Construction of a Hierarchical Gene Regulatory Network to Reveal the Drought Tolerance Mechanism of Shanxin Poplar
Abstract
:1. Introduction
2. Results
2.1. Dynamic Changes of Poplar Transcriptome in Response to Drought Stress
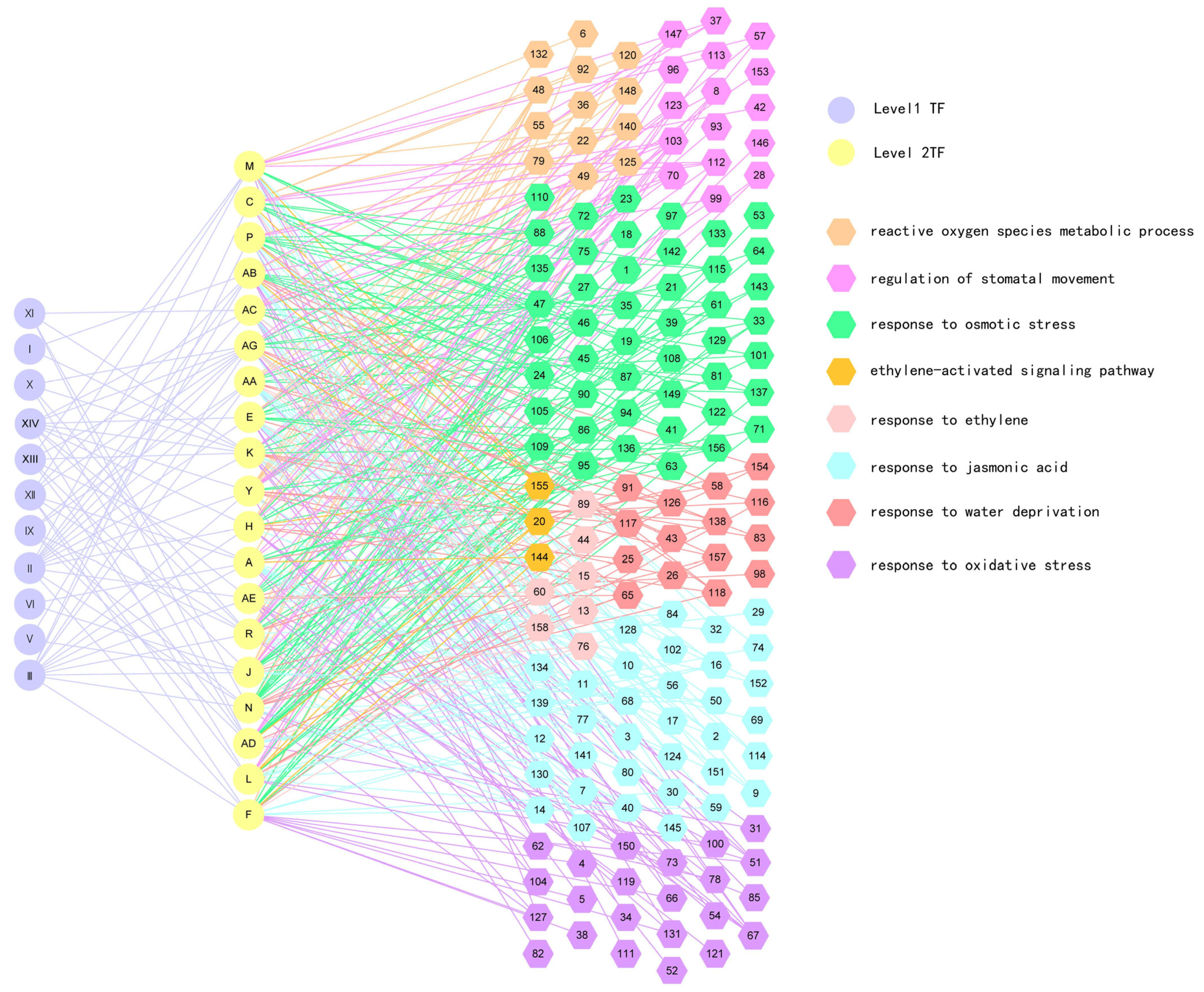
2.2. Construction of Gene Regulatory Network (GRN) in Response to Drought Stress
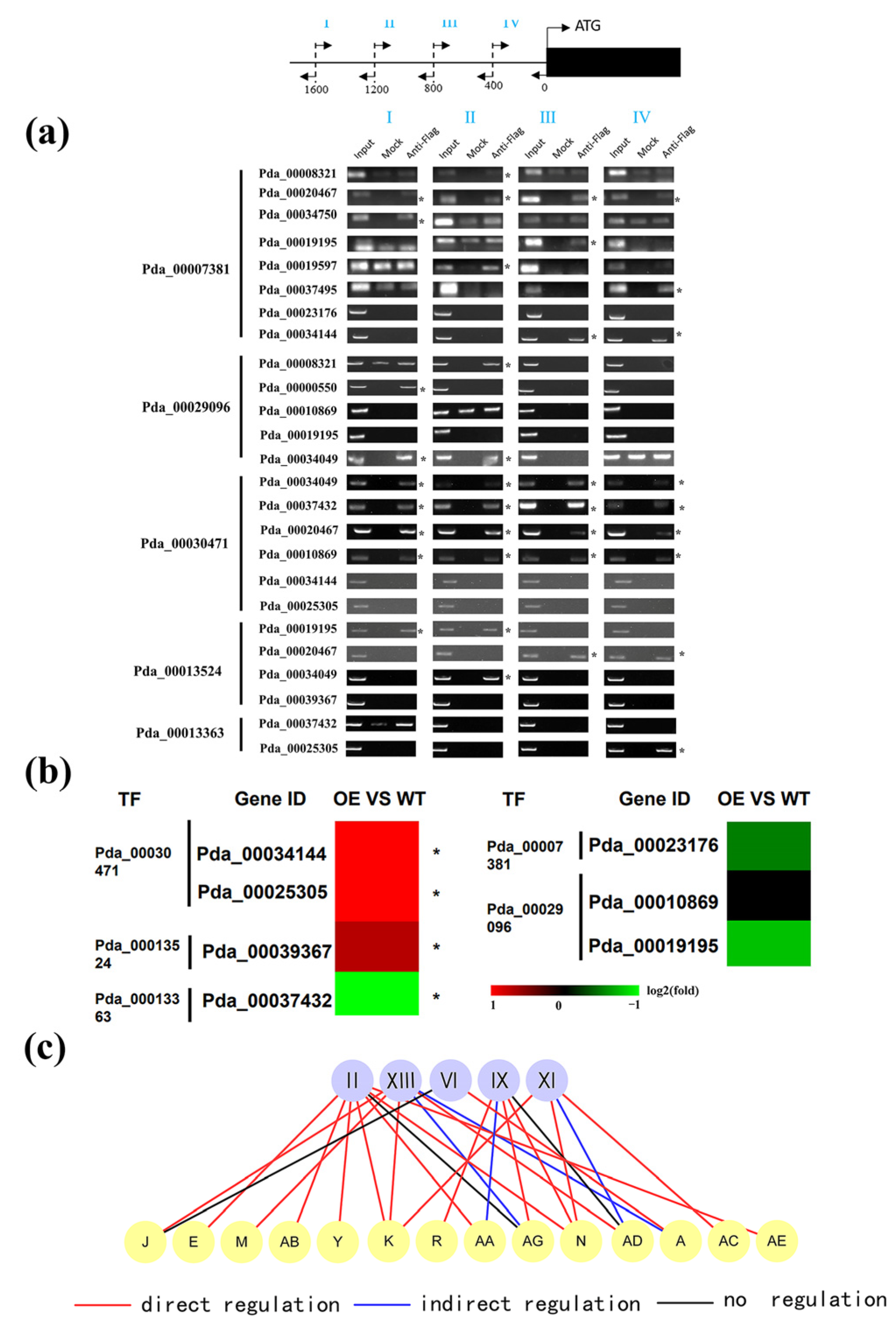
2.3. Validation of the Regulatory Relationship between First and Second Layers in GRN
2.4. Verification of the Regulatory Relationship between the Second and Third Layers
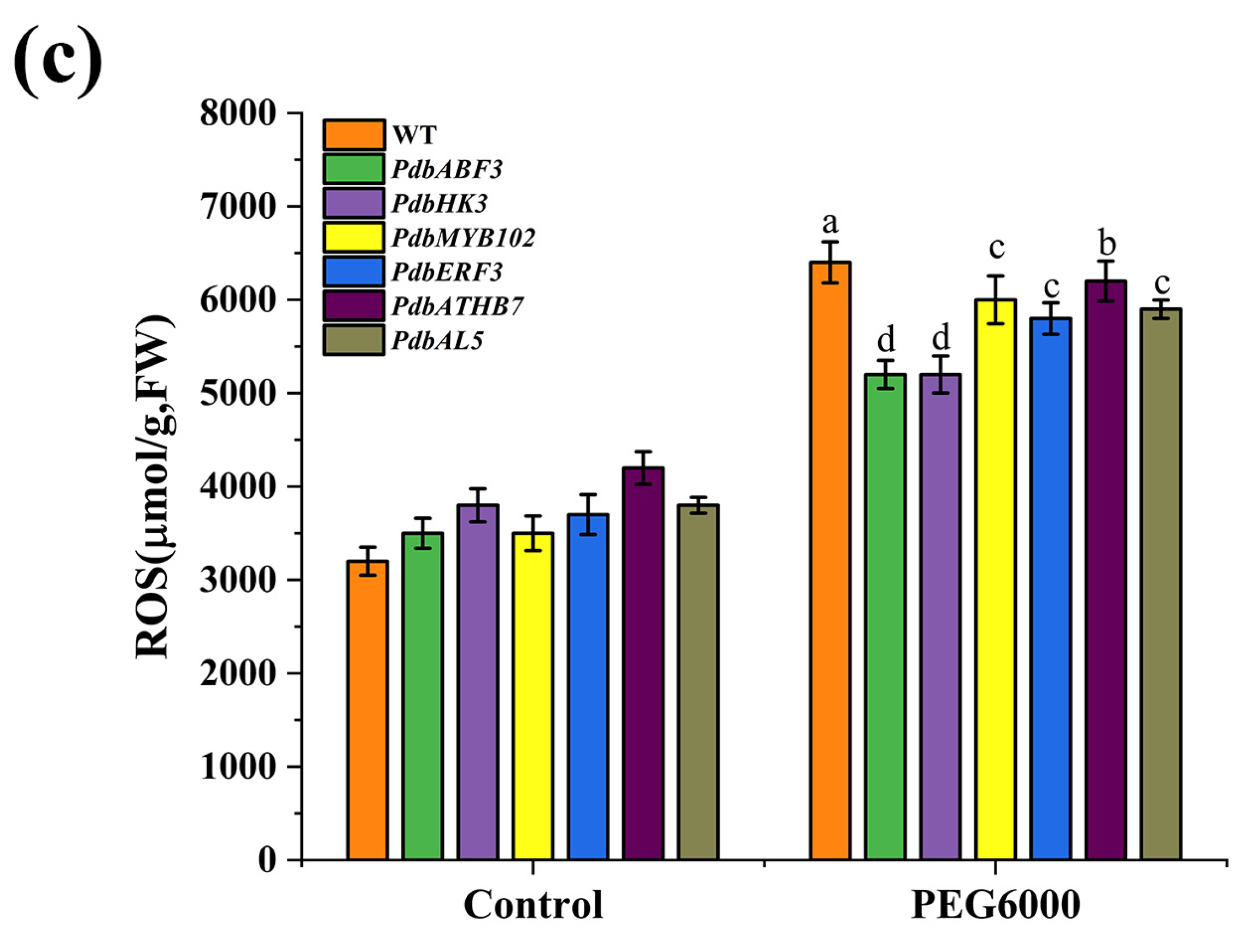
2.5. Determination of Drought Tolerance of the Regulator Genes in the First Layer of GRN
3. Discussion
3.1. PdbERF3 Is the Most Important Regulator in Response to Drought Stress
3.2. Hormone Signaling Plays an Important Role in Drought Stress Response
3.3. Stomatal Movements Play a Role in Drought Tolerance
3.4. Scavenging ROS Is Necessary for a Drought Response in Shanxin Poplar
4. Materials and Methods
4.1. Plant Materials and Drought Treatments
4.2. Library Preparation and Differential Expression Analysis
4.3. Identification of Biological Processes Involved in Drought Stress
4.4. Construction of Multilayered Hierarchical Gene Regulatory Network in Response to Drought Stress
4.5. Plant Expression Vector Construction and Transient Transformation
4.6. Validation of the Regulatory Relationship in GRN
4.7. Chromatin Immunoprecipitation (ChIP)
4.8. Real-Time RT-PCR
4.9. Physiological Change Analysis
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Huang, J.; Li, Y.; Fu, C.; Chen, F.; Fu, Q.; Dai, A.; Wang, G. Dryland climate change: Recent progress and challenges. Rev. Geophy. 2017, 55, 719–778. [Google Scholar] [CrossRef]
- Thirumalaikumar, V.P.; Devkar, V.; Mehterov, N.; Ali, S.; Ozgur, R.; Turkan, I.; Mueller-Roeber, B.; Balazadeh, S. NAC transcription factor JUNGBRUNNEN1 enhances drought tolerance in tomato. Plant Biotechnol. J. 2018, 16, 354–366. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Segal, E.; Shapira, M.; Regev, A.; Pe’er, D.; Botstein, D.; Koller, D.; Friedman, N. Module networks: Identifying regulatory modules and their condition-specific regulators from gene expression data. Nat. Genet. 2003, 34, 166–176. [Google Scholar] [CrossRef] [PubMed]
- Margolin, A.A.; Nemenman, I.; Basso, K.; Wiggins, C.; Stolovitzky, G.; Dalla Favera, R.; Califano, A. ARACNE: An algorithm for the reconstruction of gene regulatory networks in a mammalian cellular context. BMC Bioinform. 2006, 7, S7. [Google Scholar] [CrossRef] [Green Version]
- Kumari, S.; Deng, W.; Gunasekara, C.; Chiang, V.; Chen, H.S.; Ma, H.; Davis, X.; Wei, H. Bottom-up GGM algorithm for constructing multilayered hierarchical gene regulatory networks that govern biological pathways or processes. BMC Bioinform. 2016, 17, 132. [Google Scholar] [CrossRef] [Green Version]
- Deng, W.; Zhang, K.; Busov, V.; Wei, H. Recursive random forest algorithm for constructing multilayered hierarchical gene regulatory networks that govern biological pathways. PLoS ONE 2017, 12, e0171532. [Google Scholar] [CrossRef] [Green Version]
- Gunasekara, C.; Zhang, K.; Deng, W.; Brown, L.; Wei, H. TGMI: An efficient algorithm for identifying pathway regulators through evaluation of triple-gene mutual interaction. Nucleic Acids Res. 2018, 46, e67. [Google Scholar] [CrossRef]
- Huynh-Thu, V.A.; Geurts, P. dynGENIE3: Dynamical GENIE3 for the inference of gene networks from time series expression data. Sci. Rep. 2018, 8, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Moerman, T.; Santos, S.A.; González-Blas, C.B.; Simm, J.; Moreau, Y.; Aerts, J.; Aerts, S. GRNBoost2 and Arboreto: Efficient and scalable inference of gene regulatory networks. Bioinformatics 2019, 35, 2159–2161. [Google Scholar] [CrossRef]
- Liao, Z.; Dong, F.; Liu, J.; Xu, L.; Marshall-Colon, A.; Ming, R. Gene regulation network analyses of pistil development in papaya. BMC Genom. 2022, 23, 8. [Google Scholar] [CrossRef]
- Jia, Y.; Niu, Y.; Zhao, H.; Wang, Z.; Gao, C.; Wang, C.; Wang, Y. Hierarchical transcription factor and regulatory network for drought response in Betula platyphylla. Hortic. Res. 2022, 9, uhac040. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Wang, Y.; Yu, L.; Zheng, T.; Wang, S.; Yue, Z.; Jiang, J.; Kumari, S.; Zheng, C.; Tang, H.; et al. Genome sequence and evolution of Betula platyphylla. Hortic. Res. 2021, 8, 37. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Zhao, G.; Xu, J.R.; Kistler, H.C.; Gao, L.; Ma, L.J. Compartmentalized gene regulatory network of the pathogenic fungus Fusarium graminearum. New Phytol. 2016, 211, 527–541. [Google Scholar] [CrossRef] [PubMed]
- Vermeirssen, V.; De Clercq, I.; Van Parys, T.; Van Breusegem, F.; Van de Peer, Y. Arabidopsis ensemble Reverse-Engineered gene regulatory network discloses interconnected transcription factors in oxidative stress. Plant Cell. 2015, 26, 4656–4679. [Google Scholar] [CrossRef]
- Chen, H.; Wang, J.P.; Liu, H.; Li, H.; Lin, Y.-C.J.; Shi, R.; Yang, C.; Gao, J.; Zhou, C.; Li, Q.; et al. Hierarchical transcription factor and chromatin binding network for wood formation in Populus trichocarpa. Plant Cell 2019, 31, 602–626. [Google Scholar] [CrossRef] [Green Version]
- Shi, J.; Zhao, B.; Zheng, S.; Zhang, X.; Wang, X.; Dong, W.; Wang, E. A phosphate starvation response-centered network regulates mycorrhizal symbiosis. Cell 2021, 184, 5527–5540. [Google Scholar] [CrossRef]
- Mantiri, F.; Kurdyukov, S.; Lohar, D.P.; Sharopova, N.; Saeed, N.A.; Wang, X.D.; VandenBosch, K.A.; Rose, R.J. The transcription factor MtSERF1 of the ERF subfamily identified by transcriptional profiling is required for somatic embryogenesis induced by auxin plus cytokinin in Medicago truncatula. Plant Physiol. 2008, 146, 1622–1636. [Google Scholar] [CrossRef] [Green Version]
- Dossa, K.; Wei, X.; Li, D.; Fonceka, D.; Zhang, Y.; Wang, L.; Yu, J.; Boshou, L.; Diouf, D.; Cissé, N. Insight into the AP2/ERF transcription factor superfamily in sesame and expression profiling of DREB subfamily under drought stress. BMC Plant Biol. 2016, 16, 171. [Google Scholar] [CrossRef] [Green Version]
- Xing, H.; Jiang, Y.; Zou, Y.; Long, X.; Wu, X.; Ren, Y.; Li, Y. Genome-wide investigation of the AP2/ERF gene family in ginger: Evolution and expression profiling during development and abiotic stresses. BMC Plant Biol. 2021, 21, 561. [Google Scholar] [CrossRef]
- Büttner, M.; Singh, K.B. Arabidopsis thaliana ethylene-responsive element binding protein (AtEBP), an ethylene-inducible, GCC box DNA-binding protein interacts with an ocs element binding protein. Proc. Natl. Acad. Sci. USA 1997, 94, 5961–5966. [Google Scholar] [CrossRef]
- Xu, Z.S.; Xia, L.Q.; Chen, M.; Cheng, X.G.; Zhang, R.Y.; Li, L.C.; Zhao, Y.X.; Lu, Y.; Ni, Z.Y.; Liu, L. Isolation and molecular characterization of the Triticum aestivum L. ethylene-responsive factor 1 (TaERF1) that increases multiple stress tolerance. Plant Mol. Biol. 2007, 65, 719–732. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Xie, J.; Du, Q.; Song, F.; Xiao, L.; Quan, M.; Zhang, D. Transcription factors involved in the regulatory networks governing the Calvin-Benson-Bassham cycle. Tree Physiol. 2019, 39, 1159–1172. [Google Scholar] [CrossRef]
- Zhang, W.; Yang, G.; Mu, D.; Li, H.; Zang, D.; Xu, H.; Zou, X.; Wang, Y. An ethylene-responsive factor BpERF11 negatively modulates salt and osmotic tolerance in Betula platyphylla. Sci. Rep. 2016, 6, 23085. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raza, A.; Charagh, S.; Zahid, Z.; Mubarik, M.; Javed, R.; Siddiqui, M.H.; Hasanuzzaman, M. Jasmonic acid: A key frontier in conferring abiotic stress tolerance in plants. Plant Cell Rep. 2021, 40, 513–1541. [Google Scholar] [CrossRef] [PubMed]
- Kamal, A.H.M.; Komatsu, S. Jasmonic acid induced protein response to biophoton emissions and flooding stress in soybean. J. Proteom. 2016, 133, 33–47. [Google Scholar] [CrossRef] [PubMed]
- Mariotti, L.; Fambrini, M.; Pugliesi, C.; Scartazza, A. The gibberellin-deficient dwarf2 mutant of sunflower shows a high constitutive level of jasmonic and salicylic acids and an elevated energy dissipation capacity in well-watered and drought conditions. Environ. Exp. Bot. 2022, 194, 104697. [Google Scholar] [CrossRef]
- Bawa, G.; Chen, G.; Shi, J.; Ping, C.; Feng, L.; Pu, T.; Yang, H.; Chen, H.; Kai, S.; Hu, Y.; et al. Further insights into how low-light signaling delays leaf senescence in soybean under high-temperature. Environ. Exp. Bot. 2021, 188, 104516. [Google Scholar] [CrossRef]
- Dubois, M.; Van den Broeck, L.; Inzé, D. The pivotal role of ethylene in plant growth. Trends Plant Sci. 2018, 23, 311–323. [Google Scholar] [CrossRef] [Green Version]
- Tao, J.J.; Chen, H.W.; Ma, B.; Zhang, W.K.; Chen, S.Y.; Zhang, J.S. The role of ethylene in plants under salinity stress. Front. Plant Sci. 2015, 6, 1059. [Google Scholar] [CrossRef] [Green Version]
- Xu, B.; Long, Y.; Feng, X.; Zhu, X.; Sai, N.; Chirkova, L.; Betts, A.; Herrmann, J.; Edwards, E.J.; Okamoto, M.; et al. GABA signalling modulates stomatal opening to enhance plant water use efficiency and drought resilience. Nat. Commun. 2021, 12, 1952. [Google Scholar] [CrossRef]
- Benešová, M.; Holá, D.; Fischer, L.; Jedelský, P.L.; Hnilička, F.; Wilhelmová, N.; Rothová, O.; Kočová, M.; Procházková, D.; Honnerová, J.; et al. The physiology and proteomics of drought tolerance in maize: Early stomatal closure as a cause of lower tolerance to short-term dehydration? PLoS ONE 2012, 7, e38017. [Google Scholar] [CrossRef] [PubMed]
- Wojcik, K.A.; Kaminska, A.; Blasiak, J.; Szaflik, J.; Szaflik, J.P. Oxidative stress in the pathogenesis of keratoconus and fuchs endothelial corneal dystrophy. Int. J. Mol. Sci. 2013, 14, 19294–19308. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Finkel, T. Signal transduction by reactive oxygen species. J. Cell Biol. 2011, 194, 7–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wood, Z.A.; Poole, L.B.; Karplus, P.A. Peroxiredoxin evolution and the regulation of hydrogen peroxide signaling. Science 2003, 300, 650–653. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Q.; Cao, C.; Shen, T.; Zhong, L.; He, H.; Chen, X. Comprehensive metabolomic and proteomic analysis in biochemical metabolic pathways of rice spikes under drought and submergence stress. Biochim. Biophys. Acta Proteins Proteom. 2019, 1867, 237–247. [Google Scholar] [CrossRef]
- Zhou, Z.; Tang, J.; Cao, Q.; Li, Z.; Ma, D. Differential response of physiology and metabolic response to drought stress in different sweetpotato cultivars. PLoS ONE 2022, 17, e0264847. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wang, L.; Meng, H.; Wen, H.; Fan, Y.; Zhao, J. Maize ABP9 enhances tolerance to multiple stresses in transgenic Arabidopsis by modulating ABA signaling and cellular levels of reactive oxygen species. Plant Mol. Biol. 2011, 75, 365–378. [Google Scholar] [CrossRef] [Green Version]
- Lei, X.J.; Liu, Z.; Xie, Q.; Fang, J.; Wang, C.; Li, J.; Wang, C.; Gao, C. Construction of two regulatory networks related to salt stress and lignocellulosic synthesis under salt stress based on a Populus davidiana × P. bolleana transcriptome analysis. Plant Mol. Biol. 2022, 109, 1–14. [Google Scholar] [CrossRef]
- Liu, Z.Y.; Xie, Q.; Tang, F.; Wu, J.; Dong, W.; Wang, C.; Gao, C. The ThSOS3 gene improves the salt tolerance of transgenic Tamarix hispida and Arabidopsis thaliana. Front. Plant Sci. 2021, 11, 597480. [Google Scholar] [CrossRef]
- Liu, Z.Y.; Li, X.-P.; Zhang, T.-Q.; Wang, Y.-Y.; Wang, C.; Gao, C.-Q. Overexpression of ThMYB8 mediates salt stress tolerance by directly activating stress-responsive gene expression. Plant Sci. 2021, 302, 110668. [Google Scholar] [CrossRef]
- Behringer, D.; Zimmermann, H.; Ziegenhagen, B.; Liepelt, S. Differential gene expression reveals candidate genes for drought stress response in Abies alba (Pinaceae). PLoS ONE 2015, 10, e0124564. [Google Scholar] [CrossRef] [PubMed]
- Benny, J.; Pisciotta, A.; Caruso, T.; Martinelli, F. Identification of key genes and its chromosome regions linked to drought responses in leaves across different crops through meta-analysis of RNA-Seq data. BMC Plant Biol. 2019, 19, 194. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An ultra-fast all-in-one fastq preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [Green Version]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Florea, L.; Song, L.; Salzberg, S.L. Thousands of exons skipping events differentiate among splicing patterns in sixteen human tissues. F1000Research 2013, 2, 188. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boratyn, G.M.; Thierry-Mieg, J.; Thierry-Mieg, D.; Busby, B.; Madden, T.L. Magic-BLAST, an accurate RNA-seq aligner for long and short reads. BMC Bioinform. 2019, 20, 405. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Zhi, F.; Li, X.; Shen, W.; Yan, M.; He, J.; Bao, C.; Fan, T.; Zhou, S.; Ma, F.; et al. Zinc-finger protein MdBBX7/MdCOL9, a target of MdMIEL1 E3 ligase, confers drought tolerance in apple. Plant Physiol. 2021, 188, 540–559. [Google Scholar] [CrossRef]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Mukaka, M.M. A guide to appropriate use of correlation coefficient in medical research. Malawi Med. J. 2012, 24, 69–71. [Google Scholar]
- Zang, D.; Wang, L.; Zhang, Y.; Zhao, H.; Wang, Y. ThDof1.4 and ThZFP1 constitute a transcriptional regulatory cascade involved in salt or osmotic stress in Tamarix hispida. Plant Mol. Biol. 2017, 94, 495–507. [Google Scholar] [CrossRef]
- Zhao, H.; Li, H.; Jia, Y.; Wen, X.; Guo, H.; Xu, H.; Wang, Y. Building a robust chromatin immunoprecipitation method with substantially improved efficiency. Plant Physiol. 2020, 183, 1026–1034. [Google Scholar] [CrossRef] [PubMed]
- Dionisio-Sese, M.L.; Tobita, S. Antioxidant responses of rice seedlings to salinity stress. Plant Sci. 1998, 135, 1–9. [Google Scholar] [CrossRef]
- Li, D.D.; Yang, J.L.; Pak, S.; Zeng, M.Z.; Sun, J.L.; Yu, S.; He, Y.T.; Li, C.H. PuC3H35 confers drought tolerance by enhancing lignin and proanthocyanidin biosynthesis in the roots of Populus ussuriensis. New Phytol. 2022, 233, 390–408. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, P.; Wang, J.; Sun, X.; Yang, X.; Sun, S.; Han, X.; Li, D.; Wang, Y. Construction of a Hierarchical Gene Regulatory Network to Reveal the Drought Tolerance Mechanism of Shanxin Poplar. Int. J. Mol. Sci. 2023, 24, 384. https://doi.org/10.3390/ijms24010384
Wang P, Wang J, Sun X, Yang X, Sun S, Han X, Li D, Wang Y. Construction of a Hierarchical Gene Regulatory Network to Reveal the Drought Tolerance Mechanism of Shanxin Poplar. International Journal of Molecular Sciences. 2023; 24(1):384. https://doi.org/10.3390/ijms24010384
Chicago/Turabian StyleWang, Pengyu, Jingxin Wang, Xiaomeng Sun, Xue Yang, Shilin Sun, Xue Han, Dandan Li, and Yucheng Wang. 2023. "Construction of a Hierarchical Gene Regulatory Network to Reveal the Drought Tolerance Mechanism of Shanxin Poplar" International Journal of Molecular Sciences 24, no. 1: 384. https://doi.org/10.3390/ijms24010384
APA StyleWang, P., Wang, J., Sun, X., Yang, X., Sun, S., Han, X., Li, D., & Wang, Y. (2023). Construction of a Hierarchical Gene Regulatory Network to Reveal the Drought Tolerance Mechanism of Shanxin Poplar. International Journal of Molecular Sciences, 24(1), 384. https://doi.org/10.3390/ijms24010384





