Quick and Spontaneous Transformation between [3Fe–4S] and [4Fe–4S] Iron–Sulfur Clusters in the tRNA-Thiolation Enzyme TtuA
Abstract
1. Introduction
2. Results
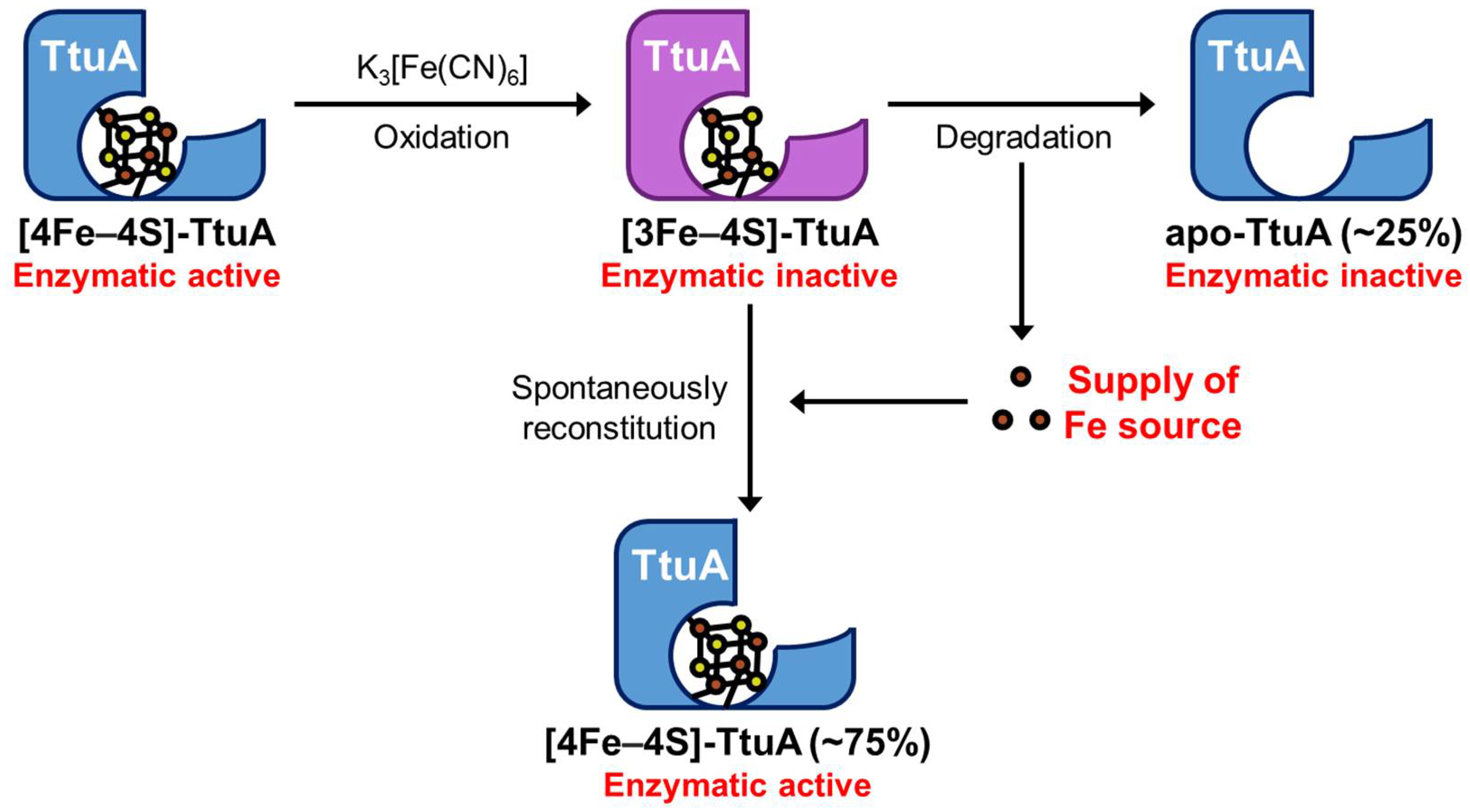
2.1. [3Fe–4S] Cluster in TtuA was Spontaneously Transformed into the [4Fe–4S] Cluster
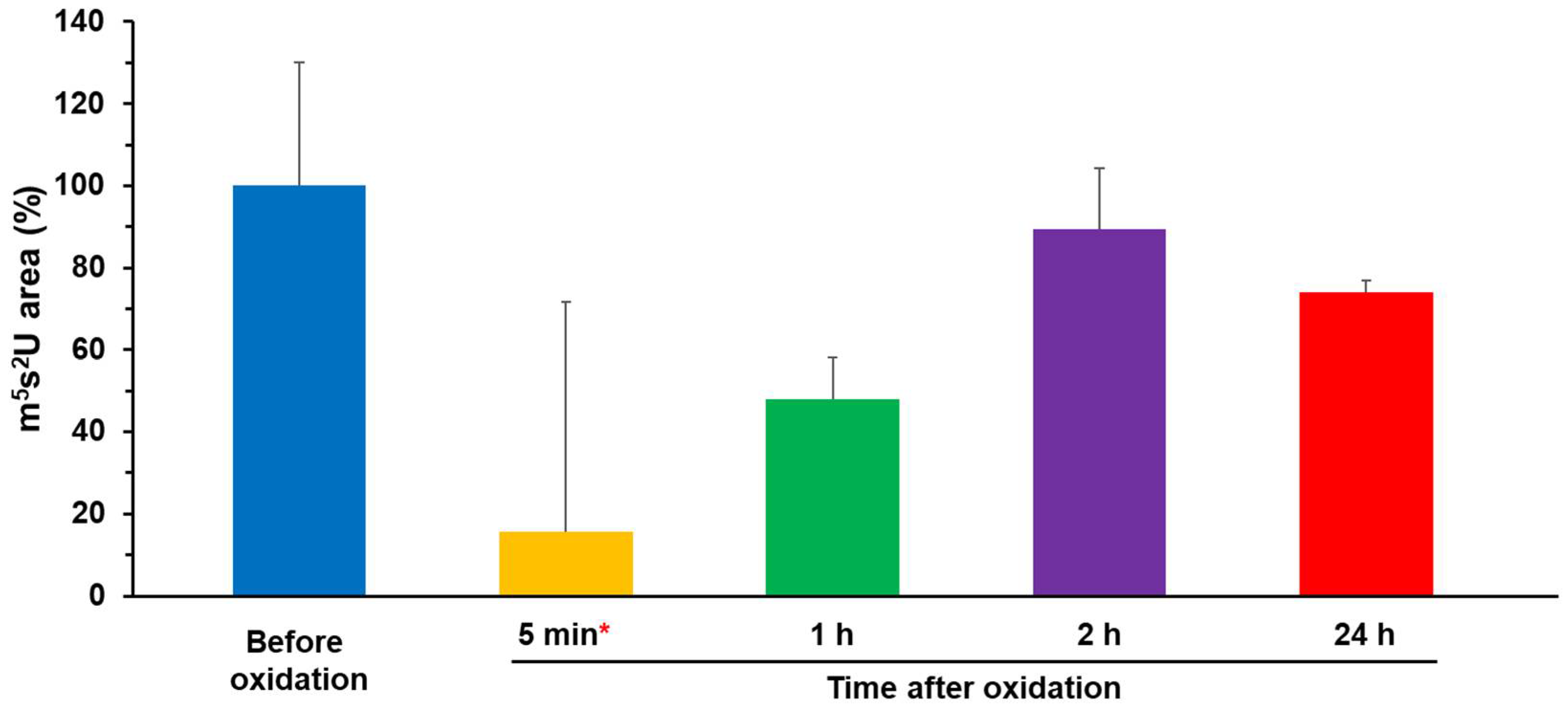
2.2. Enzymatic Activity of Oxidized TtuA was Recovered by Reconstitution of [4Fe–4S]-TtuA
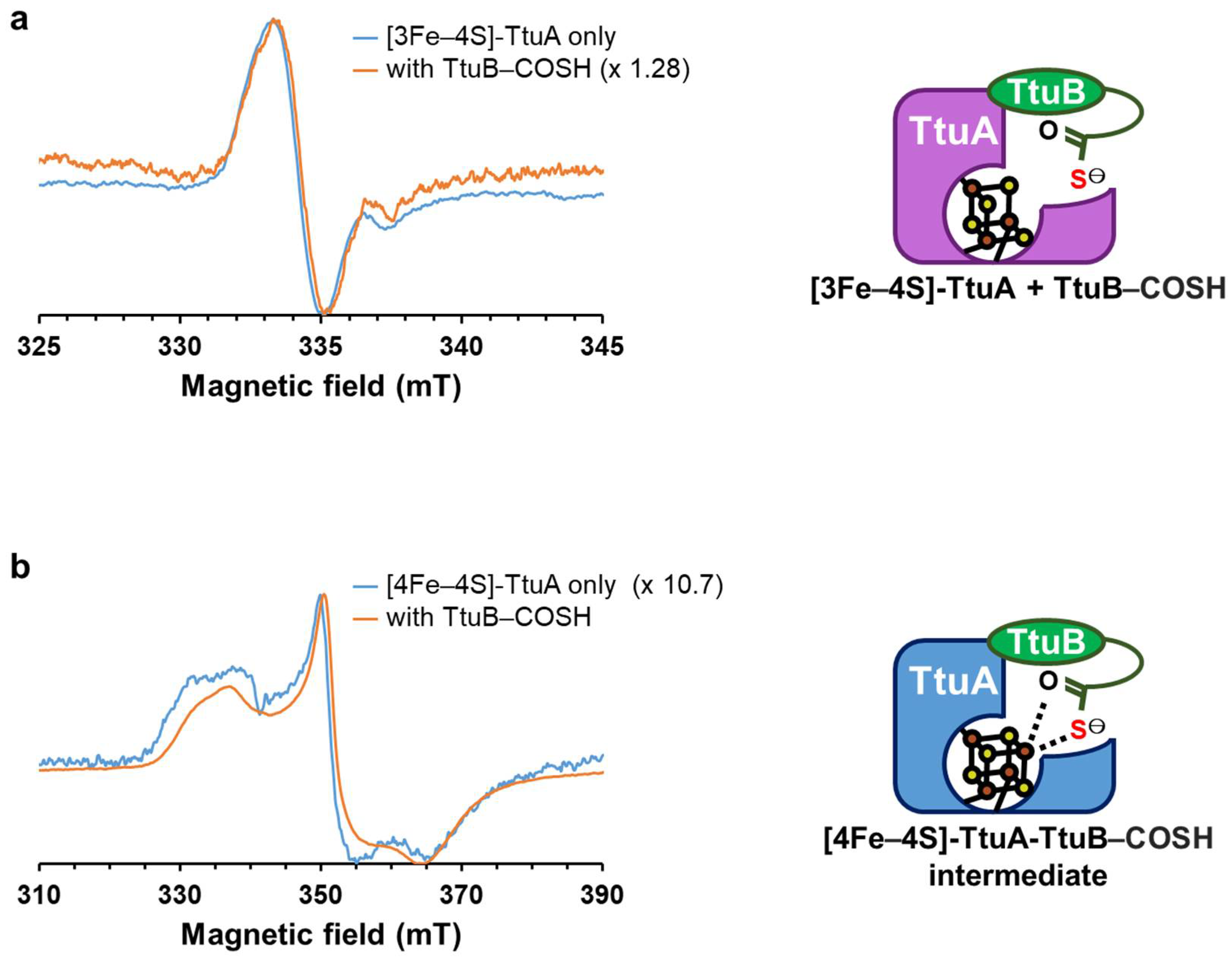
2.3. The Unique Fe in [4Fe–4S]-TtuA Is Required for its Binding to the Sulfur Donor
3. Discussion
3.1. Formation of a Functional Fe–S Cluster Bound to TtuA
3.2. The Active form of Fe–S Clusters in tRNA-Thiolation Enzymes
3.3. The Change of Fe–S Clusters under Strict Regulation of Oxidation–Reduction
4. Materials and Methods
4.1. Expression of TtuA
4.2. Purification of TtuA
4.3. Reconstitution of [4Fe–4S]-TtuA and [3Fe–4S]-TtuA
4.4. Expression and Purification of TtuB–COSH
4.5. EPR Spectroscopy of the Fe–S Cluster Bound to TtuA
4.6. Activity Assay for [3Fe–4S]-TtuA and [4Fe–4S]-TtuA
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Beinert, H.; Holm, R.H.; Münck, E. Iron-sulfur clusters: Nature’s modular, multipurpose structures. Science 1997, 277, 653–659. [Google Scholar] [CrossRef] [PubMed]
- Arnon, D.I.; Whatley, F.R.; Allen, M.B. Triphosphopyridine nucleotide as a catalyst of photosynthetic phosphorylation. Nature 1957, 180, 182–185. [Google Scholar] [CrossRef] [PubMed]
- Beinert, H.; Sands, R.H. Studies on succinic and DPNH dehydrogenase preparations by paramagnetic resonance (EPR) spectroscopy. Biochem. Biophys. Res. Commun. 1960, 3, 41–46. [Google Scholar] [CrossRef]
- Mortenson, L.E. Nitrogen fixation: Role of ferredoxin in anaerobic metabolism. Annu. Rev. Microbiol. 1963, 17, 115–138. [Google Scholar] [CrossRef]
- Beinert, H.; Kiley, P.J. Fe-S proteins in sensing and regulatory functions. Curr. Opin. Chem. Biol. 1999, 3, 152–157. [Google Scholar] [CrossRef] [PubMed]
- Netz, D.J.A.; Stith, C.M.; Stümpfig, M.; Köpf, G.; Vogel, D.; Genau, H.M.; Stodola, J.L.; Lill, R.; Burgers, P.M.J.; Pierik, A.J. Eukaryotic DNA polymerases require an iron-sulfur cluster for the formation of active complexes. Nat. Chem. Biol. 2012, 8, 125–132. [Google Scholar] [CrossRef]
- Rudolf, J.; Makrantoni, V.; Ingledew, W.J.; Stark, M.J.R.; White, M.F. The DNA repair helicases XPD and FancJ have essential iron-sulfur domains. Mol. Cell 2006, 23, 801–808. [Google Scholar] [CrossRef]
- Maio, N.; Lafont, B.A.P.; Sil, D.; Li, Y.; Bollinger, J.M.; Krebs, C.; Pierson, T.C.; Linehan, W.M.; Rouault, T.A. Fe-S cofactors in the SARS-CoV-2 RNA-dependent RNA polymerase are potential antiviral targets. Science 2021, 373, 236–241. [Google Scholar] [CrossRef]
- Shigi, N. Recent advances in our understanding of the biosynthesis of sulfur modifications in tRNAs. Front. Microbiol. 2018, 9, 2679. [Google Scholar] [CrossRef]
- Caserta, G.; Zuccarello, L.; Barbosa, C.; Silveira, C.M.; Moe, E.; Katz, S.; Hildebrandt, P.; Zebger, I.; Todorovic, S. Unusual structures and unknown roles of FeS clusters in metalloenzymes seen from a resonance Raman spectroscopic perspective. Coord. Chem. Rev. 2022, 452, 214287. [Google Scholar] [CrossRef]
- Nicolet, Y.; Rohac, R.; Martin, L.; Fontecilla-Camps, J.C. X-ray snapshots of possible intermediates in the time course of synthesis and degradation of protein-bound Fe4S4 clusters. Proc. Natl. Acad. Sci. USA 2013, 110, 7188–7192. [Google Scholar] [CrossRef] [PubMed]
- Crack, J.C.; Green, J.; Thomson, A.J.; Le Brun, N.E. Iron–sulfur clusters as biological sensors: The chemistry of reactions with molecular oxygen and nitric oxide. Acc. Chem. Res. 2014, 47, 3196–3205. [Google Scholar] [CrossRef] [PubMed]
- Lloyd, S.J.; Lauble, H.; Prasad, G.S.; Stout, C.D. The mechanism of aconitase: 1.8 Å resolution crystal structure of the S642A:citrate complex. Protein Sci. 1999, 8, 2655–2662. [Google Scholar] [CrossRef] [PubMed]
- Vey, J.L.; Yang, J.; Li, M.; Broderick, W.E.; Broderick, J.B.; Drennan, C.L. Structural basis for glycyl radical formation by pyruvate formate-lyase activating enzyme. Proc. Natl. Acad. Sci. USA 2008, 105, 16137–16141. [Google Scholar] [CrossRef] [PubMed]
- Gräwert, T.; Span, I.; Eisenreich, W.; Rohdich, F.; Eppinger, J.; Bacher, A.; Groll, M. Probing the reaction mechanism of IspH protein by x-ray structure analysis. Proc. Natl. Acad. Sci. USA 2010, 107, 1077–1081. [Google Scholar] [CrossRef] [PubMed]
- Krebs, C.; Henshaw, T.F.; Cheek, J.; Huynh, B.H.; Broderick, J.B. Conversion of 3Fe-4S to 4Fe-4S clusters in native pyruvate formate-lyase activating enzyme: Mössbauer characterization and implications for mechanism. J. Am. Chem. Soc. 2000, 122, 12497–12506. [Google Scholar] [CrossRef]
- Emptage, M.H.; Dreyers, J.L.; Kennedy, M.C.; Beinert, H. Optical and EPR characterization of different species of active and inactive aconitase. J. Biol. Chem. 1983, 258, 11106–11111. [Google Scholar] [CrossRef]
- Shigi, N. Biosynthesis and degradation of sulfur modifications in tRNAs. Int. J. Mol. Sci. 2021, 22, 11937. [Google Scholar] [CrossRef]
- Yarian, C.; Townsend, H.; Czestkowski, W.; Sochacka, E.; Malkiewicz, A.J.; Guenther, R.; Miskiewicz, A.; Agris, P.F. Accurate translation of the genetic code depends on tRNA modified nucleosides. J. Biol. Chem. 2002, 277, 16391–16395. [Google Scholar] [CrossRef]
- Shigi, N.; Sakaguchi, Y.; Suzuki, T.; Watanabe, K. Identification of two tRNA thiolation genes required for cell growth at extremely high temperatures. J. Biol. Chem. 2006, 281, 14296–14306. [Google Scholar] [CrossRef]
- Mueller, E. Identification of a gene involved in the generation of 4-thiouridine in tRNA. Nucleic Acids Res. 1998, 26, 2606–2610. [Google Scholar] [CrossRef]
- Yasukawa, T. Wobble modification defect in tRNA disturbs codon-anticodon interaction in a mitochondrial disease. EMBO J. 2001, 20, 4794–4802. [Google Scholar] [CrossRef] [PubMed]
- Barbieri, I.; Kouzarides, T. Role of RNA modifications in cancer. Nat. Rev. Cancer 2020, 20, 303–322. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Ishizaka, M.; Narai, S.; Horitani, M.; Shigi, N.; Yao, M.; Tanaka, Y. The [4Fe-4S] cluster of sulfurtransferase TtuA desulfurizes TtuB during tRNA modification in Thermus thermophilus. Commun. Biol. 2020, 3, 168. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Asai, S.; Narai, S.; Nambu, S.; Omura, N.; Sakaguchi, Y.; Suzuki, T.; Ikeda-Saito, M.; Watanabe, K.; Yao, M.; et al. Biochemical and structural characterization of oxygen-sensitive 2-thiouridine synthesis catalyzed by an iron-sulfur protein TtuA. Proc. Natl. Acad. Sci. USA 2017, 114, 4954–4959. [Google Scholar] [CrossRef] [PubMed]
- Arragain, S.; Bimai, O.; Legrand, P.; Caillat, S.; Ravanat, J.-L.; Touati, N.; Binet, L.; Atta, M.; Fontecave, M.; Golinelli-Pimpaneau, B. Nonredox thiolation in tRNA occurring via sulfur activation by a [4Fe-4S] cluster. Proc. Natl. Acad. Sci. USA 2017, 114, 7355–7360. [Google Scholar] [CrossRef]
- Bouvier, D.; Labessan, N.; Clémancey, M.; Latour, J.-M.; Ravanat, J.-L.; Fontecave, M.; Atta, M. TtcA a new tRNA-thioltransferase with an Fe-S cluster. Nucleic Acids Res. 2014, 42, 7960–7970. [Google Scholar] [CrossRef] [PubMed]
- Bimai, O.; Arragain, S.; Golinelli-Pimpaneau, B. Structure-based mechanistic insights into catalysis by tRNA thiolation enzymes. Curr. Opin. Struct. Biol. 2020, 65, 69–78. [Google Scholar] [CrossRef]
- Liu, Y.; Vinyard, D.J.; Reesbeck, M.E.; Suzuki, T.; Manakongtreecheep, K.; Holland, P.L.; Brudvig, G.W.; Söll, D. A [3Fe-4S] cluster is required for tRNA thiolation in archaea and eukaryotes. Proc. Natl. Acad. Sci. USA 2016, 113, 12703–12708. [Google Scholar] [CrossRef]
- Shigi, N.; Horitani, M.; Miyauchi, K.; Suzuki, T.; Kuroki, M. An ancient type of MnmA protein is an iron-sulfur cluster-dependent sulfurtransferase for tRNA anticodons. RNA 2020, 26, 240–250. [Google Scholar] [CrossRef]
- Zhou, J.; Lénon, M.; Ravanat, J.-L.; Touati, N.; Velours, C.; Podskoczyj, K.; Leszczynska, G.; Fontecave, M.; Barras, F.; Golinelli-Pimpaneau, B. Iron–sulfur biology invades tRNA modification: The case of U34 sulfuration. Nucleic Acids Res. 2021, 49, 3997–4007. [Google Scholar] [CrossRef] [PubMed]
- Numata, T.; Ikeuchi, Y.; Fukai, S.; Suzuki, T.; Nureki, O. Snapshots of tRNA sulphuration via an adenylated intermediate. Nature 2006, 442, 419–424. [Google Scholar] [CrossRef] [PubMed]
- Broderick, J.B.; Duffus, B.R.; Duschene, K.S.; Shepard, E.M. Radical S-adenosylmethionine enzymes. Chem. Rev. 2014, 114, 4229–4317. [Google Scholar] [CrossRef] [PubMed]
- Shigi, N.; Asai, S.; Watanabe, K. Identification of a rhodanese-like protein involved in thiouridine biosynthesis in Thermus thermophilus tRNA. FEBS Lett. 2016, 590, 4628–4637. [Google Scholar] [CrossRef] [PubMed]
- Dewez, M.; Bauer, F.; Dieu, M.; Raes, M.; Vandenhaute, J.; Hermand, D. The conserved wobble uridine tRNA thiolase Ctu1-Ctu2 is required to maintain genome integrity. Proc. Natl. Acad. Sci. USA 2008, 105, 5459–5464. [Google Scholar] [CrossRef]
- Schlieker, C.D.; Van Der Veen, A.G.; Damon, J.R.; Spooner, E.; Ploegh, H.L. A functional proteomics approach links the ubiquitin-related modifier Urm1 to a tRNA modification pathway. Proc. Natl. Acad. Sci. USA 2008, 105, 18255–18260. [Google Scholar] [CrossRef]
- Nakagawa, H.; Kuratani, M.; Goto-Ito, S.; Ito, T.; Katsura, K.; Terada, T.; Shirouzu, M.; Sekine, S.; Shigi, N.; Yokoyama, S. Crystallographic and mutational studies on the tRNA thiouridine synthetase TtuA. Proteins Struct. Funct. Bioinform. 2013, 81, 1232–1244. [Google Scholar] [CrossRef]
- Bak, D.W.; Elliott, S.J. Alternative FeS cluster ligands: Tuning redox potentials and chemistry. Curr. Opin. Chem. Biol. 2014, 19, 50–58. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Golinelli-Pimpaneau, B. Prediction of the iron–sulfur binding sites in proteins using the highly accurate three-dimensional models calculated by AlphaFold and RoseTTAFold. Inorganics 2022, 10, 2. [Google Scholar] [CrossRef]
- Wehrspan, Z.J.; McDonnell, R.T.; Elcock, A.H. Identification of iron-sulfur (Fe-S) cluster and zinc (Zn) binding sites within proteomes predicted by DeepMind’s AlphaFold2 program dramatically expands the metalloproteome. J. Mol. Biol. 2022, 434, 167377. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef] [PubMed]
- Chenna, R.; Sugawara, H.; Koike, T.; Lopez, R.; Gibson, T.J.; Higgins, D.G.; Thompson, J.D. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003, 31, 3497–3500. [Google Scholar] [CrossRef]
- Robert, X.; Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014, 42, 320–324. [Google Scholar] [CrossRef]
- Neumann, P.; Lakomek, K.; Naumann, P.-T.; Erwin, W.M.; Lauhon, C.T.; Ficner, R. Crystal structure of a 4-thiouridine synthetase–RNA complex reveals specificity of tRNA U8 modification. Nucleic Acids Res. 2014, 42, 6673–6685. [Google Scholar] [CrossRef]
- Combet, C.; Blanchet, C.; Geourjon, C.; Deléage, G. NPS@: Network protein sequence analysis. Trends Biochem. Sci. 2000, 25, 147–150. [Google Scholar] [CrossRef]
- Apweiler, R.; Bairoch, A.; Wu, C.H.; Barker, W.C.; Boeckmann, B.; Ferro, S.; Gasteiger, E.; Huang, H.; Lopez, R.; Magrane, M.; et al. NPS@: Network protein sequence analysis. Trends Biochem. Sci. 2000, 25, 147–150. [Google Scholar] [CrossRef]
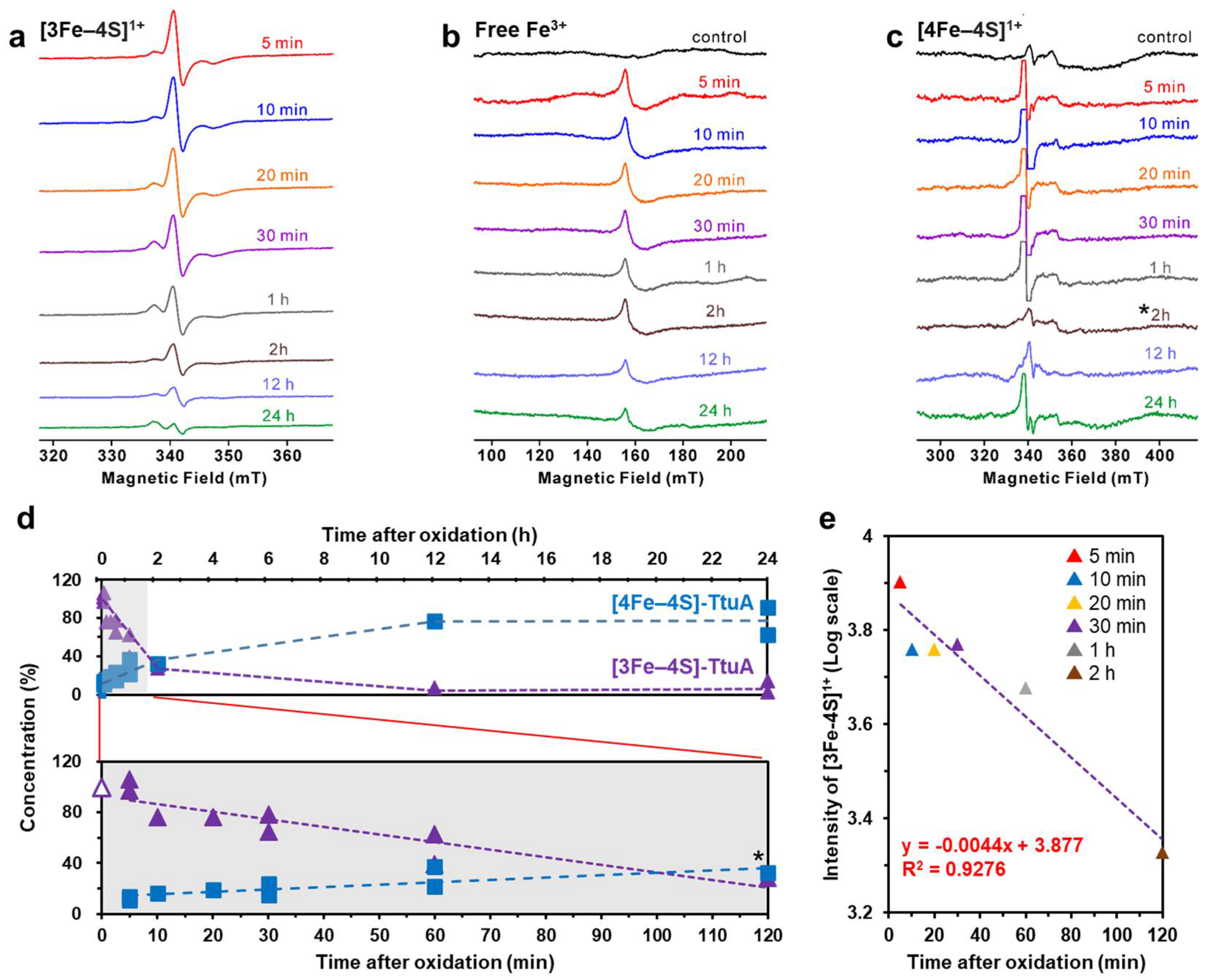
| Time after Oxidation | [3Fe–4S] | [4Fe–4S] |
|---|---|---|
| Before oxidation | ― | 100% |
| Immediately after oxidation (0 min) | (100%) | ― |
| 5 min † | 99% ± 7% | 12% ± 1% |
| 10 min | 76% | 16% |
| 20 min | 76% | 19% |
| 30 min † | 70% ± 8% | 20% ± 4% |
| 1 h † | 50% ± 13% | 30% ± 8% |
| 2 h * | 28% | 33% |
| 12 h | 7% | 77% |
| 24 h † | 9% ± 6% | 77% ± 14% |
| Time after Oxidation | Enzymatic Activity without K3[Fe(CN)6] |
|---|---|
| Before oxidation | 100% ± 30% |
| 5 min * | 16% ± 56% |
| 1 h | 48% ± 10% |
| 2 h | 89% ± 15% |
| 24 h | 74% ± 3% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ishizaka, M.; Chen, M.; Narai, S.; Tanaka, Y.; Ose, T.; Horitani, M.; Yao, M. Quick and Spontaneous Transformation between [3Fe–4S] and [4Fe–4S] Iron–Sulfur Clusters in the tRNA-Thiolation Enzyme TtuA. Int. J. Mol. Sci. 2023, 24, 833. https://doi.org/10.3390/ijms24010833
Ishizaka M, Chen M, Narai S, Tanaka Y, Ose T, Horitani M, Yao M. Quick and Spontaneous Transformation between [3Fe–4S] and [4Fe–4S] Iron–Sulfur Clusters in the tRNA-Thiolation Enzyme TtuA. International Journal of Molecular Sciences. 2023; 24(1):833. https://doi.org/10.3390/ijms24010833
Chicago/Turabian StyleIshizaka, Masato, Minghao Chen, Shun Narai, Yoshikazu Tanaka, Toyoyuki Ose, Masaki Horitani, and Min Yao. 2023. "Quick and Spontaneous Transformation between [3Fe–4S] and [4Fe–4S] Iron–Sulfur Clusters in the tRNA-Thiolation Enzyme TtuA" International Journal of Molecular Sciences 24, no. 1: 833. https://doi.org/10.3390/ijms24010833
APA StyleIshizaka, M., Chen, M., Narai, S., Tanaka, Y., Ose, T., Horitani, M., & Yao, M. (2023). Quick and Spontaneous Transformation between [3Fe–4S] and [4Fe–4S] Iron–Sulfur Clusters in the tRNA-Thiolation Enzyme TtuA. International Journal of Molecular Sciences, 24(1), 833. https://doi.org/10.3390/ijms24010833







