. [Google Scholar] [CrossRef] [PubMed]
- Begum, N.; Qin, C.; Ahanger, M.A.; Raza, S.; Khan, M.I.; Ashraf, M.; Ahmed, N.; Zhang, L. Role of arbuscular mycorrhizal fungi in plant growth regulation: Implications in abiotic stress tolerance. Front. Plant Sci. 2019, 10, 1068. [Google Scholar] [CrossRef] [PubMed]
- Pimprikar, P.; Gutjahr, C. Transcriptional regulation of arbuscular mycorrhiza development. Plant Cell Physiol. 2018, 59, 678–695. [Google Scholar] [CrossRef] [PubMed]
- Pfeffer, P.E.; Douds, D.D.; Bécard, G.; Shachar-Hill, Y. Carbon uptake and the metabolism and transport of lipids in an arbuscular mycorrhiza. Plant Physiol. 1999, 120, 587–598. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.E.; Smith, F.A. Roles of arbuscular mycorrhizas in plant nutrition and growth: New paradigms from cellular to ecosystem scales. Annu. Rev. Plant Biol. 2011, 62, 227–250. [Google Scholar] [CrossRef]
- Manck-Götzenberger, J.; Requena, N. Arbuscular mycorrhiza symbiosis induces a major transcriptional reprogramming of the potato SWEET sugar transporter family. Front. Plant Sci. 2016, 7, 487. [Google Scholar] [CrossRef]
- Bravo, A.; Brands, M.; Wewer, V.; Dörmann, P.; Harrison, M.J. Arbuscular mycorrhiza-specific enzymes FatM and RAM2 fine-tune lipid biosynthesis to promote development of arbuscular mycorrhiza. New Phytol. 2017, 214, 1631–1645. [Google Scholar] [CrossRef]
- Taylor, T.N.; Remy, W.; Hass, H.; Kerp, H. Fossil arbuscular mycorrhizae from the Early Devonian. Mycologia 1995, 87, 560–573. [Google Scholar] [CrossRef]
- Maillet, F.; Poinsot, V.; André, O.; Puech-Pagès, V.; Haouy, A.; Gueunier, M.; Cromer, L.; Giraudet, D.; Formey, D.; Niebel, A.; et al. Fungal lipochitooligosaccharide symbiotic signals in arbuscular mycorrhiza. Nature 2011, 469, 58–63. [Google Scholar] [CrossRef]
- Pradhan, A.; Ghosh, S.; Sahoo, D.; Jha, G. Fungal effectors, the double edge sword of phytopathogens. Curr. Genet. 2021, 67, 27–40. [Google Scholar] [CrossRef]
- Bent, A.F.; Mackey, D. Elicitors, effectors, and R genes: The new paradigm and a lifetime supply of questions. Annu. Rev. Phytopathol. 2007, 45, 399–436. [Google Scholar] [CrossRef] [PubMed]
- Jwa, N.-S.; Hwang, B.K. Convergent evolution of pathogen effectors toward reactive oxygen species signaling networks in plants. Front. Plant Sci. 2017, 8, 1687. [Google Scholar] [CrossRef] [PubMed]
- Plett, J.M.; Martin, F. Reconsidering mutualistic plant-fungal interactions through the lens of effector biology. Curr. Opin. Plant Biol. 2015, 26, 45–50. [Google Scholar] [CrossRef] [PubMed]
- Plett, J.M.; Martin, F.M. Know your enemy, embrace your friend: Using omics to understand how plants respond differently to pathogenic and mutualistic microorganisms. Plant J. 2018, 93, 729–746. [Google Scholar] [CrossRef]
- Qiao, Y.; Shi, J.; Zhai, Y.; Hou, Y.; Ma, W. Phytophthora effector targets a novel component of small RNA pathway in plants to promote infection. Proc. Natl. Acad. Sci. USA 2015, 112, 5850–5855. [Google Scholar] [CrossRef]
- Gui, X.; Zhang, P.; Wang, D.; Ding, Z.; Wu, X.; Shi, J.; Shen, Q.-H.; Xu, Y.-Z.; Ma, W.; Qiao, Y. Phytophthora effector PSR1 hijacks the host pre-mRNA splicing machinery to modulate small RNA biogenesis and plant immunity. Plant Cell 2022, 34, 3443–3459. [Google Scholar] [CrossRef]
- Lo Presti, L.; Lanver, D.; Schweizer, G.; Tanaka, S.; Liang, L.; Tollot, M.; Zuccaro, A.; Reissmann, S.; Kahmann, R. Fungal effectors and plant susceptibility. Annu. Rev. Plant Biol. 2015, 66, 513–545. [Google Scholar] [CrossRef]
- Tisserant, E.; Kohler, A.; Dozolme-Seddas, P.; Balestrini, R.; Benabdellah, K.; Colard, A.; Croll, D.; da Silva, C.; Gomez, S.K.; Koul, R.; et al. The transcriptome of the arbuscular mycorrhizal fungus Glomus intraradices (DAOM 197198) reveals functional tradeoffs in an obligate symbiont. New Phytol. 2012, 193, 755–769. [Google Scholar] [CrossRef]
- Tisserant, E.; Malbreil, M.; Kuo, A.; Kohler, A.; Symeonidi, A.; Balestrini, R.; Charron, P.; Duensing, N.; Frei Dit Frey, N.; Gianinazzi-Pearson, V.; et al. Genome of an arbuscular mycorrhizal fungus provides insight into the oldest plant symbiosis. Proc. Natl. Acad. Sci. USA 2013, 110, 20117–20122. [Google Scholar] [CrossRef]
- Lin, K.; Limpens, E.; Zhang, Z.; Ivanov, S.; Saunders, D.G.O.; Mu, D.; Pang, E.; Cao, H.; Cha, H.; Lin, T.; et al. Single nucleus genome sequencing reveals high similarity among nuclei of an endomycorrhizal fungus. PLoS Genet. 2014, 10, e1004078. [Google Scholar] [CrossRef]
- Sȩdzielewska Toro, K.; Brachmann, A. The effector candidate repertoire of the arbuscular mycorrhizal fungus Rhizophagus clarus. BMC Genom. 2016, 17, 101. [Google Scholar] [CrossRef] [PubMed]
- Maeda, T.; Kobayashi, Y.; Kameoka, H.; Okuma, N.; Takeda, N.; Yamaguchi, K.; Bino, T.; Shigenobu, S.; Kawaguchi, M. Evidence of non-tandemly repeated rDNAs and their intragenomic heterogeneity in Rhizophagus irregularis. Nat. Commun. 2018, 1, 87. [Google Scholar] [CrossRef]
- Kamel, L.; Tang, N.; Malbreil, M.; San Clemente, H.; Le Marquer, M.; Roux, C.; Frei dit Frey, N. The comparison of expressed candidate secreted proteins from two arbuscular mycorrhizal fungi unravels common and specific molecular tools to invade different host plants. Front. Plant Sci. 2017, 8, 124. [Google Scholar] [CrossRef]
- Morin, E.; Miyauchi, S.; San Clemente, H.; Chen, E.C.H.; Pelin, A.; de la Providencia, I.; Ndikumana, S.; Beaudet, D.; Hainaut, M.; Drula, E.; et al. Comparative genomics of Rhizophagus irregularis, R. cerebriforme, R. diaphanus and Gigaspora rosea highlights specific genetic features in Glomeromycotina. New Phytol. 2019, 222, 1584–1598. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, Y.; Maeda, T.; Yamaguchi, K.; Kameoka, H.; Tanaka, S.; Ezawa, T.; Shigenobu, S.; Kawaguchi, M. The genome of Rhizophagus clarus HR1 reveals a common genetic basis for auxotrophy among arbuscular mycorrhizal fungi. BMC Genom. 2018, 19, 465. [Google Scholar] [CrossRef]
- Zeng, T.; Holmer, R.; Hontelez, J.; te Lintel-Hekkert, B.; Marufu, L.; de Zeeuw, T.; Wu, F.; Schijlen, E.; Bisseling, T.; Limpens, E. Host- and stage-dependent secretome of the arbuscular mycorrhizal fungus Rhizophagus irregularis. Plant J. 2018, 94, 411–425. [Google Scholar] [CrossRef]
- Kloppholz, S.; Kuhn, H.; Requena, N. A secreted fungal effector of Glomus intraradices promotes symbiotic biotrophy. Curr. Biol. 2011, 21, 1204–1209. [Google Scholar] [CrossRef] [PubMed]
- Tsuzuki, S.; Handa, Y.; Takeda, N.; Kawaguchi, M. Strigolactone-induced putative secreted protein 1 is required for the establishment of symbiosis by the arbuscular mycorrhizal fungus Rhizophagus irregularis. Mol. Plant-Microbe Interact. 2016, 29, 277–286. [Google Scholar] [CrossRef]
- Voß, S.; Betz, R.; Heidt, S.; Corradi, N.; Requena, N. RiCRN1, a crinkler effector from the arbuscular mycorrhizal fungus Rhizophagus irregularis, functions in arbuscule development. Front. Microbiol. 2018, 9, 2068. [Google Scholar] [CrossRef]
- Zeng, T.; Rodriguez-Moreno, L.; Mansurkhodzaev, A.; Wang, P.; van den Berg, W.; Gasciolli, V.; Cottaz, S.; Fort, S.; Thomma, B.P.H.J.; Bono, J.-J.; et al. A lysin motif effector subverts chitin-triggered immunity to facilitate arbuscular mycorrhizal symbiosis. New Phytol. 2020, 225, 448–460. [Google Scholar] [CrossRef]
- Wang, P.; Jiang, H.; Boeren, S.; Dings, H.; Kulikova, O.; Bisseling, T.; Limpens, E. A nuclear-targeted effector of Rhizophagus irregularis interferes with histone 2B mono-ubiquitination to promote arbuscular mycorrhization. New Phytol. 2021, 230, 1142–1155. [Google Scholar] [CrossRef] [PubMed]
- Irieda, H.; Inoue, Y.; Mori, M.; Yamada, K.; Oshikawa, Y.; Saitoh, H.; Uemura, A.; Terauchi, R.; Kitakura, S.; Kosaka, A.; et al. Conserved fungal effector suppresses PAMP-triggered immunity by targeting plant immune kinases. Proc. Natl. Acad. Sci. USA 2019, 116, 496–505. [Google Scholar] [CrossRef]
- Zhu, C.; Liu, J.H.; Zhao, J.H.; Liu, T.; Chen, Y.Y.; Wang, C.H.; Zhang, Z.H.; Guo, H.S.; Duan, C.G. A fungal effector suppresses the nuclear export of AGO1–miRNA complex to promote infection in plants. Proc. Natl. Acad. Sci. USA 2022, 119, e2114583119. [Google Scholar] [CrossRef] [PubMed]
- Tsuda, K.; Katagiri, F. Comparing signaling mechanisms engaged in pattern-triggered and effector-triggered immunity. Curr. Opin. Plant Biol. 2010, 13, 459–465. [Google Scholar] [CrossRef] [PubMed]
- Vidhyasekaran, P. PAMP signaling in plant innate immunity. In PAMP Signals in Plant Innate Immunity: Signal Perception and Transduction; Vidhyasekaran, P., Ed.; Signaling and Communication in Plants; Springer: Dordrecht, The Netherlands, 2014; Volume 21, pp. 17–161. [Google Scholar]
- Jones, J.D.G.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef]
- Dodds, P.N.; Rathjen, J.P. Plant immunity: Towards an integrated view of plant-pathogen interactions. Nat. Rev. Genet. 2010, 11, 539–548. [Google Scholar] [CrossRef]
- Naveed, Z.A.; Wei, X.; Chen, J.; Mubeen, H.; Ali, G.S. The PTI to ETI continuum in Phytophthora-plant interactions. Front. Plant Sci. 2020, 11, 593905. [Google Scholar] [CrossRef]
- Cui, H.; Tsuda, K.; Parker, J.E. Effector-triggered immunity: From pathogen perception to robust defense. Annu. Rev. Plant Biol. 2015, 66, 487–511. [Google Scholar] [CrossRef]
- Selin, C.; de Kievit, T.R.; Belmonte, M.F.; Fernando, W.G.D. Elucidating the role of effectors in plant-fungal interactions: Progress and challenges. Front. Microbiol. 2016, 7, 600. [Google Scholar] [CrossRef]
- Liu, T.; Song, T.; Zhang, X.; Yuan, H.; Su, L.; Li, W.; Xu, J.; Liu, S.; Chen, L.; Chen, T.; et al. Unconventionally secreted effectors of two filamentous pathogens target plant salicylate biosynthesis. Nat. Commun. 2014, 5, 4686. [Google Scholar] [CrossRef]
- Kim, S.; Kim, C.-Y.; Park, S.-Y.; Kim, K.-T.; Jeon, J.; Chung, H.; Choi, G.; Kwon, S.; Choi, J.; Jeon, J.; et al. Two nuclear effectors of the rice blast fungus modulate host immunity via transcriptional reprogramming. Nat. Commun. 2020, 11, 5845. [Google Scholar] [CrossRef] [PubMed]
- Stam, R.; Jupe, J.; Howden, A.J.M.; Morris, J.A.; Boevink, P.C.; Hedley, P.E.; Huitema, E. Identification and characterisation CRN effectors in Phytophthora capsici shows modularity and functional diversity. PLoS ONE 2013, 8, e59517. [Google Scholar] [CrossRef]
- Mak, A.N.-S.; Bradley, P.; Bogdanove, A.J.; Stoddard, B.L. TAL effectors: Function, structure, engineering and applications. Curr. Opin. Struct. Biol. 2013, 23, 93–99. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.Q.; Hou, B.H.; Lalonde, S.; Takanaga, H.; Hartung, M.L.; Qu, X.Q.; Guo, W.J.; Kim, J.G.; Underwood, W.; Chaudhuri, B.; et al. Sugar Transporters for Intercellular Exchange and Nutrition of Pathogens. Nature 2010, 468, 527–532. [Google Scholar] [CrossRef]
- Streubel, J.; Pesce, C.; Hutin, M.; Koebnik, R.; Boch, J.; Szurek, B. Five phylogenetically close rice SWEET genes confer TAL effector-mediated susceptibility to Xanthomonas oryzae. New Phytol. 2013, 200, 808–819. [Google Scholar] [CrossRef]
- Chong, J.; Piron, M.-C.; Meyer, S.; Merdinoglu, D.; Bertsch, C.; Mestre, P. The SWEET family of sugar transporters in grapevine: VvSWEET4 is involved in the interaction with Botrytis cinerea. J. Exp. Bot. 2014, 65, 6589–6601. [Google Scholar] [CrossRef] [PubMed]
- Gebauer, P.; Korn, M.; Engelsdorf, T.; Sonnewald, U.; Koch, C.; Voll, L.M. Sugar accumulation in leaves of Arabidopsis sweet11/sweet12 double mutants enhances priming of the salicylic acid-mediated defense response. Front. Plant Sci. 2017, 8, 1378. [Google Scholar] [CrossRef]
- Gao, Y.; Zhang, C.; Han, X.; Wang, Z.Y.; Ma, L.; Yuan, D.P.; Wu, J.N.; Zhu, X.F.; Liu, J.M.; Li, D.P.; et al. Inhibition of OsSWEET11 function in mesophyll cells improves resistance of rice to sheath blight disease. Mol. Plant Pathol. 2018, 19, 2149–2161. [Google Scholar] [CrossRef]
- Sperschneider, J.; Dodds, P.N. EffectorP 3.0: Prediction of apoplastic and cytoplasmic effectors in fungi and oomycetes. Mol. Plant-Microbe Interact. 2022, 35, 146–156. [Google Scholar] [CrossRef]
- Jaswal, R.; Kiran, K.; Rajarammohan, S.; Dubey, H.; Singh, P.K.; Sharma, Y.; Deshmukh, R.; Sonah, H.; Gupta, N.; Sharma, T.R. Effector biology of biotrophic plant fungal pathogens: Current advances and future prospects. Microbiol. Res. 2020, 241, 126567. [Google Scholar] [CrossRef]
- Tzelepis, G.; Dölfors, F.; Holmquist, L.; Dixelius, C. Plant mitochondria and chloroplasts are targeted by the Rhizoctonia solani RsCRP1 effector. Biochem. Biophys. Res. Commun. 2021, 544, 86–90. [Google Scholar] [CrossRef] [PubMed]
- Shen, Q.; Liu, Y.; Naqvi, N.I. Fungal effectors at the crossroads of phytohormone signaling. Curr. Opin. Microbiol. 2018, 46, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Seong, K.; Krasileva, K.V. Prediction of effector protein structures from fungal phytopathogens enables evolutionary analyses. Nat. Microbiol. 2023, 8, 174–187. [Google Scholar] [CrossRef]
- Ahmed, M.B.; Gonçalves dos Santos, K.C.; Sanchez, I.B.; Petre, B.; Lorrain, C.; Plourde, M.B.; Duplessis, S.; Desgagné-Penix, I.; Germain, H. A rust fungal effector binds plant DNA and modulates transcription. Sci. Rep. 2018, 8, 14718. [Google Scholar] [CrossRef] [PubMed]
- Tang, C.; Xu, Q.; Zhao, J.; Yue, M.; Wang, J.; Wang, X.; Kang, Z.; Wang, X. A rust fungus effector directly binds plant pre-mRNA splice site to reprogram alternative splicing and suppress host immunity. Plant Biotechnol. J. 2022, 20, 1167–1181. [Google Scholar] [CrossRef]
- Bonfante, P.; Genre, A. Arbuscular mycorrhizal dialogues: Do you speak ‘plantish’ or ‘fungish’? Trends Plant Sci. 2015, 20, 150–154. [Google Scholar] [CrossRef] [PubMed]
- Brundrett, M.C.; Tedersoo, L. Evolutionary history of mycorrhizal symbioses and global host plant diversity. New Phytol. 2018, 220, 1108–1115. [Google Scholar] [CrossRef] [PubMed]
- López-Ráez, J.A.; Shirasu, K.; Foo, E. Strigolactones in plant interactions with beneficial and detrimental organisms: The yin and yang. Trends Plant Sci. 2017, 22, 527–537. [Google Scholar] [CrossRef]
- Gutjahr, C.; Parniske, M. Cell and developmental biology of arbuscular mycorrhiza symbiosis. Annu. Rev. Cell Dev. Biol. 2013, 29, 593–617. [Google Scholar] [CrossRef]
- Kobae, Y.; Hata, S. Dynamics of periarbuscular membranes visualized with a fluorescent phosphate transporter in arbuscular mycorrhizal roots of rice. Plant Cell Physiol. 2010, 51, 341–353. [Google Scholar] [CrossRef]
- Keymer, A.; Pimprikar, P.; Wewer, V.; Huber, C.; Brands, M.; Bucerius, S.L.; Delaux, P.-M.; Klingl, V.; von Röpenack-Lahaye, E.; Wang, T.L.; et al. Lipid transfer from plants to arbuscular mycorrhiza fungi. eLife 2017, 6, e29107. [Google Scholar] [CrossRef] [PubMed]
- López-Pedrosa, A.; González-Guerrero, M.; Valderas, A.; Azcón-Aguilar, C.; Ferrol, N. GintAMT1 encodes a functional high-affinity ammonium transporter that is expressed in the extraradical mycelium of Glomus intraradices. Fungal Genet. Biol. 2006, 43, 102–110. [Google Scholar] [CrossRef]
- Jung, S.C.; Martinez-Medina, A.; Lopez-Raez, J.A.; Pozo, M.J. Mycorrhiza-induced resistance and priming of plant defenses. J. Chem. Ecol. 2012, 38, 651–664. [Google Scholar] [CrossRef] [PubMed]
- García-Garrido, J.M.; Ocampo, J.A. Regulation of the plant defence response in arbuscular mycorrhizal symbiosis. J. Exp. Bot. 2002, 53, 1377–1386. [Google Scholar] [CrossRef]
- Hossain, M.A.; Bhattacharjee, S.; Armin, S.-M.; Qian, P.; Xin, W.; Li, H.-Y.; Burritt, D.J.; Fujita, M.; Tran, L.-S.P. Hydrogen peroxide priming modulates abiotic oxidative stress tolerance: Insights from ROS detoxification and scavenging. Front. Plant Sci. 2015, 6, 420. [Google Scholar] [CrossRef] [PubMed]
- Blilou, I.; Bueno, P.; Ocampo, J.A.; García-Garrido, J.M. Induction of catalase and ascorbate peroxidase activities in tobacco roots inoculated with the arbuscular mycorrhizal Glomus mosseae. Mycol. Res. 2000, 104, 722–725. [Google Scholar] [CrossRef]
- Salzer, P.; Corbière, H.; Boller, T. Hydrogen peroxide accumulation in Medicago truncatula roots colonized by the arbuscular mycorrhiza-forming fungus Glomus intraradices. Planta 1999, 208, 319–325. [Google Scholar] [CrossRef]
- Klessig, D.F.; Durner, J.; Noad, R.; Navarre, D.A.; Wendehenne, D.; Kumar, D.; Zhou, J.M.; Shah, J.; Zhang, S.; Kachroo, P.; et al. Nitric oxide and salicylic acid signaling in plant defense. Proc. Natl. Acad. Sci. USA 2000, 97, 8849–8855. [Google Scholar] [CrossRef]
- Martínez-Medina, A.; Pescador, L.; Fernández, I.; Rodríguez-Serrano, M.; García, J.M.; Romero-Puertas, M.C.; Pozo, M.J. Nitric oxide and phytoglobin PHYTOGB1 are regulatory elements in the Solanum lycopersicum–Rhizophagus irregularis mycorrhizal symbiosis. New Phytol. 2019, 223, 1560–1574. [Google Scholar] [CrossRef]
- Garcia Romera, I.; Garcia Garrido, J.M.; Ocampo, J.A. Pectolytic enzymes in the vesicular-arbuscular mycorrhizal fungus Glomus mosseae. FEMS Microbiol. Lett. 1991, 78, 343–346. [Google Scholar] [CrossRef]
- Maharshi, A.; Kumar, G.; Mukherjee, A.; Raghuwanshi, R.; Singh, H.B.; Sarma, B.K. Arbuscular mycorrhizal colonization and activation of plant defense responses against phytopathogens. In Microbial Interventions in Agriculture and Environment: Volume 1: Research Trends, Priorities and Prospects; Singh, D.P., Gupta, V.K., Prabha, R., Eds.; Springer: Singapore, 2019; pp. 219–240. [Google Scholar] [CrossRef]
- Navazio, L.; Moscatiello, R.; Genre, A.; Novero, M.; Baldan, B.; Bonfante, P.; Mariani, P. A diffusible signal from arbuscular mycorrhizal fungi elicits a transient cytosolic calcium elevation in host plant cells. Plant Physiol. 2007, 144, 673–681. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Parniske, M. Activation of calcium- and calmodulin-dependent protein kinase (CCaMK), the central regulator of plant root endosymbiosis. Curr. Opin. Plant Biol. 2012, 15, 444–453. [Google Scholar] [CrossRef]
- Takeda, N.; Maekawa, T.; Hayashi, M. Nuclear-localized and deregulated calcium- and calmodulin-dependent protein kinase activates rhizobial and mycorrhizal responses in Lotus japonicus. Plant Cell 2012, 24, 810–822. [Google Scholar] [CrossRef]
- Siciliano, V.; Genre, A.; Balestrini, R.; Cappellazzo, G.; deWit, P.J.G.M.; Bonfante, P. Transcriptome analysis of arbuscular mycorrhizal roots during development of the prepenetration apparatus. Plant Physiol. 2007, 144, 1455–1466. [Google Scholar] [CrossRef]
- Foucher, F.; Kondorosi, E. Cell cycle regulation in the course of nodule organogenesis in Medicago. Plant Mol. Biol. 2000, 43, 773–786. [Google Scholar] [CrossRef]
- Vieira, P.; Kyndt, T.; Gheysen, G.; de Almeida Engler, J. An insight into critical endocycle genes for plant-parasitic nematode feeding sites establishment. Plant Signal. Behav. 2013, 8, e24223. [Google Scholar] [CrossRef]
- de Almeida Engler, J.; De Vleesschauwer, V.; Burssens, S.; Celenza, J.L.; Inzé, D.; Van Montagu, M.; Engler, G.; Gheysen, G. Molecular markers and cell cycle inhibitors show the importance of cell cycle progression in nematode-induced galls and syncytia. Plant Cell 1999, 11, 793–807. [Google Scholar] [CrossRef] [PubMed]
- Coelho, R.R.; Vieira, P.; Antonino de Souza Júnior, J.D.; Martin-Jimenez, C.; De Veylder, L.; Cazareth, J.; Engler, G.; Grossi-de-Sa, M.F.; de Almeida Engler, J. Exploiting cell cycle inhibitor genes of the KRP family to control root-knot nematode induced feeding sites in plants. Plant Cell Environ. 2017, 40, 1174–1188. [Google Scholar] [CrossRef] [PubMed]
- Russo, G.; Genre, A. Divide and be conquered—Cell cycle reactivation in arbuscular mycorrhizal symbiosis. Front. Plant Sci. 2021, 12, 753265. [Google Scholar] [CrossRef] [PubMed]
- Russo, G.; Carotenuto, G.; Fiorilli, V.; Volpe, V.; Chiapello, M.; Van Damme, D.; Genre, A. Ectopic activation of cortical cell division during the accommodation of arbuscular mycorrhizal fungi. New Phytol. 2019, 221, 1036–1048. [Google Scholar] [CrossRef]
- Carotenuto, G.; Volpe, V.; Russo, G.; Politi, M.; Sciascia, I.; de Almeida-Engler, J.; Genre, A. Local endoreduplication as a feature of intracellular fungal accommodation in arbuscular mycorrhizas. New Phytol. 2019, 223, 430–446. [Google Scholar] [CrossRef]
- An, J.; Zeng, T.; Ji, C.; de Graaf, S.; Zheng, Z.; Xiao, T.T.; Deng, X.; Xiao, S.; Bisseling, T.; Limpens, E.; et al. A Medicago truncatula SWEET transporter implicated in arbuscule maintenance during arbuscular mycorrhizal symbiosis. New Phytol. 2019, 224, 396–408. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.-H.; Eo, J.-K.; Ka, K.-H.; Eom, A.-H. Diversity of arbuscular mycorrhizal fungi and their roles in ecosystems. Mycobiology 2013, 41, 121–125. [Google Scholar] [CrossRef]
- Chen, E.C.H.; Morin, E.; Beaudet, D.; Noel, J.; Yildirir, G.; Ndikumana, S.; Charron, P.; St-Onge, C.; Giorgi, J.; Krüger, M.; et al. High intraspecific genome diversity in the model arbuscular mycorrhizal symbiont Rhizophagus irregularis. New Phytol. 2018, 220, 1161–1171. [Google Scholar] [CrossRef]
- Tang, N.; San Clemente, H.; Roy, S.; Bécard, G.; Zhao, B.; Roux, C. A survey of the gene repertoire of Gigaspora rosea unravels conserved features among glomeromycota for obligate biotrophy. Front. Microbiol. 2016, 7, 233. [Google Scholar] [CrossRef]
- Venice, F.; Ghignone, S.; Salvioli di Fossalunga, A.; Amselem, J.; Novero, M.; Xianan, X.; Sędzielewska Toro, K.; Morin, E.; Lipzen, A.; Grigoriev, I.V.; et al. At the nexus of three kingdoms: The genome of the mycorrhizal fungus Gigaspora margarita provides insights into plant, endobacterial and fungal interactions. Environ. Microbiol. 2020, 22, 122–141. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Chen, W.; Ivanov, S.; MacLean, A.M.; Wight, H.; Ramaraj, T.; Mudge, J.; Harrison, M.J.; Fei, Z. Genome and evolution of the arbuscular mycorrhizal fungus Diversispora epigaea (formerly Glomus versiforme) and its bacterial endosymbionts. New Phytol. 2019, 221, 1556–1573. [Google Scholar] [CrossRef] [PubMed]
- Montoliu-Nerin, M.; Sánchez-García, M.; Bergin, C.; Kutschera, V.E.; Johannesson, H.; Bever, J.D.; Rosling, A. In-depth phylogenomic analysis of arbuscular mycorrhizal fungi based on a comprehensive set of de novo genome assemblies. Front. Fungal Biol. 2021, 2, 716385. [Google Scholar] [CrossRef]
- Sadhana, B. Arbuscular Mycorrhizal Fungi (AMF) as a Biofertilizer-a Review. Int. J. Curr. Microbiol. Appl. Sci. 2014, 3, 384–400. [Google Scholar]
- Sugiura, Y.; Akiyama, R.; Tanaka, S.; Yano, K.; Kameoka, H.; Marui, S.; Saito, M.; Kawaguchi, M.; Akiyama, K.; Saito, K. Myristate can be used as a carbon and energy source for the asymbiotic growth of arbuscular mycorrhizal fungi. Proc. Natl. Acad. Sci. USA 2020, 117, 25779–25788. [Google Scholar] [CrossRef]
- Danesh, Y.R.; Goltapeh, E.M.; Alizadeh, A.; Sanavy, M.M. Optimizing carrot hairy root production for monoxenic culture of arbuscular mycorrhizal fungi in Iran. J. Biol. Sci. 2006, 6, 87–91. [Google Scholar] [CrossRef]
- Petrovská, B.; Šebela, M.; Doležel, J. Inside a plant nucleus: Discovering the proteins. J. Exp. Bot. 2015, 66, 1627–1640. [Google Scholar] [CrossRef] [PubMed]
- Belair, M.; Restrepo-Leal, J.D.; Praz, C.; Fontaine, F.; Rémond, C.; Fernandez, O.; Besaury, L. Botryosphaeriaceae gene machinery: Correlation between diversity and virulence. Fungal Biol. 2023, 127, 1010–1031. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhao, S.; Zhang, B.; Ma, H.-Y.; Fang, W.-H.; Sheng, W.-Q.; Yang, L.-G.; Li, X.-C. A novel ML domain-containing protein (SpMD2) functions as a potential LPS receptor involved in anti-Vibrio immune response. Dev. Comp. Immunol. 2020, 103, 103529. [Google Scholar] [CrossRef]
- Cord-Landwehr, S.; Melcher, R.L.J.; Kolkenbrock, S.; Moerschbacher, B.M. A chitin deacetylase from the endophytic fungus Pestalotiopsis sp. efficiently inactivates the elicitor activity of chitin oligomers in rice cells. Sci. Rep. 2016, 6, 38018. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Kim, J.; Kim, M.-S.; Min, C.W.; Kim, S.T.; Choi, S.-B.; Lee, J.H.; Choi, D. A Phytophthora nucleolar effector, Pi23226, targets to host ribosome biogenesis for necrotrophic cell death. Plant. Commun. 2023, 4, 100606. [Google Scholar] [CrossRef]
- Rona, G.B.; Eleutherio, E.C.A.; Pinheiro, A.S. PWWP domains and their modes of sensing DNA and histone methylated lysines. Biophys. Rev. 2016, 8, 63–74. [Google Scholar] [CrossRef]
- Paysan-Lafosse, T.; Blum, M.; Chuguransky, S.; Grego, T.; Pinto, B.L.; Salazar, G.A.; Bileschi, M.L.; Bork, P.; Bridge, A.; Colwell, L.; et al. InterPro in 2022. Nucleic Acids Res. 2023, 51, D418–D427. [Google Scholar] [CrossRef]
- Desai, N.A.; Shankar, V. Single-strand-specific nucleases. FEMS Microbiol. Rev. 2003, 26, 457–491. [Google Scholar] [CrossRef]
- Jain, B.P.; Pandey, S. WD40 repeat proteins: Signalling scaffold with diverse functions. Protein J. 2018, 37, 391–406. [Google Scholar] [CrossRef]
- Huh, S.U.; Paek, K.-H. APUM5, encoding a Pumilio RNA binding protein, negatively regulates abiotic stress responsive gene expression. BMC Plant Biol. 2014, 14, 75. [Google Scholar] [CrossRef] [PubMed]
- Klug, A. Zinc finger peptides for the regulation of gene expression. J. Mol. Biol. 1999, 293, 215–218. [Google Scholar] [CrossRef] [PubMed]
- Daou, M.; Faulds, C.B. Glyoxal oxidases: Their nature and properties. World J. Microbiol. Biotechnol. 2017, 33, 87. [Google Scholar] [CrossRef]
- Navazio, L.; Mariani, P. Calcium opens the dialogue between plants and arbuscular mycorrhizal fungi. Plant Signal. Behav. 2008, 3, 229–230. [Google Scholar] [CrossRef] [PubMed]
- Mohanty, S.K.; Arthikala, M.-K.; Nanjareddy, K.; Lara, M. Plant-symbiont interactions: The functional role of expansins. Symbiosis 2018, 74, 1–10. [Google Scholar] [CrossRef]
- Balestrini, R.; Cosgrove, D.J.; Bonfante, P. Differential location of α-expansin proteins during the accommodation of root cells to an arbuscular mycorrhizal fungus. Planta 2005, 220, 889–899. [Google Scholar] [CrossRef]
- Journet, E.-P.; El-Gachtouli, N.; Vernoud, V.; De Billy, F.; Pichon, M.; Dedieu, A.; Arnould, C.; Morandi, D.; Barker, D.G.; Gianinazzi-Pearson, V. Medicago truncatula ENOD11: A novel RPRP-encoding early nodulin gene expressed during mycorrhization in arbuscule-containing cells. Mol. Plant-Microbe Interact. 2007, 14, 737–748. [Google Scholar] [CrossRef]
- Rich, M.K.; Schorderet, M.; Reinhardt, D. The role of the cell wall compartment in mutualistic symbioses of plants. Front. Plant Sci. 2014, 5, 238. [Google Scholar] [CrossRef]
- Arthikala, M.-K.; Nanjareddy, K.; Blanco, L.; Alvarado-Affantranger, X.; Lara, M. Target of rapamycin, PvTOR, is a key regulator of arbuscule development during mycorrhizal symbiosis in Phaseolus. Sci. Rep. 2021, 11, 11319. [Google Scholar] [CrossRef]
- Helber, N.; Requena, N. Expression of the fluorescence markers DsRed and GFP fused to a nuclear localization signal in the arbuscular mycorrhizal fungus Glomus intraradices. New Phytol. 2008, 177, 537–548. [Google Scholar] [CrossRef]
- Harrier, L.A.; Millam, S.; Franken, P. Biolistic transformation of arbuscular mycorrhizal fungi: Advances and applications. In Mycorrhizal Technology in Agriculture: From Genes to Bioproducts; Gianinazzi, S., Schüepp, H., Barea, J.M., Haselwandter, K., Eds.; Birkhäuser: Basel, Switzerland, 2002; pp. 59–70. [Google Scholar]
- Testa, A.C.; Hane, J.K.; Ellwood, S.R.; Oliver, R.P. CodingQuarry: Highly accurate hidden Markov model gene prediction in fungal genomes using RNA-seq transcripts. BMC Genom. 2015, 16, 170. [Google Scholar] [CrossRef] [PubMed]
- Almagro Armenteros, J.J.; Tsirigos, K.D.; Sønderby, C.K.; Petersen, T.N.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019, 37, 420–423. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.; Park, J.; Kim, D.; Jung, K.; Kang, S.; Lee, Y.-H. Fungal secretome database: Integrated platform for annotation of fungal secretomes. BMC Genom. 2010, 11, 105. [Google Scholar] [CrossRef] [PubMed]
- Lum, G.; Min, X.J. FunSecKB: The Fungal Secretome KnowledgeBase. Database 2011, 2011, bar001. [Google Scholar] [CrossRef] [PubMed]
- Sperschneider, J.; Williams, A.H.; Hane, J.K.; Singh, K.B.; Taylor, J.M. Evaluation of secretion prediction highlights differing approaches needed for oomycete and fungal effectors. Front. Plant Sci. 2015, 6, 1168. [Google Scholar] [CrossRef]
- Wang, S.; Boevink, P.C.; Welsh, L.; Zhang, R.; Whisson, S.C.; Birch, P.R.J. Delivery of cytoplasmic and apoplastic effectors from Phytophthora infestans haustoria by distinct secretion pathways. New Phytol. 2017, 216, 205–215. [Google Scholar] [CrossRef]
- Reindl, M.; Hänsch, S.; Weidtkamp-Peters, S.; Schipper, K. A potential lock-type mechanism for unconventional secretion in fungi. Int. J. Mol. Sci. 2019, 20, 460. [Google Scholar] [CrossRef]
- Stuer, N.; Van Damme, P.; Goormachtig, S.; Van Dingenen, J. Seeking the interspecies crosswalk for filamentous microbe effectors. Trends Plant Sci. 2023, 28. [Google Scholar] [CrossRef]
- Sonah, H.; Deshmukh, R.K.; Bélanger, R.R. Computational prediction of effector proteins in fungi: Opportunities and challenges. Front. Plant Sci. 2016, 7, 126. [Google Scholar] [CrossRef]
- Petersen, T.N.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods 2011, 8, 785–786. [Google Scholar] [CrossRef]
- Käll, L.; Krogh, A.; Sonnhammer, E.L.L. A combined transmembrane topology and signal peptide prediction method. J. Mol. Biol. 2004, 338, 1027–1036. [Google Scholar] [CrossRef]
- Sperschneider, J.; Gardiner, D.M.; Dodds, P.N.; Tini, F.; Covarelli, L.; Singh, K.B.; Manners, J.M.; Taylor, J.M. EffectorP: Predicting fungal effector proteins from secretomes using machine learning. New Phytol. 2016, 210, 743–761. [Google Scholar] [CrossRef] [PubMed]
- Bendtsen, J.D.; Jensen, L.J.; Blom, N.; Von Heijne, G.; Brunak, S. Feature-based prediction of non-classical and leaderless protein secretion. Protein Eng. Des. Sel. 2004, 17, 349–356. [Google Scholar] [CrossRef]
- Sperschneider, J.; Dodds, P.N.; Gardiner, D.M.; Manners, J.M.; Singh, K.B.; Taylor, J.M. Advances and challenges in computational prediction of effectors from plant pathogenic fungi. PLoS Pathog. 2015, 11, e1004806. [Google Scholar] [CrossRef]
- Carreón-Anguiano, K.G.; Islas-Flores, I.; Vega-Arreguín, J.; Sáenz-Carbonell, L.; Canto-Canché, B. EffHunter: A tool for prediction of effector protein candidates in fungal proteomic databases. Biomolecules 2020, 10, 712. [Google Scholar] [CrossRef] [PubMed]
- Sperschneider, J.; Catanzariti, A.-M.; DeBoer, K.; Petre, B.; Gardiner, D.M.; Singh, K.B.; Dodds, P.N.; Taylor, J.M. LOCALIZER: Subcellular localization prediction of both plant and effector proteins in the plant cell. Sci. Rep. 2017, 7, 44598. [Google Scholar] [CrossRef]
- Emanuelsson, O.; Nielsen, H.; Brunak, S.; von Heijne, G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 2000, 300, 1005–1016. [Google Scholar] [CrossRef]
- Almagro Armenteros, J.J.; Sønderby, C.K.; Sønderby, S.K.; Nielsen, H.; Winther, O. DeepLoc: Prediction of protein subcellular localization using deep learning. Bioinformatics 2017, 33, 4049. [Google Scholar] [CrossRef]
- Feldman, D.; Yarden, O.; Hadar, Y. Seeking the roles for fungal small-secreted proteins in affecting saprophytic lifestyles. Front. Microbiol. 2020, 11, 455. [Google Scholar] [CrossRef]
- Marín, M.; Uversky, V.N.; Ott, T. Intrinsic disorder in pathogen effectors: Protein flexibility as an evolutionary hallmark in a molecular arms race. Plant Cell 2013, 25, 3153–3157. [Google Scholar] [CrossRef] [PubMed]
- Shen, D.; Li, Q.; Sun, P.; Zhang, M.; Dou, D. Intrinsic disorder is a common structural characteristic of RxLR effectors in oomycete pathogens. Fungal Biol. 2017, 121, 911–919. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.-N.; Liu, H.; Duan, G.-H.; Huang, Y.-M.; Liu, S.; Fang, Z.-G.; Wu, E.-J.; Shang, L.; Zhan, J. The Phytophthora infestans AVR2 effector escapes R2 recognition through effector disordering. Mol. Plant-Microbe Interact. 2020, 33, 921–931. [Google Scholar] [CrossRef] [PubMed]
- Bondos, S.E.; Dunker, A.K.; Uversky, V.N. On the roles of intrinsically disordered proteins and regions in cell communication and signaling. Cell Commun. Signal. 2021, 19, 88. [Google Scholar] [CrossRef] [PubMed]
- Chakrabarti, P.; Chakravarty, D. Intrinsically disordered proteins/regions and insight into their biomolecular interactions. Biophys. Chem. 2022, 283, 106769. [Google Scholar] [CrossRef]
- Ward, J.J.; McGuffin, L.J.; Bryson, K.; Buxton, B.F.; Jones, D.T. The DISOPRED server for the prediction of protein disorder. Bioinformatics 2004, 20, 2138–2139. [Google Scholar] [CrossRef]
- De Bekker, C.; Bruning, O.; Jonker, M.J.; Breit, T.M.; Wösten, H.A.B. Single cell transcriptomics of neighboring hyphae of Aspergillus niger. Genome Biol. 2011, 12, R71. [Google Scholar] [CrossRef] [PubMed]
- Lieben, L. Plant genetics: Spatial transcriptomics in plants. Nat. Rev. Genet. 2017, 18, 394. [Google Scholar] [CrossRef]
- Bai, Y.; Liu, H.; Lyu, H.; Su, L.; Xiong, J.; Cheng, Z.-M.M. Development of a single-cell atlas for woodland strawberry (Fragaria vesca) leaves during early Botrytis cinerea infection using single-cell RNA-seq. Hortic. Res. 2022, 9, uhab055. [Google Scholar] [CrossRef]
- Andrews, T.S.; Kiselev, V.Y.; McCarthy, D.; Hemberg, M. Tutorial: Guidelines for the computational analysis of single-cell RNA sequencing data. Nat. Protoc. 2021, 16, 1–9. [Google Scholar] [CrossRef]
- Wang, L.; Zhou, Y.; Li, R.; Liang, J.; Tian, T.; Ji, J.; Chen, R.; Zhou, Y.; Fan, Q.; Ning, G.; et al. Single cell-type transcriptome profiling reveals genes that promote nitrogen fixation in the infected and uninfected cells of legume nodules. Plant Biotechnol. J. 2022, 20, 616–618. [Google Scholar] [CrossRef]
- Lee, S.-J.; Kim, B.-D.; Rose, J.K.C. Identification of eukaryotic secreted and cell surface proteins using the yeast secretion trap screen. Nat. Protoc. 2006, 1, 2439–2447. [Google Scholar] [CrossRef]
- Lee, S.-J.; Rose, J.K.C. A yeast secretion trap assay for identification of secreted proteins from eukaryotic phytopathogens and their plant hosts. Methods Mol. Biol. 2012, 835, 519–530. [Google Scholar] [CrossRef] [PubMed]
- Huisman, R.; Bouwmeester, K.; Brattinga, M.; Govers, F.; Bisseling, T.; Limpens, E. Haustorium formation in Medicago truncatula roots infected by Phytophthora palmivora does not involve the common endosymbiotic program shared by arbuscular mycorrhizal fungi and rhizobia. Mol. Plant-Microbe Interact. 2015, 28, 1271–1280. [Google Scholar] [CrossRef]
- Zhang, S.; Xu, J.R. Effectors and Effector Delivery in Magnaporthe oryzae. PLoS Pathog. 2014, 10, e1003826. [Google Scholar] [CrossRef]
- Sugio, A.; Maclean, A.M.; Hogenhout, S.A. The small phytoplasma virulence effector SAP11 contains distinct domains required for nuclear targeting and CIN-TCP binding and destabilization. New Phytol. 2014, 202, 838–848. [Google Scholar] [CrossRef] [PubMed]
- Maes, E.; Cools, N.; Willems, H.; Baggerman, G. FACS-based Proteomics Enables Profiling of Proteins in Rare Cell Populations. Int. J. Mol. Sci. 2020, 21, 6557. [Google Scholar] [CrossRef]
- Ho-Plágaro, T.; Huertas, R.; Tamayo-Navarrete, M.I.; Ocampo, J.A.; García-Garrido, J.M. An improved method for Agrobacterium rhizogenes-mediated transformation of tomato suitable for the study of arbuscular mycorrhizal symbiosis. Plant Methods 2018, 14, 34. [Google Scholar] [CrossRef]
- Boisson-Dernier, A.; Chabaud, M.; Garcia, F.; Bécard, G.; Rosenberg, C.; Barker, D.G. Agrobacterium rhizogenes-transformed roots of Medicago truncatula for the study of nitrogen-fixing and endomycorrhizal symbiotic associations. Mol. Plant-Microbe Interact. 2001, 14, 695–700. [Google Scholar] [CrossRef]
- Küster, H. Medicago truncatula. In Brenner’s Encyclopedia of Genetics, 2nd ed.; Maloy, S., Hughes, K., Eds.; Elsevier: Amsterdam, The Netherlands, 2013; Volume 4, pp. 335–337. [Google Scholar]
- Rosa, C.; Kuo, Y.-W.; Wuriyanghan, H.; Falk, B.W. RNA interference mechanisms and applications in plant pathology. Annu. Rev. Phytopathol. 2018, 56, 581–610. [Google Scholar] [CrossRef] [PubMed]
- Koch, A.; Wassenegger, M. Host-induced gene silencing—Mechanisms and applications. New Phytol. 2021, 231, 54–59. [Google Scholar] [CrossRef]
- Potze, J. VIGS, HIGS and INTACT to Analyse Function and Translocalizion of Arbuscular Mycorrhizal Effectors; Wageningen University: Wageningen, The Netherlands, 2017; p. 31. [Google Scholar]
- Huang, G.; Allen, R.; Davis, E.L.; Baum, T.J.; Hussey, R.S. Engineering broad root-knot resistance in transgenic plants by RNAi silencing of a conserved and essential root-knot nematode parasitism gene. Proc. Natl. Acad. Sci. USA 2006, 103, 14302–14306. [Google Scholar] [CrossRef] [PubMed]
- Karimi, H.Z.; Innes, R.W. Molecular mechanisms underlying host-induced gene silencing. Plant Cell 2022, 34, 3183–3199. [Google Scholar] [CrossRef] [PubMed]
- Trouvelot, A.; Kough, J.L.; Gianinazzi-Pearson, V. Mesure du taux de mycorhization VA d’un système radiculaire. Recherche de méthode d’estimation ayant une signification fonctionnelle. In Physiological and Genetical Aspects of Mycorrhizae, Proceedings of the 1st European Symposium on Mycorrhizae, Dijon, France, 1–5 July 1985; Gianinazzi, S., Gianinazzi-Pearson, V., Eds.; Institut National de la Recherche Agronomique: Paris, France, 1986; pp. 217–221. [Google Scholar]
- McGonigle, T.P.; Miller, M.H.; Evans, D.G.; Fairchild, G.L.; Swan, J.A. A new method which gives an objective measure of colonization of roots by vesicular—Arbuscular mycorrhizal fungi. New Phytol. 1990, 115, 495–501. [Google Scholar] [CrossRef] [PubMed]
- Kokkoris, V.; Pogiatzis, A.; Hart, M.M. Contrasting common measures of arbuscular mycorrhizal fungal root colonization. J. Microbiol. Methods 2019, 167, 105727. [Google Scholar] [CrossRef]
- Evangelisti, E.; Turner, C.; McDowell, A.; Shenhav, L.; Yunusov, T.; Gavrin, A.; Servante, E.K.; Quan, C.; Schornack, S. Deep learning-based quantification of arbuscular mycorrhizal fungi in plant roots. New Phytol. 2021, 232, 2207–2219. [Google Scholar] [CrossRef]
- Timoneda, A.; Yunusov, T.; Quan, C.; Gavrin, A.; Brockington, S.F.; Schornack, S. MycoRed: Betalain pigments enable in vivo real-time visualisation of arbuscular mycorrhizal colonisation. PLoS Biol. 2021, 19, e3001326. [Google Scholar] [CrossRef]
- Ivanov, S.; Harrison, M.J. Accumulation of phosphoinositides in distinct regions of the periarbuscular membrane. New Phytol. 2019, 221, 2213–2227. [Google Scholar] [CrossRef]
- Harrison, M.J.; Dewbre, G.R.; Liu, J. A phosphate transporter from Medicago truncatula involved in the acquisition of phosphate released by arbuscular mycorrhizal fungi. Plant Cell 2002, 14, 2413–2429. [Google Scholar] [CrossRef]
- Zhang, Q.; Blaylock, L.A.; Harrison, M.J. Two Medicago truncatula half-ABC transporters are essential for arbuscule development in arbuscular mycorrhizal symbiosis. Plant Cell 2010, 22, 1483–1497. [Google Scholar] [CrossRef]
- Pumplin, N.; Mondo, S.J.; Topp, S.; Starker, C.G.; Gantt, J.S.; Harrison, M.J. Medicago truncatula Vapyrin is a novel protein required for arbuscular mycorrhizal symbiosis. Plant J. 2010, 61, 482–494. [Google Scholar] [CrossRef]
- Aceves-García, P.; Álvarez-Buylla, E.R.; Garay-Arroyo, A.; García-Ponce, B.; Muñoz, R.; de la Paz Sánchez, M. Root architecture diversity and meristem dynamics in different populations of Arabidopsis thaliana. Front. Plant Sci. 2016, 7, 858. [Google Scholar] [CrossRef]
- Muñoz, A.; Pillot, J.-P.; Cubas, P.; Rameau, C. Methods for phenotyping shoot branching and testing strigolactone bioactivity for shoot branching in Arabidopsis and pea. Methods Mol. Biol. 2021, 2309, 115–127. [Google Scholar] [CrossRef] [PubMed]
- Cosme, M.; Fernández, I.; Van der Heijden, M.G.A.; Pieterse, C.M.J. Non-mycorrhizal plants: The exceptions that prove the rule. Trends Plant Sci. 2018, 23, 577–587. [Google Scholar] [CrossRef]
- Cosme, M.; Fernández, I.; Declerck, S.; van der Heijden, M.G.A.; Pieterse, C.M.J. A coumarin exudation pathway mitigates arbuscular mycorrhizal incompatibility in Arabidopsis thaliana. Plant Mol. Biol. 2021, 106, 319–334. [Google Scholar] [CrossRef] [PubMed]
- Brückner, A.; Polge, C.; Lentze, N.; Auerbach, D.; Schlattner, U. Yeast two-hybrid, a powerful tool for systems biology. Int. J. Mol. Sci. 2009, 10, 2763–2788. [Google Scholar] [CrossRef] [PubMed]
- Erffelinck, M.-L.; Ribeiro, B.; Perassolo, M.; Pauwels, L.; Pollier, J.; Storme, V.; Goossens, A. A user-friendly platform for yeast two-hybrid library screening using next generation sequencing. PLoS ONE 2018, 13, e0201270. [Google Scholar] [CrossRef]
- Wanamaker, S.A.; Garza, R.M.; MacWilliams, A.; Nery, J.R.; Bartlett, A.; Castanon, R.; Goubil, A.; Feeney, J.; O’Malley, R.; Huang, S.-s.C.; et al. CrY2H-seq: A massively-multiplexed assay for deep coverage interactome mapping. Nat. Methods 2017, 14, 819. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, Y.; Yang, X.; Wen, Z.; Nagalakshmi, U.; Dinesh-Kumar, S.P. TurboID-based proximity labeling for in planta identification of protein-protein interaction networks. J. Vis. Exp. 2020, 159, e60728. [Google Scholar] [CrossRef]
- Petre, B.; Saunders, D.G.O.; Sklenar, J.; Lorrain, C.; Win, J.; Duplessis, S.; Kamoun, S. Candidate effector proteins of the rust pathogen Melampsora larici-populina target diverse plant cell compartments. Mol. Plant-Microbe Interact. 2015, 28, 689–700. [Google Scholar] [CrossRef]
- Struk, S.; Jacobs, A.; Sánchez Martín-Fontecha, E.; Gevaert, K.; Cubas, P.; Goormachtig, S. Exploring the protein–protein interaction landscape in plants. Plant Cell Environ. 2019, 42, 387–409. [Google Scholar] [CrossRef]
- De Ryck, J.; Van Damme, P.; Goormachtig, S. From prediction to function: Current practices and challenges towards the functional characterization of type III effectors. Front. Microbiol. 2023, 14, 1113442. [Google Scholar] [CrossRef] [PubMed]
- Kanja, C.; Hammond-Kosack, K.E. Proteinaceous effector discovery and characterization in filamentous plant pathogens. Mol. Plant Pathol. 2020, 21, 1353–1376. [Google Scholar] [CrossRef] [PubMed]
- Mair, A.; Bergmann, D.C. Advances in enzyme-mediated proximity labeling and its potential for plant research. Plant Physiol. 2022, 188, 756–768. [Google Scholar] [CrossRef]
- Kubitz, L.; Bitsch, S.; Zhao, X.; Schmitt, K.; Deweid, L.; Roehrig, A.; Barazzone, E.C.; Valerius, O.; Kolmar, H.; Béthune, J. Engineering of ultraID, a compact and hyperactive enzyme for proximity-dependent biotinylation in living cells. Commun. Biol. 2022, 5, 657. [Google Scholar] [CrossRef] [PubMed]
- Lin, Q.; Zhou, Z.; Luo, W.; Fang, M.; Li, M.; Li, H. Screening of proximal and interacting proteins in rice protoplasts by proximity-dependent biotinylation. Front. Plant Sci. 2017, 8, 749. [Google Scholar] [CrossRef]
- Miltenburg, M.G.; Bonner, C.; Hepworth, S.; Huang, M.; Rampitsch, C.; Subramaniam, R. Proximity-dependent biotinylation identifies a suite of candidate effector proteins from Fusarium graminearum. Plant J. 2022, 112, 369–382. [Google Scholar] [CrossRef]
- Xu, G.; Zhong, X.; Shi, Y.; Liu, Z.; Jiang, N.; Liu, J.; Ding, B.; Li, Z.; Kang, H.; Ning, Y.; et al. A fungal effector targets a heat shock-dynamin protein complex to modulate mitochondrial dynamics and reduce plant immunity. Sci. Adv. 2020, 6, eabb7719. [Google Scholar] [CrossRef]
- Grefen, C.; Blatt, M.R. A 2in1 cloning system enables ratiometric bimolecular fluorescence complementation (rBiFC). Biotechniques 2012, 53, 311–314. [Google Scholar] [CrossRef]
- Tebo, A.G.; Gautier, A. A split fluorescent reporter with rapid and reversible complementation. Nat. Commun. 2019, 10, 2822. [Google Scholar] [CrossRef] [PubMed]
- Li, J.-F.; Bush, J.; Xiong, Y.; Li, L.; McCormack, M. Large-scale protein-protein interaction analysis in Arabidopsis mesophyll protoplasts by split firefly luciferase complementation. PLoS ONE 2011, 6, e27364. [Google Scholar] [CrossRef]
- Chen, H.; Zou, Y.; Shang, Y.; Lin, H.; Wang, Y.; Cai, R.; Tang, X.; Zhou, J.M. Firefly Luciferase Complementation Imaging Assay for Protein-Protein Interactions in Plants. Plant Physiol. 2008, 146, 323–324. [Google Scholar] [CrossRef] [PubMed]
- Liu, E.T.; Pott, S.; Huss, M. Q&A: ChIP-seq technologies and the study of gene regulation. BMC Biol. 2010, 8, 56. [Google Scholar] [CrossRef]
- John, E.; Singh, K.B.; Oliver, R.P.; Soyer, J.L.; Muria-Gonzalez, J.; Soo, D.; Jacques, S.; Tan, K.-C. Chromatin-immunoprecipitation reveals the PnPf2 transcriptional network controlling effector-mediated virulence in a fungal pathogen of wheat. bioRxiv 2022. [Google Scholar] [CrossRef]
- Tollot, M.; Assmann, D.; Becker, C.; Altmüller, J.; Dutheil, J.Y.; Wegner, C.E.; Kahmann, R. The WOPR protein Ros1 is a master regulator of sporogenesis and late effector gene expression in the maize pathogen Ustilago maydis. PLoS Pathog. 2016, 12, e1005697. [Google Scholar] [CrossRef] [PubMed]
- Burjoski, V.; Reddy, A.S.N. The landscape of RNA-protein interactions in plants: Approaches and current status. Int. J. Mol. Sci. 2021, 22, 2845. [Google Scholar] [CrossRef] [PubMed]
- Gagliardi, M.; Matarazzo, M.R. RIP: RNA immunoprecipitation. Methods Mol. Biol. 2016, 1480, 73–86. [Google Scholar] [CrossRef]
- Huppertz, I.; Attig, J.; D’Ambrogio, A.; Easton, L.E.; Sibley, C.R.; Sugimoto, Y.; Tajnik, M.; König, J.; Ule, J. iCLIP: Protein–RNA interactions at nucleotide resolution. Methods 2014, 65, 274–287. [Google Scholar] [CrossRef]

| R. irregularis Effector ID a | InterProScan Protein Domains |
|---|---|
| RirG175680 | RlpA-like protein, double-psi beta-barrel domain/expansin |
| RirG165580 | Nitrogen permease regulator 3/negative regulation of Target Of Rapamycin signaling |
| RirG263220 | Phytocyanin domain/Early nodulin-like protein domain |
| RirG200050 | WD40 repeat-containing domain superfamily/Armadillo-like helical protein |
| jgi.p|Gloin1|346360 | RING-H2 Zinc finger C3HC4 |
| RirG013260 | S1/P1 nuclease domain superfamily |
| RirG267270 | RNA-binding protein/Plant self-incompatibility S1/Pumilio homolog 15-like |
| jgi.p|Gloin1|154898 | Calcium/lipid-binding domain/tricalbin |
| RirG043250 | EF-hand domain/Calcium-binding protein |
| RirG045350 | Calreticulin/calnexin calcium-binding ER chaperones |
| RirG101100 | Calcium-dependent phosphotriesterase |
| RirG043650, RirG257590, RirG187640, RirG180400, jgi.p|Gloin1|161262 | Glyoxal oxidase |
| EP | Secretion | Subcellular Localization | Plant Target | Ref. | |||
|---|---|---|---|---|---|---|---|
| Method | Results | Method | Results | Method | Results | ||
| SP7 | YST and M. oryzae SP7-mediated secretion | Secreted and translocated to the cell nucleus | Transient expression; Agrobacterium-mediated infiltration of N. benthamiana leaves | Nuclear | Y2H screening | ERF19 | [27] |
| SIS1 | Not tested | Not tested | Not tested | Not tested | Not tested | Not tested | [28] |
| RiCRN1 | Not tested | Not tested | Transient expression; Agrobacterium-mediated infiltration of N. benthamiana leaves | Nuclear bodies | Not tested | Not tested | [29] |
| RiLSM | YST | Secreted | Not tested | Not tested | Microscale thermophoresis | R. irregularis CO 4, 5, and 7 | [30] |
| RiNLE1 | YST | Secreted | Ectopic expression in M. truncatula composite plants | Nucleolar and nuclear bodies | IP-LC MS/MS on M. truncatula arbusculated cells | MtH2B | [31] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aparicio Chacón, M.V.; Van Dingenen, J.; Goormachtig, S. Characterization of Arbuscular Mycorrhizal Effector Proteins. Int. J. Mol. Sci. 2023, 24, 9125. https://doi.org/10.3390/ijms24119125
Aparicio Chacón MV, Van Dingenen J, Goormachtig S. Characterization of Arbuscular Mycorrhizal Effector Proteins. International Journal of Molecular Sciences. 2023; 24(11):9125. https://doi.org/10.3390/ijms24119125
Chicago/Turabian StyleAparicio Chacón, María V., Judith Van Dingenen, and Sofie Goormachtig. 2023. "Characterization of Arbuscular Mycorrhizal Effector Proteins" International Journal of Molecular Sciences 24, no. 11: 9125. https://doi.org/10.3390/ijms24119125
APA StyleAparicio Chacón, M. V., Van Dingenen, J., & Goormachtig, S. (2023). Characterization of Arbuscular Mycorrhizal Effector Proteins. International Journal of Molecular Sciences, 24(11), 9125. https://doi.org/10.3390/ijms24119125






