Matched Analyses of Brain Metastases versus Primary Non-Small Cell Lung Cancer Reveal a Unique microRNA Signature
Abstract
1. Introduction
2. Results
2.1. Patient Characteristics
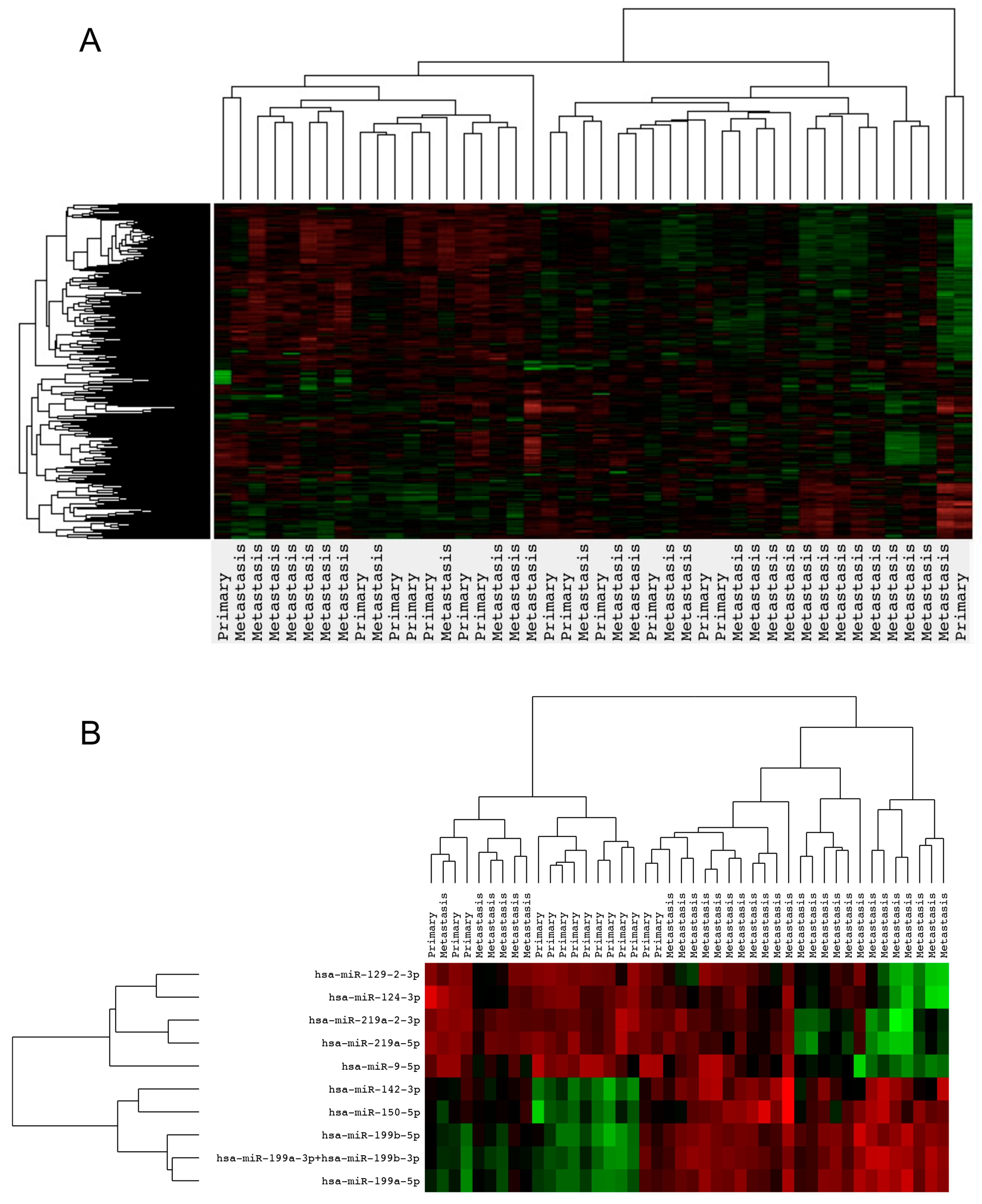
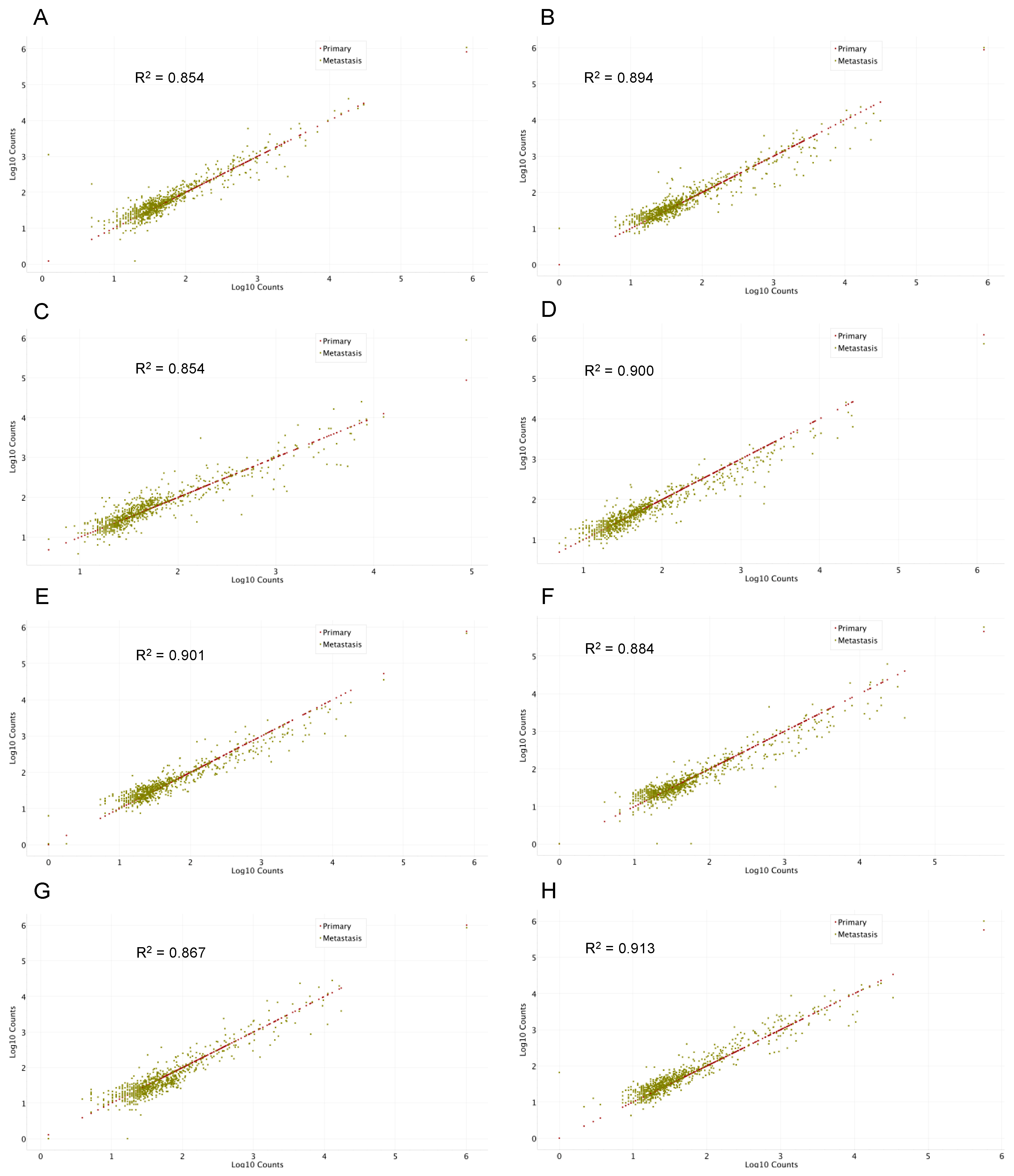
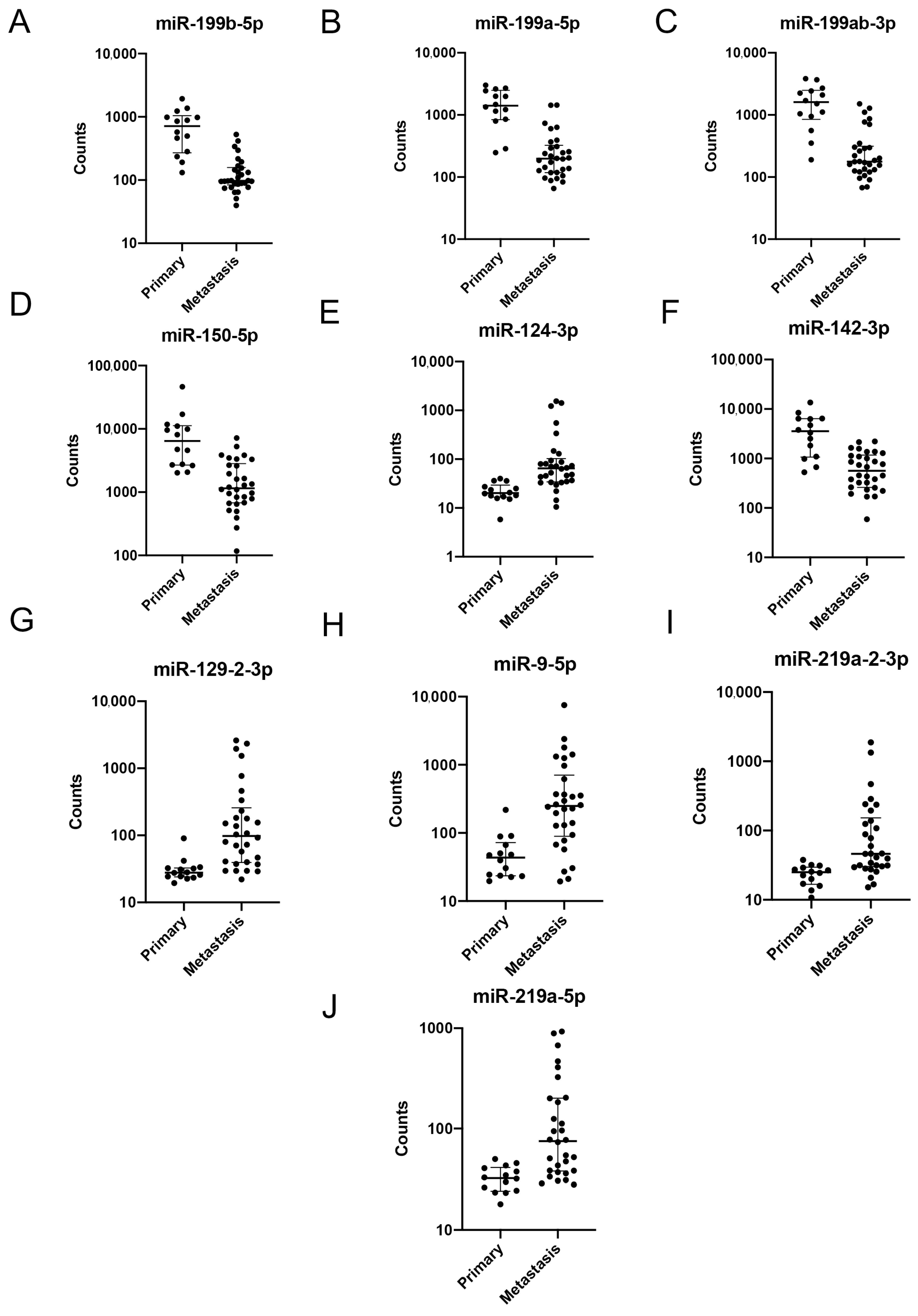
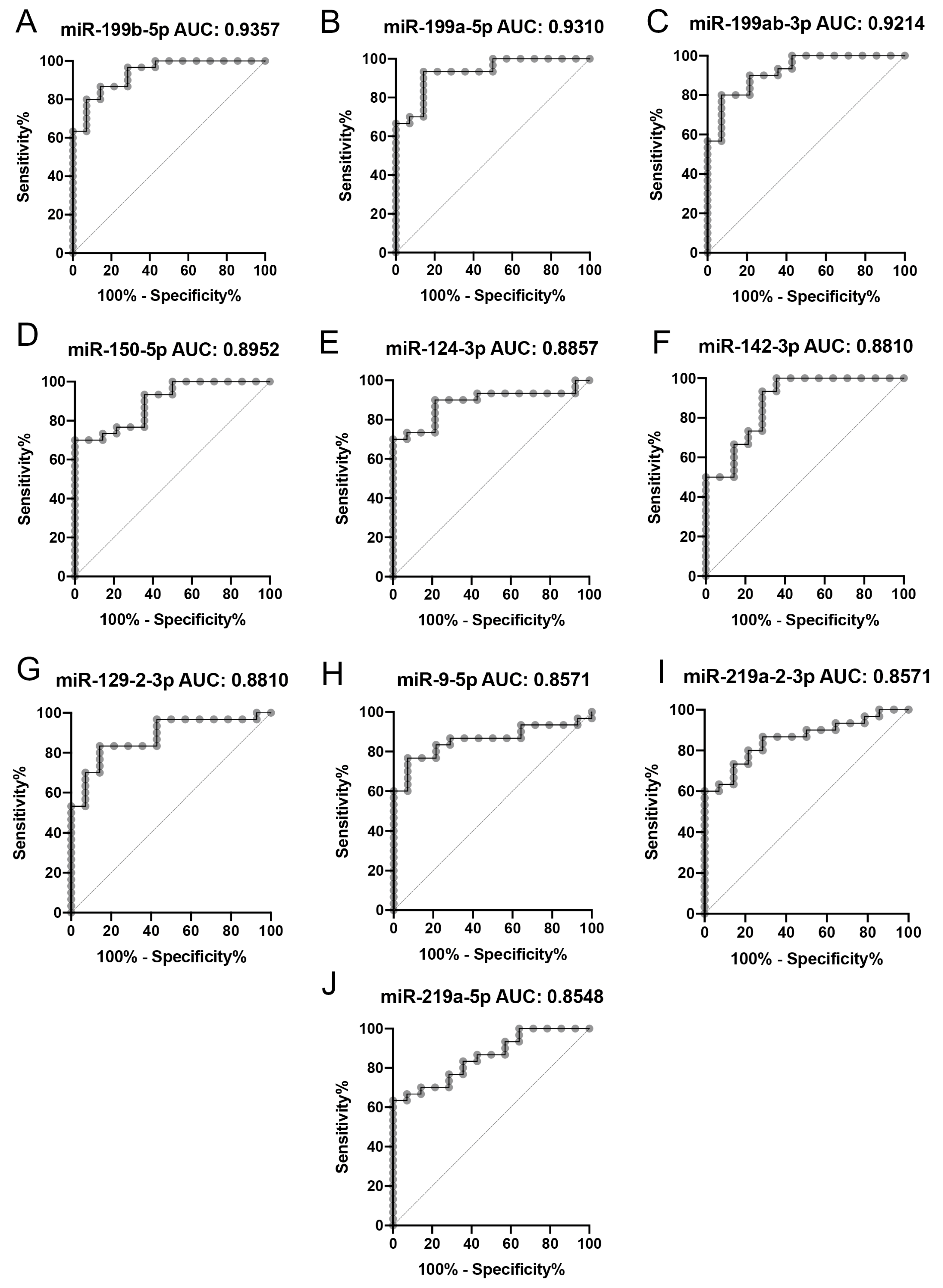
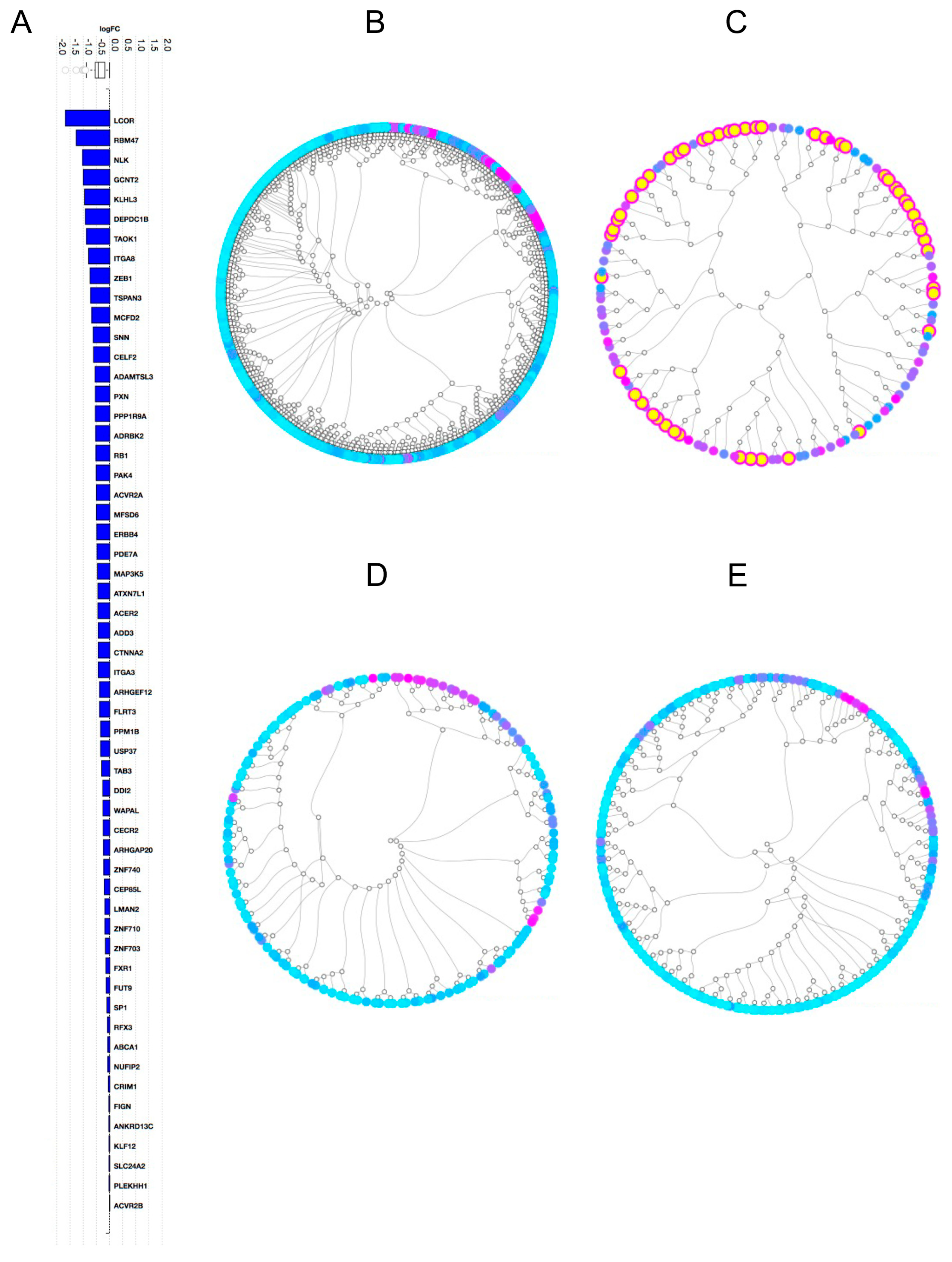
2.2. Gene Expression Analysis
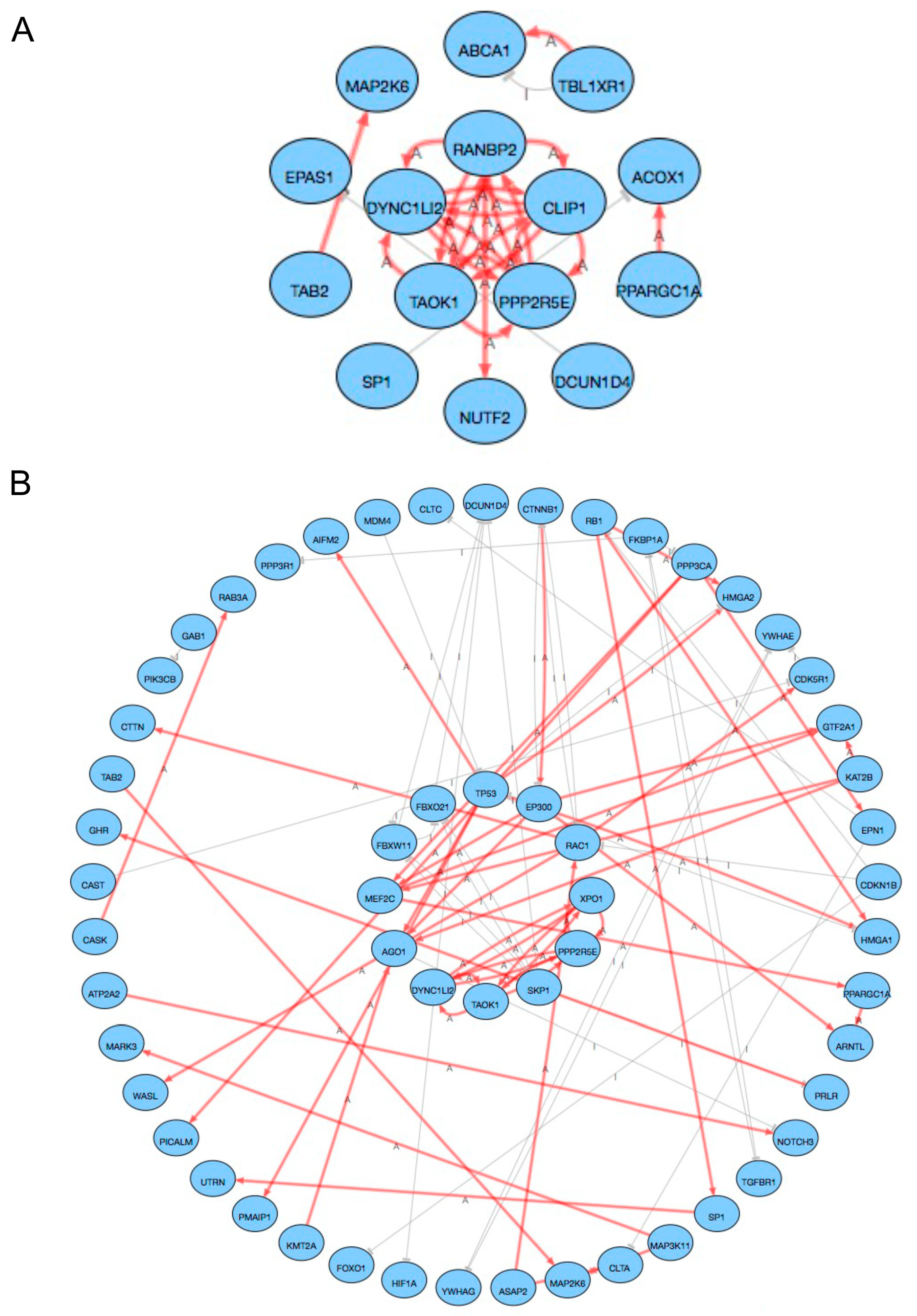
2.3. Pathway Analysis
3. Discussion
4. Materials and Methods
4.1. Patient Cohort
4.2. Sample Preparation
4.3. miRNA Expression and Pathway Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Ge, M.; Zhuang, Y.; Zhou, X.; Huang, R.; Liang, X.; Zhan, Q. High probability and frequency of EGFR mutations in non-small cell lung cancer with brain metastases. J. Neurooncol. 2017, 135, 413–418. [Google Scholar] [CrossRef] [PubMed]
- Doebele, R.C.; Lu, X.; Sumey, C.; Maxson, D.A.; Weickhardt, A.J.; Oton, A.B.; Bunn, P.A., Jr.; Barón, A.E.; Franklin, W.A.; Aisner, D.L.; et al. Oncogene status predicts patterns of metastatic spread in treatment-naive nonsmall cell lung cancer. Cancer 2012, 118, 4502–4511. [Google Scholar] [CrossRef] [PubMed]
- Patil, T.; Smith, D.E.; Bunn, P.A.; Aisner, D.L.; Le, A.T.; Hancock, M.; Purcell, W.T.; Bowles, D.W.; Camidge, D.R.; Doebele, R.C. The Incidence of Brain Metastases in Stage IV ROS1-Rearranged Non-Small Cell Lung Cancer and Rate of Central Nervous System Progression on Crizotinib. J. Thorac. Oncol. 2018, 13, 1717–1726. [Google Scholar] [CrossRef] [PubMed]
- Rusthoven, C.G.; Doebele, R.C. Management of Brain Metastases in ALK-Positive Non-Small-Cell Lung Cancer. J. Clin. Oncol. 2016, 34, 2814–2819. [Google Scholar] [CrossRef]
- Tsakonas, G.; Kamali, C.; De Petris, L.; Friesland, S.; Lewensohn, R.; Ekman, S. ALK-Brain Prognostic Index-Preliminary Study of a Prognostic Tool for Patients with ALK-Rearranged, Non-small Cell Lung Cancer and Brain Metastases. Cancers 2020, 12, 1804. [Google Scholar] [CrossRef]
- Gandhi, L.; Rodriguez-Abreu, D.; Gadgeel, S.; Esteban, E.; Felip, E.; De Angelis, F.; Domine, M.; Clingan, P.; Hochmair, M.J.; Powell, S.F.; et al. Pembrolizumab plus Chemotherapy in Metastatic Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2018, 378, 2078–2092. [Google Scholar] [CrossRef]
- Paz-Ares, L.; Ciuleanu, T.E.; Cobo, M.; Schenker, M.; Zurawski, B.; Menezes, J.; Richardet, E.; Bennouna, J.; Felip, E.; Juan-Vidal, O.; et al. First-line nivolumab plus ipilimumab combined with two cycles of chemotherapy in patients with non-small-cell lung cancer (CheckMate 9LA): An international, randomised, open-label, phase 3 trial. Lancet Oncol. 2021, 22, 198–211. [Google Scholar] [CrossRef]
- Bernardo, G.; Cuzzoni, Q.; Strada, M.R.; Bernardo, A.; Brunetti, G.; Jedrychowska, I.; Pozzi, U.; Palumbo, R. First-line chemotherapy with vinorelbine, gemcitabine, and carboplatin in the treatment of brain metastases from non-small-cell lung cancer: A phase II study. Cancer Investig. 2002, 20, 293–302. [Google Scholar] [CrossRef]
- Cortes, J.; Rodriguez, J.; Aramendia, J.M.; Salgado, E.; Gurpide, A.; Garcia-Foncillas, J.; Aristu, J.J.; Claver, A.; Bosch, A.; Lopez-Picazo, J.M.; et al. Front-line paclitaxel/cisplatin-based chemotherapy in brain metastases from non-small-cell lung cancer. Oncology 2003, 64, 28–35. [Google Scholar] [CrossRef]
- Hendriks, L.E.L.; Henon, C.; Auclin, E.; Mezquita, L.; Ferrara, R.; Audigier-Valette, C.; Mazieres, J.; Lefebvre, C.; Rabeau, A.; Le Moulec, S.; et al. Outcome of Patients with Non-Small Cell Lung Cancer and Brain Metastases Treated with Checkpoint Inhibitors. J. Thorac. Oncol. 2019, 14, 1244–1254. [Google Scholar] [CrossRef]
- Tsakonas, G.; Lewensohn, R.; Botling, J.; Ortiz-Villalon, C.; Micke, P.; Friesland, S.; Nord, H.; Lindskog, M.; Sandelin, M.; Hydbring, P.; et al. An immune gene expression signature distinguishes central nervous system metastases from primary tumours in non-small-cell lung cancer. Eur. J. Cancer 2020, 132, 24–34. [Google Scholar] [CrossRef]
- The ENCODE Project Consortium; Birney, E.; Stamatoyannopoulos, J.A.; Dutta, A.; Guigo, R.; Gingeras, T.R.; Margulies, E.H.; Weng, Z.; Snyder, M.; Dermitzakis, E.T.; et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 2007, 447, 799–816. [Google Scholar]
- Song, Y.; Kelava, L.; Zhang, L.; Kiss, I. Microarray data analysis to identify miRNA biomarkers and construct the lncRNA-miRNA-mRNA network in lung adenocarcinoma. Medicine 2022, 101, e30393. [Google Scholar] [CrossRef]
- Meng, W.; Li, Y.; Chai, B.; Liu, X.; Ma, Z. miR-199a: A Tumor Suppressor with Noncoding RNA Network and Therapeutic Candidate in Lung Cancer. Int. J. Mol. Sci. 2022, 23, 8518. [Google Scholar] [CrossRef]
- Wang, J.; Wang, L.; Zhang, C. miR-765 Acts as a Tumor Promoter and Indicates Poor Prognosis in Non-Small Cell Lung Cancer. OncoTargets Ther. 2021, 14, 4335–4343. [Google Scholar] [CrossRef]
- Doghish, A.S.; Ismail, A.; Elrebehy, M.A.; Elbadry, A.M.M.; Mahmoud, H.H.; Farouk, S.M.; Abu Serea, G.A.; Elghany, R.A.A.; El-Halwany, K.K.; Alsawah, A.O.; et al. A study of miRNAs as cornerstone in lung cancer pathogenesis and therapeutic resistance: A focus on signaling pathways interplay. Pathol. Res. Pract. 2022, 237, 154053. [Google Scholar] [CrossRef]
- Alsidawi, S.; Malek, E.; Driscoll, J.J. MicroRNAs in brain metastases: Potential role as diagnostics and therapeutics. Int. J. Mol. Sci. 2014, 15, 10508–10526. [Google Scholar] [CrossRef]
- Sun, G.; Ding, X.; Bi, N.; Wang, Z.; Wu, L.; Zhou, W.; Zhao, Z.; Wang, J.; Zhang, W.; Fan, J.; et al. Molecular predictors of brain metastasis-related microRNAs in lung adenocarcinoma. PLoS Genet. 2019, 15, e1007888. [Google Scholar] [CrossRef]
- Wang, Q.; Yang, G.; Jiang, Y.; Luo, M.; Li, C.; Zhao, Y.; Xie, Y.; Song, K.; Zhou, J. XB130, regulated by miR-203, miR-219, and miR-4782-3p, mediates the proliferation and metastasis of non-small-cell lung cancer cells. Mol. Carcinog. 2020, 59, 557–568. [Google Scholar] [CrossRef]
- Sun, X.; Xu, M.; Liu, H.; Ming, K. MicroRNA-219 is downregulated in non-small cell lung cancer and inhibits cell growth and metastasis by targeting HMGA2. Mol. Med. Rep. 2017, 16, 3557–3564. [Google Scholar] [CrossRef] [PubMed]
- Zhu, K.; Lin, J.; Chen, S.; Xu, Q. miR-9-5p Promotes Lung Adenocarcinoma Cell Proliferation, Migration and Invasion by Targeting ID4. Technol. Cancer Res. Treat. 2021, 20, 15330338211048592. [Google Scholar] [CrossRef]
- Li, G.; Wu, F.; Yang, H.; Deng, X.; Yuan, Y. MiR-9-5p promotes cell growth and metastasis in non-small cell lung cancer through the repression of TGFBR2. Biomed. Pharmacother. 2017, 96, 1170–1178. [Google Scholar] [CrossRef] [PubMed]
- Cai, J.; Huang, J.; Wang, W.; Zeng, J.; Wang, P. miR-124-3p Regulates FGF2-EGFR Pathway to Overcome Pemetrexed Resistance in Lung Adenocarcinoma Cells by Targeting MGAT5. Cancer Manag. Res. 2020, 12, 11597–11609. [Google Scholar] [CrossRef] [PubMed]
- Yan, G.; Li, Y.; Zhan, L.; Sun, S.; Yuan, J.; Wang, T.; Yin, Y.; Dai, Z.; Zhu, Y.; Jiang, Z.; et al. Decreased miR-124-3p promoted breast cancer proliferation and metastasis by targeting MGAT5. Am. J. Cancer Res. 2019, 9, 585–596. [Google Scholar] [PubMed]
- Deng, L.; Petrek, H.; Tu, M.J.; Batra, N.; Yu, A.X.; Yu, A.M. Bioengineered miR-124-3p prodrug selectively alters the proteome of human carcinoma cells to control multiple cellular components and lung metastasis in vivo. Acta Pharm. Sin. B 2021, 11, 3950–3965. [Google Scholar] [CrossRef]
- Li, X.; Li, C.; Bi, H.; Bai, S.; Zhao, L.; Zhang, J.; Qi, C. Targeting ZEB2 By microRNA-129 In Non-Small Cell Lung Cancer Suppresses Cell Proliferation, Invasion and Migration Via Regulating Wnt/β-Catenin Signaling Pathway and Epithelial-Mesenchymal Transition. OncoTargets Ther. 2019, 12, 9165–9175. [Google Scholar] [CrossRef]
- Xue, T.; Yin, G.; Yang, W.; Chen, X.; Liu, C.; Yang, W.; Zhu, J. MiR-129-5p Promotes Radio-sensitivity of NSCLC Cells by Targeting SOX4 and RUNX1. Curr. Cancer Drug Targets 2021, 21, 702–712. [Google Scholar] [CrossRef]
- Zhou, W.; Cai, C.; Lu, J.; Fan, Q. miR-129-2 upregulation induces apoptosis and promotes NSCLC chemosensitivity by targeting SOX4. Thorac. Cancer 2022, 13, 956–964. [Google Scholar] [CrossRef]
- Lee, A.S. GRP78 induction in cancer: Therapeutic and prognostic implications. Cancer Res. 2007, 67, 3496–3499. [Google Scholar] [CrossRef]
- Ahmadi, A.; Khansarinejad, B.; Hosseinkhani, S.; Ghanei, M.; Mowla, S. miR-199a-5p and miR-495 target GRP78 within UPR pathway of lung cancer. Gene 2017, 620, 15–22. [Google Scholar] [CrossRef]
- Yang, X.; Zheng, Y.; Tan, J.; Tian, R.; Shen, P.; Cai, W.; Liao, H. MiR-199a-5p–HIF-1α-STAT3 Positive Feedback Loop Contributes to the Progression of Non-Small Cell Lung Cancer. Front. Cell Dev. Biol. 2021, 8, 620615. [Google Scholar] [CrossRef]
- Li, Y.; Wang, D.; Li, X.; Shao, Y.; He, Y.; Yu, H.; Ma, Z.-L. MiR-199a-5p suppresses non-small cell lung cancer via targeting MAP3K11. J. Cancer 2019, 10, 2472–2479. [Google Scholar] [CrossRef]
- Mizuno, K.; Tanigawa, K.; Misono, S.; Suetsugu, T.; Sanada, H.; Uchida, A.; Kawano, M.; Machida, K.; Asai, S.; Moriya, S.; et al. Regulation of Oncogenic Targets by Tumor-Suppressive miR-150-3p in Lung Squamous Cell Carcinoma. Biomedicines 2021, 9, 1883. [Google Scholar] [CrossRef]
- Suetsugu, T.; Koshizuka, K.; Seki, N.; Mizuno, K.; Okato, A.; Arai, T.; Misono, S.; Uchida, A.; Kumamoto, T.; Inoue, H. Downregulation of matrix metalloproteinase 14 by the antitumor miRNA, miR-150-5p, inhibits the aggressiveness of lung squamous cell carcinoma cells. Int. J. Oncol. 2018, 52, 913–924. [Google Scholar] [CrossRef]
- Misono, S.; Seki, N.; Mizuno, K.; Yamada, Y.; Uchida, A.; Sanada, H.; Moriya, S.; Kikkawa, N.; Kumamoto, T.; Suetsugu, T.; et al. Molecular Pathogenesis of Gene Regulation by the miR-150 Duplex: miR-150-3p Regulates TNS4 in Lung Adenocarcinoma. Cancers 2019, 11, 601. [Google Scholar] [CrossRef]
- Wu, Z.; Li, W.; Li, J.; Zhang, Y.; Zhang, X.; Xu, Y.; Hu, Y.; Li, Q.; Sun, Q.; Ma, Z. Higher expression of miR-150-5p promotes tumorigenesis by suppressing LKB1 in non-small cell lung cancer. Pathol. Res. Pract. 2020, 216, 153145. [Google Scholar] [CrossRef]
- Dai, F.Q.; Li, C.R.; Fan, X.Q.; Tan, L.; Wang, R.T.; Jin, H. miR-150-5p Inhibits Non-Small-Cell Lung Cancer Metastasis and Recurrence by Targeting HMGA2 and β-Catenin Signaling. Mol. Ther. Nucleic Acids. 2019, 16, 675–685. [Google Scholar] [CrossRef]
- Li, H.; Ouyang, R.; Wang, Z.; Zhou, W.; Chen, H.; Jiang, Y.; Zhang, Y.; Liao, M.; Wang, W.; Ye, M.; et al. MiR-150 promotes cellular metastasis in non-small cell lung cancer by targeting FOXO4. Sci. Rep. 2016, 6, 39001. [Google Scholar] [CrossRef]
- Gu, X.Y.; Wang, J.; Luo, Y.Z.; Du, Q.; Li, R.R.; Shi, H.; Yu, T.-P. Down-regulation of miR-150 induces cell proliferation inhibition and apoptosis in non-small-cell lung cancer by targeting BAK1 in vitro. Tumor Biol. 2014, 35, 5287–5293. [Google Scholar] [CrossRef]
- Jiang, K.; Shen, M.; Chen, Y.; Xu, W. miR-150 promotes the proliferation and migration of non-small cell lung cancer cells by regulating the SIRT2/JMJD2A signaling pathway. Oncol. Rep. 2018, 40, 943–951. [Google Scholar] [CrossRef] [PubMed]
- Cao, M.; Hou, D.; Liang, H.; Gong, F.; Wang, Y.; Yan, X.; Jiang, X.; Wang, C.; Zhang, J.; Zen, K.; et al. miR-150 promotes the proliferation and migration of lung cancer cells by targeting SRC kinase signalling inhibitor 1. Eur. J. Cancer. 2014, 50, 1013–1024. [Google Scholar] [CrossRef] [PubMed]
- Yin, Q.W.; Sun, X.F.; Yang, G.T.; Li, X.B.; Wu, M.S.; Zhao, J. Increased expression of microRNA-150 is associated with poor prognosis in non-small cell lung cancer. Int. J. Clin. Exp. Pathol. 2015, 8, 842–846. [Google Scholar] [PubMed]
- Xiao, P.; Liu, W.L. MiR-142-3p functions as a potential tumor suppressor directly targeting HMGB1 in non-small-cell lung carcinoma. Int. J. Clin. Exp. Pathol. 2015, 8, 10800–10807. [Google Scholar] [PubMed]
- Wang, Z.; Liu, Z.; Fang, X.; Yang, H. MiR-142-5p Suppresses Tumorigenesis by Targeting PIK3CA in Non-Small Cell Lung Cancer. Cell. Physiol. Biochem. 2017, 43, 2505–2515. [Google Scholar] [CrossRef] [PubMed]
- Jin, C.; Xiao, L.; Zhou, Z.; Zhu, Y.; Tian, G.; Ren, S. MiR-142-3p suppresses the proliferation, migration and invasion through inhibition of NR2F6 in lung adenocarcinoma. Hum. Cell 2019, 32, 437–446. [Google Scholar] [CrossRef]
- Wu, X.B.; Li, Q.H.; Zhang, N.; Li, M.; Li, K. MiR-142 inhibits lung cancer cell proliferation and promotes apoptosis by targeting XIAP. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 7430–7437. [Google Scholar]
- Ye, Q.; Putila, J.; Raese, R.; Dong, C.; Qian, Y.; Dowlati, A.; Guo, N. Identification of Prognostic and Chemopredictive microRNAs for Non-Small-Cell Lung Cancer by Integrating SEER-Medicare Data. Int. J. Mol. Sci. 2021, 22, 7658. [Google Scholar] [CrossRef]
- Ahsan, S.; Draghici, S. Identifying Significantly Impacted Pathways and Putative Mechanisms with iPathwayGuide. Curr. Protoc. Bioinform. 2017, 57, 7–15. [Google Scholar] [CrossRef]
- Celia-Terrassa, T.; Liu, D.D.; Choudhury, A.; Hang, X.; Wei, Y.; Zamalloa, J.; Alfaro-Aco, R.; Chakrabarti, R.; Jiang, Y.-Z.; Koh, B.I.; et al. Normal and cancer mammary stem cells evade interferon-induced constraints through the miR-199a-LCOR axis. Nat. Cell Biol. 2017, 19, 711–723. [Google Scholar] [CrossRef]
- Izumi, H.; Matsumoto, S.; Liu, J.; Tanaka, K.; Mori, S.; Hayashi, K.; Kumagai, S.; Shibata, Y.; Hayashida, T.; Watanabe, K.; et al. The CLIP1-LTK fusion is an oncogenic driver in non-small-cell lung cancer. Nature 2021, 600, 319–323. [Google Scholar] [CrossRef]
- Uhlen, M.; Zhang, C.; Lee, S.; Sjöstedt, E.; Fagerberg, L.; Bidkhori, G.; Benfeitas, R.; Arif, M.; Liu, Z.; Edfors, F.; et al. A pathology atlas of the human cancer transcriptome. Science 2017, 357, 2507. [Google Scholar] [CrossRef]
- Human Protein Atlas. Available online: www.proteinatlas.org (accessed on 1 October 2022).
- Brambilla, E.; Brambilla, C. p53 and lung cancer. Pathol. Biol. 1997, 45, 852–863. [Google Scholar]
- Powell, E.; Piwnica-Worms, D.; Piwnica-Worms, H. Contribution of p53 to metastasis. Cancer Discov. 2014, 4, 405–414. [Google Scholar] [CrossRef]
- Liao, L.; Ji, X.; Ge, M.; Zhan, Q.; Huang, R.; Liang, X.; Zhou, X. Characterization of genetic alterations in brain metastases from non-small cell lung cancer. FEBS Open Bio 2018, 8, 1544–1552. [Google Scholar] [CrossRef]
| Variable | % (n) |
|---|---|
| Sex (male) | 64 (16) |
| Age (mean ± SD) | 62 ± 7.8 |
| WHO Performance Status | |
| 0–1 | 80 (20) |
| 2 | 16 (4) |
| 3 | 4 (1) |
| Histology (%) | |
| Adenocarcinoma | 68 (17) |
| Squamous cell | 16 (4) |
| SCLC | 4 (1) |
| Large cell | 12 (3) |
| CNS metastasis at diagnosis | 24 (6) |
| Oncologic therapy | |
| Surgery primary tumor | 52 (13) |
| Systemic therapy | 80 (20) |
| CNS radiotherapy | 92 (23) |
| GPA class group (%) | |
| 1 (0–1) | 28 (7) |
| 2 (1.5–2.5) | 56 (14) |
| 3 (3–4) | 16 (4) |
| RPA class (%) | |
| 1 | 20 (5) |
| 2 | 64 (16) |
| 3 | 16 (4) |
| Number of CNS metastases (%) | |
| 1 | 40 (10) |
| 2–3 | 28 (7) |
| >3 | 32 (8) |
| Stage at diagnosis | |
| I | 20 (5) |
| II | 8 (2) |
| III | 40 (10) |
| IV | 32 (8) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tsakonas, G.; Koulouris, A.; Kazmierczak, D.; Botling, J.; Ortiz-Villalon, C.; Nord, H.; Lindskog, M.; Sandelin, M.; Micke, P.; Hydbring, P.; et al. Matched Analyses of Brain Metastases versus Primary Non-Small Cell Lung Cancer Reveal a Unique microRNA Signature. Int. J. Mol. Sci. 2023, 24, 193. https://doi.org/10.3390/ijms24010193
Tsakonas G, Koulouris A, Kazmierczak D, Botling J, Ortiz-Villalon C, Nord H, Lindskog M, Sandelin M, Micke P, Hydbring P, et al. Matched Analyses of Brain Metastases versus Primary Non-Small Cell Lung Cancer Reveal a Unique microRNA Signature. International Journal of Molecular Sciences. 2023; 24(1):193. https://doi.org/10.3390/ijms24010193
Chicago/Turabian StyleTsakonas, Georgios, Andreas Koulouris, Dominika Kazmierczak, Johan Botling, Cristian Ortiz-Villalon, Helena Nord, Magnus Lindskog, Martin Sandelin, Patrick Micke, Per Hydbring, and et al. 2023. "Matched Analyses of Brain Metastases versus Primary Non-Small Cell Lung Cancer Reveal a Unique microRNA Signature" International Journal of Molecular Sciences 24, no. 1: 193. https://doi.org/10.3390/ijms24010193
APA StyleTsakonas, G., Koulouris, A., Kazmierczak, D., Botling, J., Ortiz-Villalon, C., Nord, H., Lindskog, M., Sandelin, M., Micke, P., Hydbring, P., & Ekman, S. (2023). Matched Analyses of Brain Metastases versus Primary Non-Small Cell Lung Cancer Reveal a Unique microRNA Signature. International Journal of Molecular Sciences, 24(1), 193. https://doi.org/10.3390/ijms24010193






