S100A8 and S100A12 Proteins as Biomarkers of High Disease Activity in Patients with Rheumatoid Arthritis That Can Be Regulated by Epigenetic Drugs
Abstract
1. Introduction
2. Results
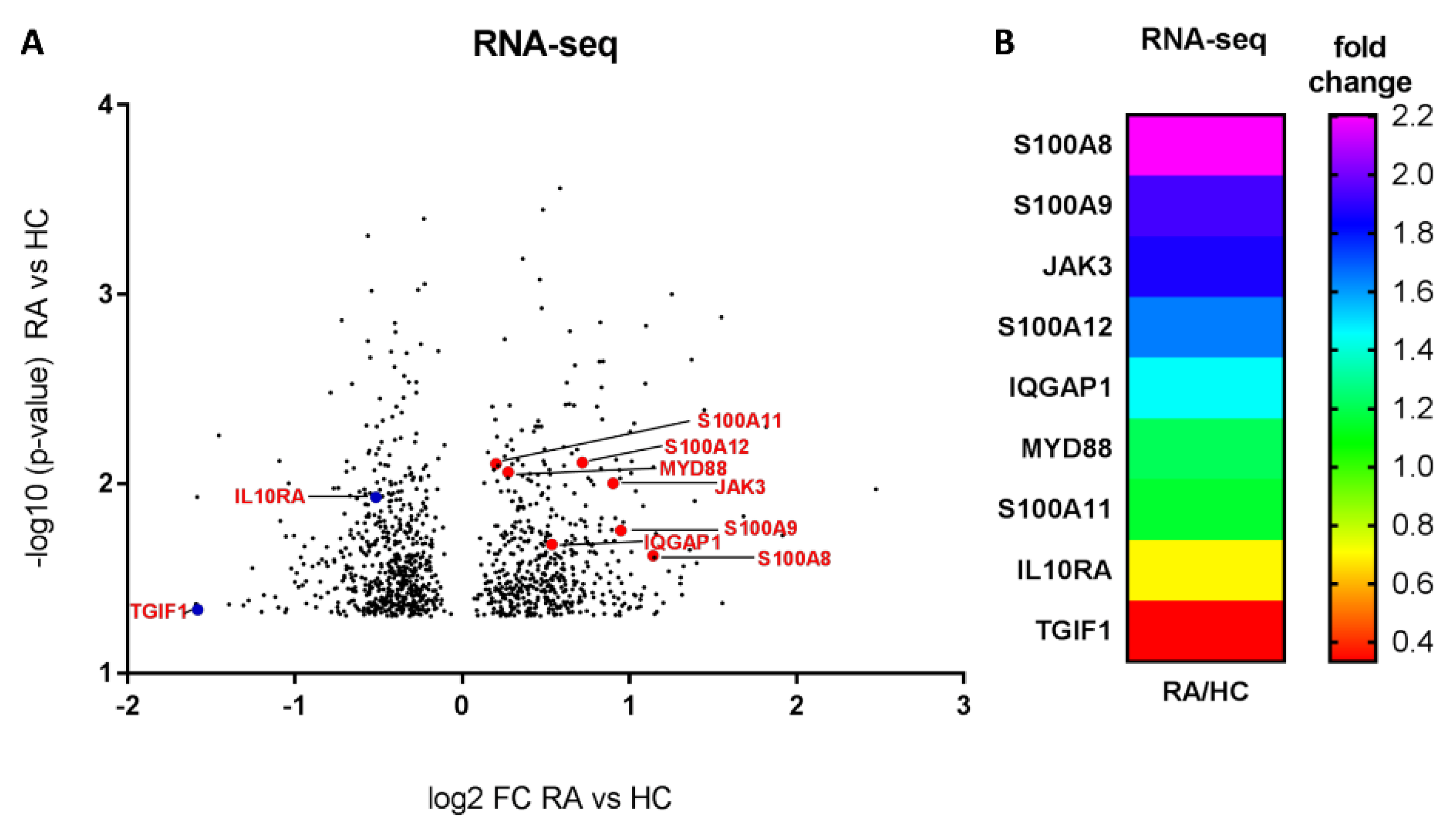
2.1. Different Expressions of Pro-Inflammatory and Anti-Inflammatory Genes in Monocytes from RA Patients Based on Global Profiling
2.2. Viability of the Monocytic THP-1 Cell Line Treated with Budesonide and RG108
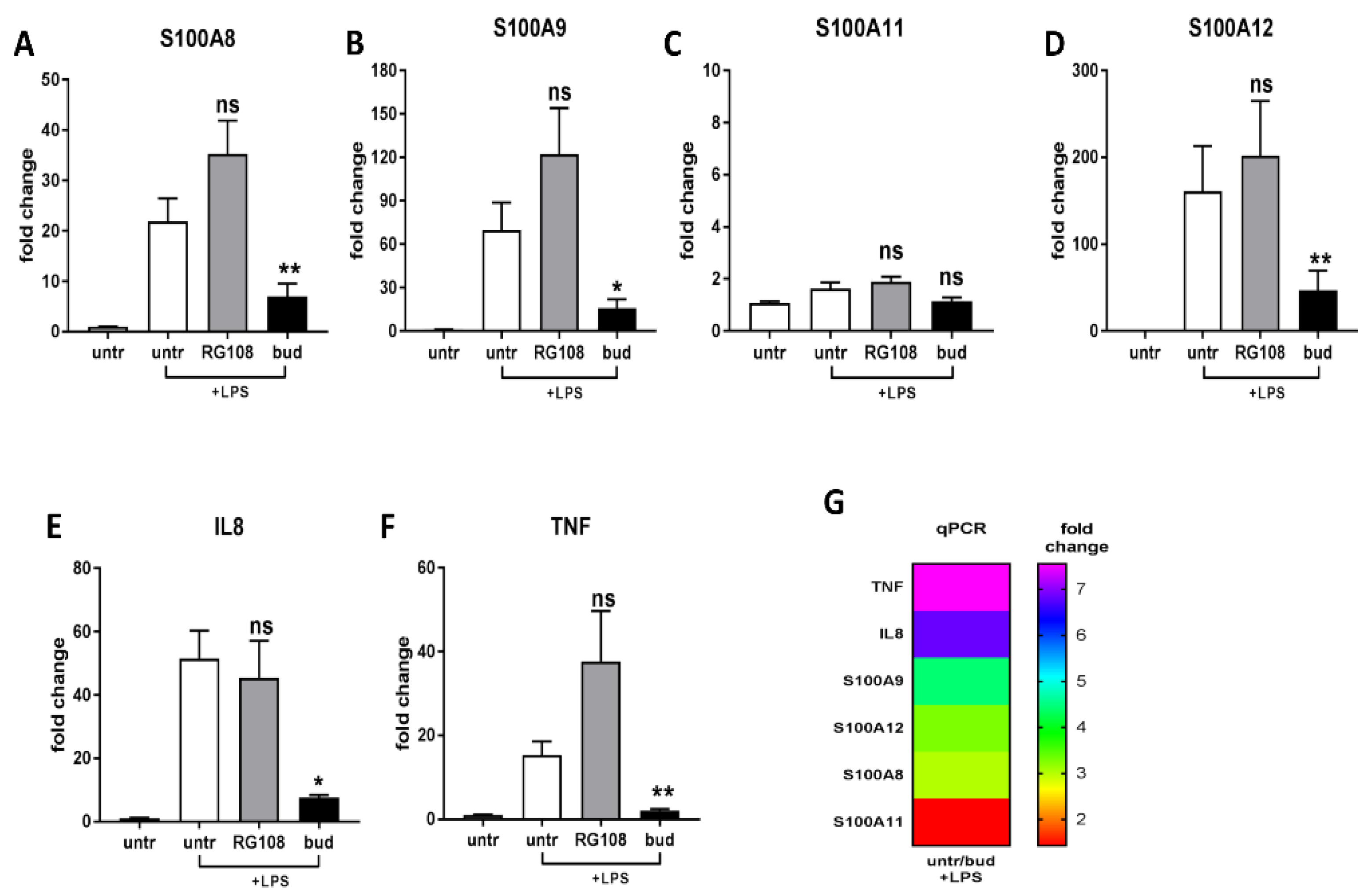
2.3. Different Expressions of Pro-Inflammatory and Anti-Inflammatory Genes
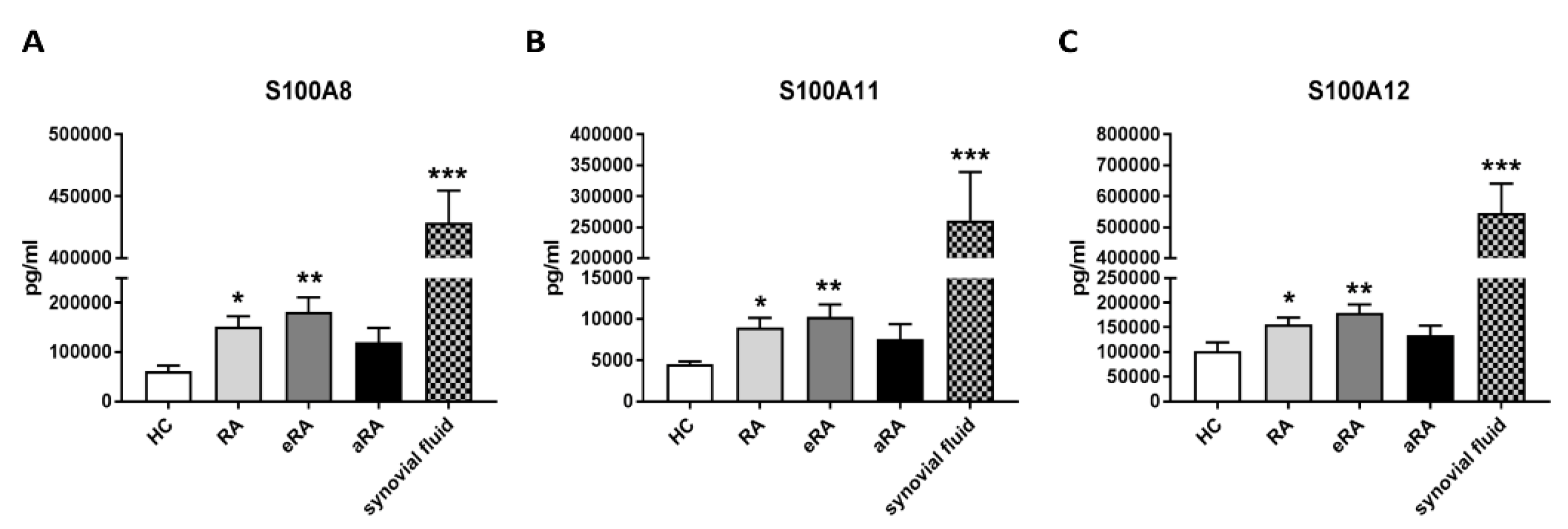
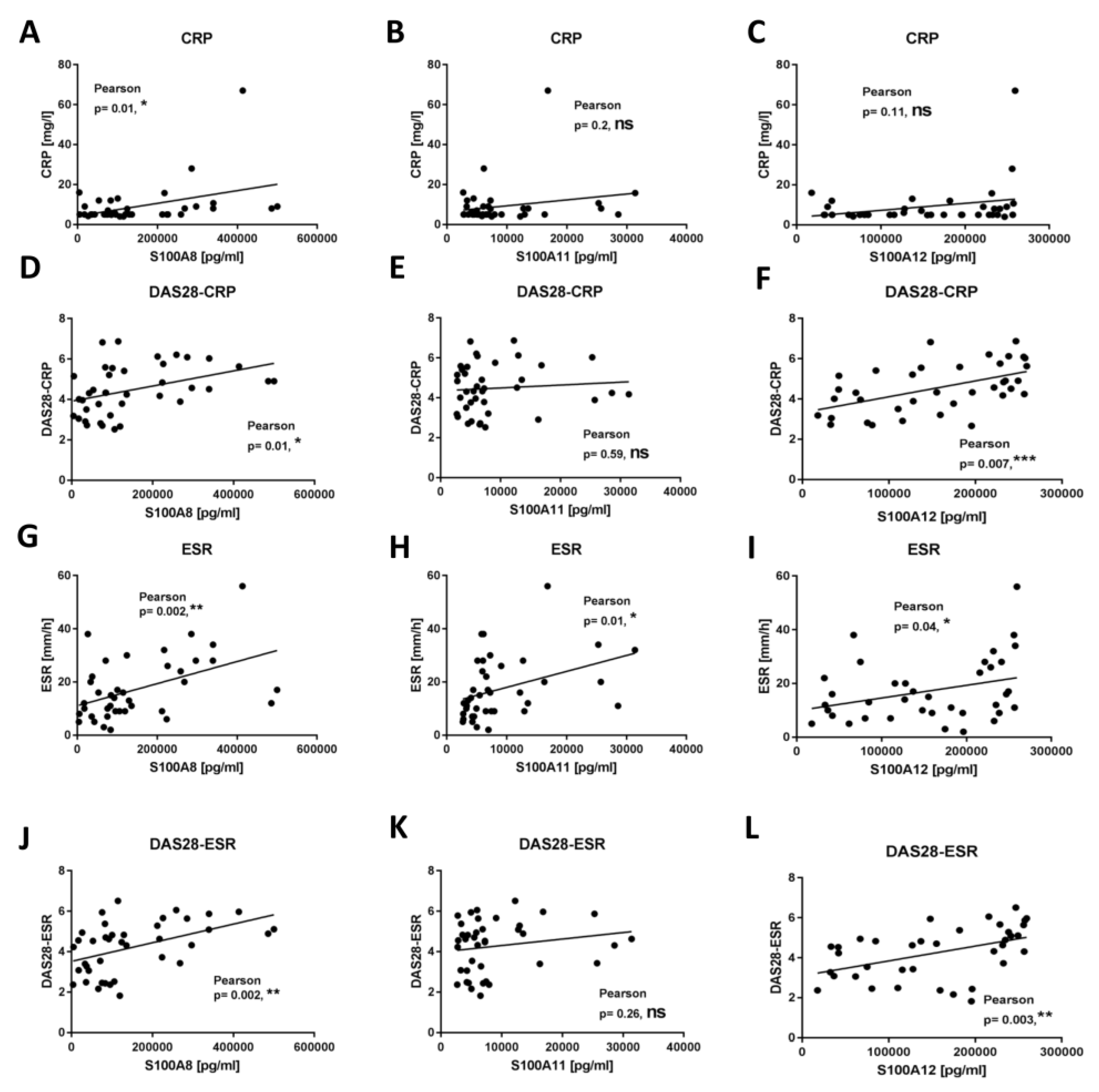
2.4. Secretion of S100 Family Proteins in Sera and Synovial Fluids
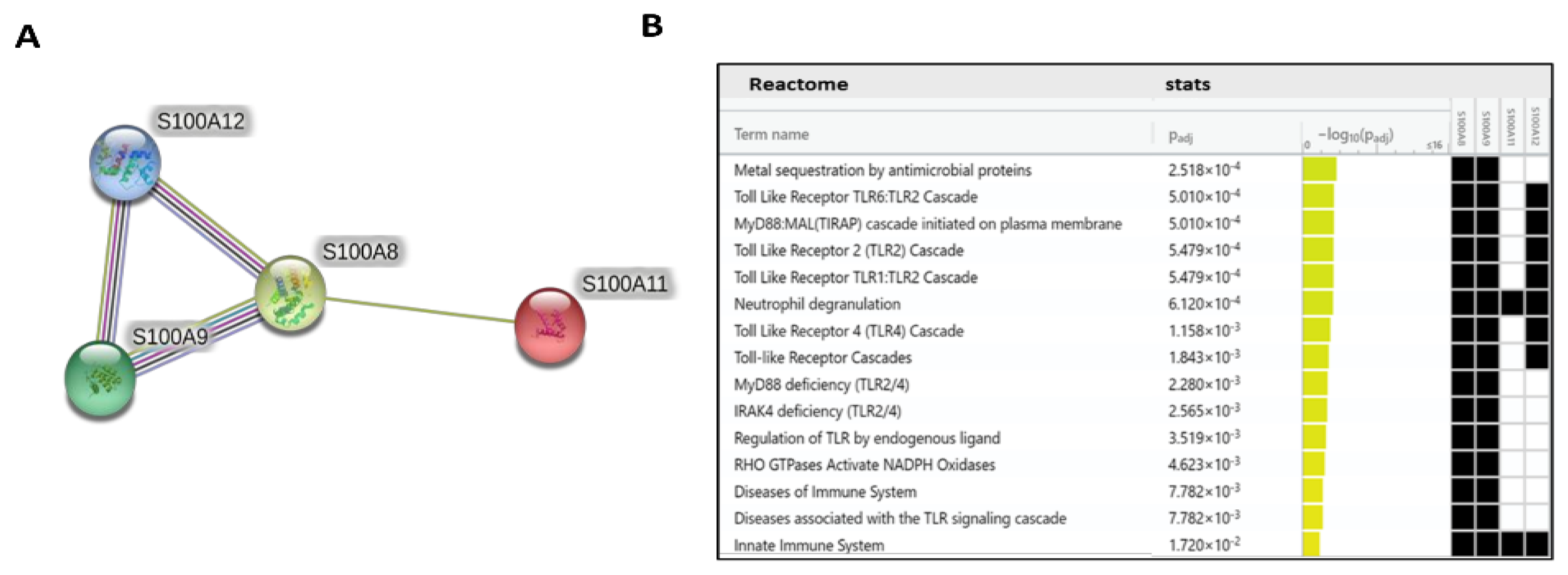
2.5. PPI Network Analysis and Biological Function Annotation
3. Discussion
4. Materials and Methods
4.1. Participants and Cell Purification
4.2. Transcriptomic Sequencing
4.3. THP-1 Cell Culture
4.4. Cell Viability
4.5. qPCR Analysis and Gene Expression
4.6. Enzyme-Linked Immunosorbent Assays (ELISAs)
4.7. Functional and Pathway Analysis
4.8. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Smolen, J.S.; Aletaha, D.; Barton, A.; Burmester, G.R.; Emery, P.; Firestein, G.S.; Kavanaugh, A.; McInnes, I.B.; Solomon, D.H.; Strand, V.; et al. Rheumatoid arthritis. Nat. Rev. Dis. Prim. 2018, 4, 18001. [Google Scholar] [CrossRef] [PubMed]
- Roszkowski, L.; Ciechomska, M. Tuning Monocytes and Macrophages for Personalized Therapy and Diagnostic Challenge in Rheumatoid Arthritis. Cells 2021, 10, 1860. [Google Scholar] [CrossRef] [PubMed]
- Kinne, R.W.; Stuhlmuller, B.; Burmester, G.R. Cells of the synovium in rheumatoid arthritis. Macrophages. Arthritis Res. 2007, 9, 224. [Google Scholar] [CrossRef] [PubMed]
- Kotsovilis, S.; Andreakos, E. Therapeutic human monoclonal antibodies in inflammatory diseases. Methods Mol. Biol. 2014, 1060, 37–59. [Google Scholar] [CrossRef]
- Gonzalez, L.L.; Garrie, K.; Turner, M.D. Role of S100 proteins in health and disease. Biochim. Biophys. Acta Mol. Cell Res. 2020, 1867, 118677. [Google Scholar] [CrossRef]
- Bird, A. Perceptions of epigenetics. Nature 2007, 447, 396–398. [Google Scholar] [CrossRef]
- Perkins, D.J.; Patel, M.C.; Blanco, J.C.; Vogel, S.N. Epigenetic Mechanisms Governing Innate Inflammatory Responses. J. Interferon Cytokine Res. 2016, 36, 454–461. [Google Scholar] [CrossRef]
- Kristensen, L.S.; Wojdacz, T.K.; Thestrup, B.B.; Wiuf, C.; Hager, H.; Hansen, L.L. Quality assessment of DNA derived from up to 30 years old formalin fixed paraffin embedded (FFPE) tissue for PCR-based methylation analysis using SMART-MSP and MS-HRM. BMC Cancer 2009, 9, 453. [Google Scholar] [CrossRef]
- Graca, I.; Sousa, E.J.; Baptista, T.; Almeida, M.; Ramalho-Carvalho, J.; Palmeira, C.; Henrique, R.; Jeronimo, C. Anti-tumoral effect of the non-nucleoside DNMT inhibitor RG108 in human prostate cancer cells. Curr. Pharm. Des. 2014, 20, 1803–1811. [Google Scholar] [CrossRef]
- Borges, S.; Doppler, H.R.; Storz, P. A combination treatment with DNA methyltransferase inhibitors and suramin decreases invasiveness of breast cancer cells. Breast Cancer Res. Treat. 2014, 144, 79–91. [Google Scholar] [CrossRef]
- Suzuki, T.; Tanaka, R.; Hamada, S.; Nakagawa, H.; Miyata, N. Design, synthesis, inhibitory activity, and binding mode study of novel DNA methyltransferase 1 inhibitors. Bioorg. Med. Chem. Lett. 2010, 20, 1124–1127. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.H.; Liu, Y.H.; Liu, J.D.; Xu, D.D.; Li, X.F.; Meng, X.M.; Ma, T.T.; Huang, C.; Li, J. MeCP2 Regulates PTCH1 Expression Through DNA Methylation in Rheumatoid Arthritis. Inflammation 2017, 40, 1497–1508. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Wang, Q.; Zhang, C.; Huang, C. Genome-wide DNA methylation analysis of articular chondrocytes identifies TRAF1, CTGF, and CX3CL1 genes as hypomethylated in osteoarthritis. Clin. Rheumatol. 2017, 36, 2335–2342. [Google Scholar] [CrossRef] [PubMed]
- Xue, Z.T.; Sjogren, H.O.; Salford, L.G.; Widegren, B. An epigenetic mechanism for high, synergistic expression of indoleamine 2,3-dioxygenase 1 (IDO1) by combined treatment with zebularine and IFN-gamma: Potential therapeutic use in autoimmune diseases. Mol. Immunol. 2012, 51, 101–111. [Google Scholar] [CrossRef] [PubMed]
- Pereira, M.A.; Tao, L.; Liu, Y.; Li, L.; Steele, V.E.; Lubet, R.A. Modulation by budesonide of DNA methylation and mRNA expression in mouse lung tumors. Int. J. Cancer 2007, 120, 1150–1153. [Google Scholar] [CrossRef] [PubMed]
- Alyaqoub, F.S.; Tao, L.; Kramer, P.M.; Steele, V.E.; Lubet, R.A.; Gunning, W.T.; Pereira, M.A. Prevention of mouse lung tumors and modulation of DNA methylation by combined treatment with budesonide and R115777 (Zarnestra MT). Carcinogenesis 2007, 28, 124–129. [Google Scholar] [CrossRef] [PubMed]
- Ciechomska, M.; Roszkowski, L.; Maslinski, W. DNA Methylation as a Future Therapeutic and Diagnostic Target in Rheumatoid Arthritis. Cells 2019, 8, 953. [Google Scholar] [CrossRef]
- Brueckner, B.; Garcia Boy, R.; Siedlecki, P.; Musch, T.; Kliem, H.C.; Zielenkiewicz, P.; Suhai, S.; Wiessler, M.; Lyko, F. Epigenetic reactivation of tumor suppressor genes by a novel small-molecule inhibitor of human DNA methyltransferases. Cancer Res. 2005, 65, 6305–6311. [Google Scholar] [CrossRef]
- Xu, W.; Li, Z.; Yu, B.; He, X.; Shi, J.; Zhou, R.; Liu, D.; Wu, Z. Effects of DNMT1 and HDAC inhibitors on gene-specific methylation reprogramming during porcine somatic cell nuclear transfer. PLoS ONE 2013, 8, e64705. [Google Scholar] [CrossRef]
- Stenzig, J.; Hirt, M.N.; Loser, A.; Bartholdt, L.M.; Hensel, J.T.; Werner, T.R.; Riemenschneider, M.; Indenbirken, D.; Guenther, T.; Muller, C.; et al. DNA methylation in an engineered heart tissue model of cardiac hypertrophy: Common signatures and effects of DNA methylation inhibitors. Basic Res. Cardiol. 2016, 111, 9. [Google Scholar] [CrossRef]
- Assis, R.I.F.; Wiench, M.; Silverio, K.G.; Da Silva, R.A.; Feltran, G.D.S.; Sallum, E.A.; Casati, M.Z.; Nociti, F.H., Jr.; Andia, D.C. RG108 increases NANOG and OCT4 in bone marrow-derived mesenchymal cells through global changes in DNA modifications and epigenetic activation. PLoS ONE 2018, 13, e0207873. [Google Scholar] [CrossRef] [PubMed]
- Oh, Y.S.; Jeong, S.G.; Cho, G.W. Anti-senescence effects of DNA methyltransferase inhibitor RG108 in human bone marrow mesenchymal stromal cells. Biotechnol. Appl. Biochem. 2015, 62, 583–590. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Zhai, Y.; Man, X.; Zhang, S.; An, X. Inhibition of DNA Methyltransferase by RG108 Promotes Pluripotency-Related Character of Porcine Bone Marrow Mesenchymal Stem Cells. Cell. Reprogram. 2020, 22, 82–89. [Google Scholar] [CrossRef] [PubMed]
- Ramakrishnan, S.; Nicolau, D.V., Jr.; Langford, B.; Mahdi, M.; Jeffers, H.; Mwasuku, C.; Krassowska, K.; Fox, R.; Binnian, I.; Glover, V.; et al. Inhaled budesonide in the treatment of early COVID-19 (STOIC): A phase 2, open-label, randomised controlled trial. Lancet Respir. Med. 2021, 9, 763–772. [Google Scholar] [CrossRef] [PubMed]
- Ethgen, O.; De Lemos Esteves, F.; Bruyere, O.; Reginster, J.Y. What do we know about the safety of corticosteroids in rheumatoid arthritis? Curr. Med. Res. Opin. 2013, 29, 1147–1160. [Google Scholar] [CrossRef]
- Orta, M.L.; Dominguez, I.; Pastor, N.; Cortes, F.; Mateos, S. The role of the DNA hypermethylating agent Budesonide in the decatenating activity of DNA topoisomerase II. Mutat. Res. 2010, 694, 45–52. [Google Scholar] [CrossRef]
- Adjakly, M.; Bosviel, R.; Rabiau, N.; Boiteux, J.P.; Bignon, Y.J.; Guy, L.; Bernard-Gallon, D. DNA methylation and soy phytoestrogens: Quantitative study in DU-145 and PC-3 human prostate cancer cell lines. Epigenomics 2011, 3, 795–803. [Google Scholar] [CrossRef]
- Bosviel, R.; Dumollard, E.; Dechelotte, P.; Bignon, Y.J.; Bernard-Gallon, D. Can soy phytoestrogens decrease DNA methylation in BRCA1 and BRCA2 oncosuppressor genes in breast cancer? OMICS 2012, 16, 235–244. [Google Scholar] [CrossRef]
- Karsli-Ceppioglu, S.; Ngollo, M.; Adjakly, M.; Dagdemir, A.; Judes, G.; Lebert, A.; Boiteux, J.P.; Penault, L.F.; Bignon, Y.J.; Guy, L.; et al. Genome-wide DNA methylation modified by soy phytoestrogens: Role for epigenetic therapeutics in prostate cancer? OMICS 2015, 19, 209–219. [Google Scholar] [CrossRef]
- Leclerc, E.; Fritz, G.; Vetter, S.W.; Heizmann, C.W. Binding of S100 proteins to RAGE: An update. Biochim. Biophys. Acta 2009, 1793, 993–1007. [Google Scholar] [CrossRef]
- Vogl, T.; Tenbrock, K.; Ludwig, S.; Leukert, N.; Ehrhardt, C.; Van Zoelen, M.A.; Nacken, W.; Foell, D.; Van der Poll, T.; Sorg, C.; et al. Mrp8 and Mrp14 are endogenous activators of Toll-like receptor 4, promoting lethal, endotoxin-induced shock. Nat. Med. 2007, 13, 1042–1049. [Google Scholar] [CrossRef] [PubMed]
- Sunahori, K.; Yamamura, M.; Yamana, J.; Takasugi, K.; Kawashima, M.; Yamamoto, H.; Chazin, W.J.; Nakatani, Y.; Yui, S.; Makino, H. The S100A8/A9 heterodimer amplifies proinflammatory cytokine production by macrophages via activation of nuclear factor kappa B and p38 mitogen-activated protein kinase in rheumatoid arthritis. Arthritis Res. Ther. 2006, 8, R69. [Google Scholar] [CrossRef] [PubMed]
- Bae, S.C.; Lee, Y.H. Calprotectin levels in rheumatoid arthritis and their correlation with disease activity: A meta-analysis. Postgrad. Med. 2017, 129, 531–537. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.Y.; Li, X.F.; Wu, S.; Niu, X.N.; Yin, S.Q.; Huang, C.; Li, J. Role of the S100 protein family in rheumatoid arthritis. Arthritis Res. Ther. 2022, 24, 35. [Google Scholar] [CrossRef] [PubMed]
- Ramirez, J.; Narvaez, J.A.; Ruiz-Esquide, V.; Hernandez-Ganan, J.; Cuervo, A.; Inciarte-Mundo, J.; Hernandez, M.V.; Sampayo-Cordero, M.; Pablos, J.L.; Sanmarti, R.; et al. Clinical and sonographic biomarkers of structural damage progression in RA patients in clinical remission: A prospective study with 12 months follow-up. Semin. Arthritis Rheum. 2017, 47, 303–309. [Google Scholar] [CrossRef]
- Garcia-Arias, M.; Pascual-Salcedo, D.; Ramiro, S.; Ueberschlag, M.E.; Jermann, T.M.; Cara, C.; Martin-Mola, E.; Balsa, A. Calprotectin in rheumatoid arthritis: Association with disease activity in a cross-sectional and a longitudinal cohort. Mol. Diagn. Ther. 2013, 17, 49–56. [Google Scholar] [CrossRef] [PubMed]
- Hurnakova, J.; Hulejova, H.; Zavada, J.; Hanova, P.; Komarc, M.; Mann, H.; Klein, M.; Sleglova, O.; Olejarova, M.; Forejtova, S.; et al. Relationship between serum calprotectin (S100A8/9) and clinical, laboratory and ultrasound parameters of disease activity in rheumatoid arthritis: A large cohort study. PLoS ONE 2017, 12, e0183420. [Google Scholar] [CrossRef] [PubMed]
- Van Lent, P.L.; Grevers, L.; Blom, A.B.; Sloetjes, A.; Mort, J.S.; Vogl, T.; Nacken, W.; Van den Berg, W.B.; Roth, J. Myeloid-related proteins S100A8/S100A9 regulate joint inflammation and cartilage destruction during antigen-induced arthritis. Ann. Rheum. Dis. 2008, 67, 1750–1758. [Google Scholar] [CrossRef] [PubMed]
- Austermann, J.; Spiekermann, C.; Roth, J. S100 proteins in rheumatic diseases. Nat. Rev. Rheumatol. 2018, 14, 528–541. [Google Scholar] [CrossRef]
- Ghoreschi, K.; Laurence, A.; O’Shea, J.J. Janus kinases in immune cell signaling. Immunol. Rev. 2009, 228, 273–287. [Google Scholar] [CrossRef]
- Leng, R.X.; Di, D.S.; Ni, J.; Wu, X.X.; Zhang, L.L.; Wang, X.F.; Liu, R.S.; Huang, Q.; Fan, Y.G.; Pan, H.F.; et al. Identification of new susceptibility loci associated with rheumatoid arthritis. Ann. Rheum. Dis. 2020, 79, 1565–1571. [Google Scholar] [CrossRef] [PubMed]
- Ramirez-Perez, S.; Oregon-Romero, E.; Reyes-Perez, I.V.; Bhattaram, P. Targeting MyD88 Downregulates Inflammatory Mediators and Pathogenic Processes in PBMC From DMARDs-Naive Rheumatoid Arthritis Patients. Front. Pharmacol. 2021, 12, 800220. [Google Scholar] [CrossRef] [PubMed]
- Sugiura, Y.; Niimi, T.; Sato, S.; Yoshinouchi, T.; Banno, S.; Naniwa, T.; Maeda, H.; Shimizu, S.; Ueda, R. Transforming growth factor beta1 gene polymorphism in rheumatoid arthritis. Ann. Rheum. Dis. 2002, 61, 826–828. [Google Scholar] [CrossRef]
- Moore, K.W.; De Waal Malefyt, R.; Coffman, R.L.; O’Garra, A. Interleukin-10 and the interleukin-10 receptor. Annu. Rev. Immunol. 2001, 19, 683–765. [Google Scholar] [CrossRef] [PubMed]
- Anderson, C.F.; Chakroun, R.W.; Su, H.; Mitrut, R.E.; Cui, H. Interface-Enrichment-Induced Instability and Drug-Loading-Enhanced Stability in Inhalable Delivery of Supramolecular Filaments. ACS Nano 2019, 13, 12957–12968. [Google Scholar] [CrossRef] [PubMed]
- Kirwan, J.R.; Hallgren, R.; Mielants, H.; Wollheim, F.; Bjorck, E.; Persson, T.; Book, C.; Bowman, S.; Byron, M.; Cox, N.; et al. A randomised placebo controlled 12 week trial of budesonide and prednisolone in rheumatoid arthritis. Ann. Rheum. Dis. 2004, 63, 688–695. [Google Scholar] [CrossRef]
- Ansari, M.M.; Ahmad, A.; Kumar, A.; Alam, P.; Khan, T.H.; Jayamurugan, G.; Raza, S.S.; Khan, R. Aminocellulose-grafted-polycaprolactone coated gelatin nanoparticles alleviate inflammation in rheumatoid arthritis: A combinational therapeutic approach. Carbohydr. Polym. 2021, 258, 117600. [Google Scholar] [CrossRef]
- Ali, H.; Weigmann, B.; Collnot, E.M.; Khan, S.A.; Windbergs, M.; Lehr, C.M. Budesonide Loaded PLGA Nanoparticles for Targeting the Inflamed Intestinal Mucosa--Pharmaceutical Characterization and Fluorescence Imaging. Pharm. Res. 2016, 33, 1085–1092. [Google Scholar] [CrossRef]
- Siedlecki, P.; Garcia Boy, R.; Musch, T.; Brueckner, B.; Suhai, S.; Lyko, F.; Zielenkiewicz, P. Discovery of two novel, small-molecule inhibitors of DNA methylation. J. Med. Chem. 2006, 49, 678–683. [Google Scholar] [CrossRef]
- Zheng, Z.; Zeng, S.; Liu, C.; Li, W.; Zhao, L.; Cai, C.; Nie, G.; He, Y. The DNA methylation inhibitor RG108 protects against noise-induced hearing loss. Cell. Biol. Toxicol. 2021, 37, 751–771. [Google Scholar] [CrossRef]
- Singh, A.K.; Halder-Sinha, S.; Clement, J.P.; Kundu, T.K. Epigenetic modulation by small molecule compounds for neurodegenerative disorders. Pharmacol. Res. 2018, 132, 135–148. [Google Scholar] [CrossRef] [PubMed]
- Matsunaga, Y.; Hashimoto, Y.; Ishiko, A. Stratum corneum levels of calprotectin proteins S100A8/A9 correlate with disease activity in psoriasis patients. J. Dermatol. 2021, 48, 1518–1525. [Google Scholar] [CrossRef] [PubMed]
- Angeles-Han, S.T.; Utz, V.M.; Thornton, S.; Schulert, G.; Rodriguez-Smith, J.; Kauffman, A.; Sproles, A.; Mwase, N.; Hennard, T.; Grom, A.; et al. S100 proteins, cytokines, and chemokines as tear biomarkers in children with juvenile idiopathic arthritis-associated uveitis. Ocul. Immunol. Inflamm. 2021, 29, 1616–1620. [Google Scholar] [CrossRef] [PubMed]
- Edwards, T.S.; Dickerhof, N.; Magon, N.J.; Paton, L.N.; Sly, P.D.; Kettle, A.J. Formation of Calprotectin-Derived Peptides in the Airways of Children with Cystic Fibrosis. J. Immunol. 2022, 208, 979–990. [Google Scholar] [CrossRef]
- Andres Cerezo, L.; Sumova, B.; Prajzlerova, K.; Veigl, D.; Damgaard, D.; Nielsen, C.H.; Pavelka, K.; Vencovsky, J.; Senolt, L. Calgizzarin (S100A11): A novel inflammatory mediator associated with disease activity of rheumatoid arthritis. Arthritis Res. Ther. 2017, 19, 79. [Google Scholar] [CrossRef]
- Cecil, D.L.; Terkeltaub, R. Transamidation by transglutaminase 2 transforms S100A11 calgranulin into a procatabolic cytokine for chondrocytes. J. Immunol. 2008, 180, 8378–8385. [Google Scholar] [CrossRef]
- Baillet, A.; Trocme, C.; Berthier, S.; Arlotto, M.; Grange, L.; Chenau, J.; Quetant, S.; Seve, M.; Berger, F.; Juvin, R.; et al. Synovial fluid proteomic fingerprint: S100A8, S100A9 and S100A12 proteins discriminate rheumatoid arthritis from other inflammatory joint diseases. Rheumatology 2010, 49, 671–682. [Google Scholar] [CrossRef]
- Foell, D.; Kane, D.; Bresnihan, B.; Vogl, T.; Nacken, W.; Sorg, C.; Fitzgerald, O.; Roth, J. Expression of the pro-inflammatory protein S100A12 (EN-RAGE) in rheumatoid and psoriatic arthritis. Rheumatology 2003, 42, 1383–1389. [Google Scholar] [CrossRef]
- Wang, Z.W.; Zhang, L.J.; Zhuang, Y.; Lv, Z.F.; Tan, Z.M. CKS2 and S100A12: Two Novel Diagnostic Biomarkers for Rheumatoid Arthritis. Dis. Markers 2022, 2022, 2431976. [Google Scholar] [CrossRef]
- Nguyen, M.V.C.; Baillet, A.; Romand, X.; Trocme, C.; Courtier, A.; Marotte, H.; Thomas, T.; Soubrier, M.; Miossec, P.; Tebib, J.; et al. Prealbumin, platelet factor 4 and S100A12 combination at baseline predicts good response to TNF alpha inhibitors in rheumatoid arthritis. Jt. Bone Spine 2019, 86, 195–201. [Google Scholar] [CrossRef]
- Aletaha, D.; Neogi, T.; Silman, A.J.; Funovits, J.; Felson, D.T.; Bingham, C.O., III; Birnbaum, N.S.; Burmester, G.R.; Bykerk, V.P.; Cohen, M.D.; et al. 2010 Rheumatoid arthritis classification criteria: An American College of Rheumatology/European League Against Rheumatism collaborative initiative. Arthritis Rheum. 2010, 62, 2569–2581. [Google Scholar] [CrossRef] [PubMed]
- Ciechomska, M.; Huigens, C.A.; Hugle, T.; Stanly, T.; Gessner, A.; Griffiths, B.; Radstake, T.R.; Hambleton, S.; O’Reilly, S.; Van Laar, J.M. Toll-like receptor-mediated, enhanced production of profibrotic TIMP-1 in monocytes from patients with systemic sclerosis: Role of serum factors. Ann. Rheum. Dis. 2013, 72, 1382–1389. [Google Scholar] [CrossRef] [PubMed]
- Ciechomska, M.; Wojtas, B.; Bonek, K.; Roszkowski, L.; Gluszko, P.; Benes, V.; Maslinski, W. Comprehensive microRNA and transcriptomic profiling of rheumatoid arthritis monocytes: Role of microRNA-146b in pro-inflammatory progression. Rheumatology 2021, 60, 5424–5435. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Franceschini, A.; Kuhn, M.; Simonovic, M.; Roth, A.; Minguez, P.; Doerks, T.; Stark, M.; Muller, J.; Bork, P.; et al. The STRING database in 2011: Functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011, 39, D561–D568. [Google Scholar] [CrossRef] [PubMed]

| Parameters of RA Patients and HCs | Early RA (n = 20) | Advanced RA (n = 23) | All RA (n = 43) | HCs (n = 19) |
|---|---|---|---|---|
| Age, years, median (range) | 45 (18–80) | 62 (25–82) | 58 (18–82) | 42 (24–67) |
| Sex F/M | 16/4 | 20/3 | 36/7 | 17/2 |
| Disease duration, years, median (range) | 0 | 9 (2–22) | 2 (0–22) | N/A |
| Anti-CCP Abs, % (n) | 75% (n = 15) | 69% (n = 16) | 72% (n = 31) | N/A |
| RF, % (n) | 60% (n = 12) | 60.9% (n = 14) | 60.5% (n = 26) | N/A |
| CRP, mg/L, median (range) | 6 (4–67) | 5 (5–125) | 5.5 (4–125) | N/A |
| ESR, mm/h, median (range) | 15.5 (3–56) | 17 (2–56) | 16 (2–56) | N/A |
| DAS28-ESR, median (range) | 4.66 (2.16–6.51) | 4.32 (1.82–6.25) | 4.56 (1.82–6.51) | N/A |
| DAS28-CRP, median (range) | 4.675 (2.52–6.87) | 4.33 (2.66–6.44) | 4.51 (2.52–6.87) | N/A |
| Treatment | ||||
| Methotrexate, % (n) | 45% (n = 9) | 65.2% (n = 15) | 55.8% (n = 24) | N/A |
| Leflunomide, % (n) | 0% (n = 0) | 4.3% (n = 1) | 2.3% (n = 1) | N/A |
| Sulfasalazine, % (n) | 20% (n = 4) | 13% (n = 3) | 16.3% (n = 7) | N/A |
| Hydroxychloroquine, % (n) | 10% (n = 2) | 13% (n = 3) | 11.6% (n = 5) | N/A |
| Methylprednisolone, % (n) | 15% (n = 3) | 43.5% (n = 10) | 30.2% (n = 13) | N/A |
| Without treatment, % (n) | 40% (n = 8) | 0% (n = 0) | 18.6% (n = 8) | N/A |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Roszkowski, L.; Jaszczyk, B.; Plebańczyk, M.; Ciechomska, M. S100A8 and S100A12 Proteins as Biomarkers of High Disease Activity in Patients with Rheumatoid Arthritis That Can Be Regulated by Epigenetic Drugs. Int. J. Mol. Sci. 2023, 24, 710. https://doi.org/10.3390/ijms24010710
Roszkowski L, Jaszczyk B, Plebańczyk M, Ciechomska M. S100A8 and S100A12 Proteins as Biomarkers of High Disease Activity in Patients with Rheumatoid Arthritis That Can Be Regulated by Epigenetic Drugs. International Journal of Molecular Sciences. 2023; 24(1):710. https://doi.org/10.3390/ijms24010710
Chicago/Turabian StyleRoszkowski, Leszek, Bożena Jaszczyk, Magdalena Plebańczyk, and Marzena Ciechomska. 2023. "S100A8 and S100A12 Proteins as Biomarkers of High Disease Activity in Patients with Rheumatoid Arthritis That Can Be Regulated by Epigenetic Drugs" International Journal of Molecular Sciences 24, no. 1: 710. https://doi.org/10.3390/ijms24010710
APA StyleRoszkowski, L., Jaszczyk, B., Plebańczyk, M., & Ciechomska, M. (2023). S100A8 and S100A12 Proteins as Biomarkers of High Disease Activity in Patients with Rheumatoid Arthritis That Can Be Regulated by Epigenetic Drugs. International Journal of Molecular Sciences, 24(1), 710. https://doi.org/10.3390/ijms24010710






