Towards Understanding the Gene-Specific Roles of GATA Factors in Heart Development: Does GATA4 Lead the Way?
Abstract
1. Introduction
2. The Requirement of Cardiogenic Gata Genes for Heart Development
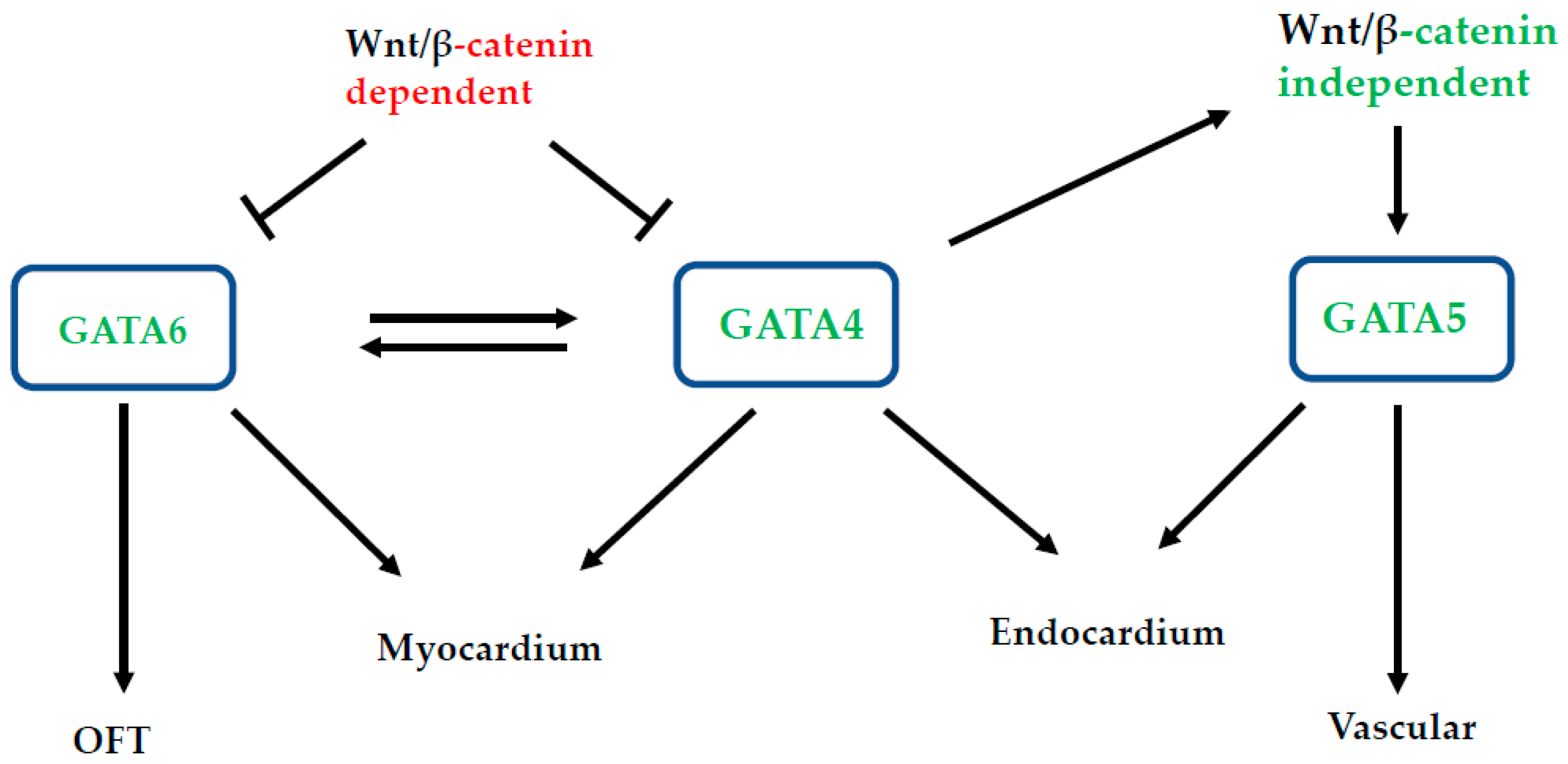
3. Redundancy and Functional Interactions among Cardiogenic GATA Genes
4. Direct and Indirect Roles of Gata6 in Cardiac Development: Is Gata6 the Junior Partner?
5. Direct and Indirect Roles of gata5 in Cardiac Development: Listen to the Fish!
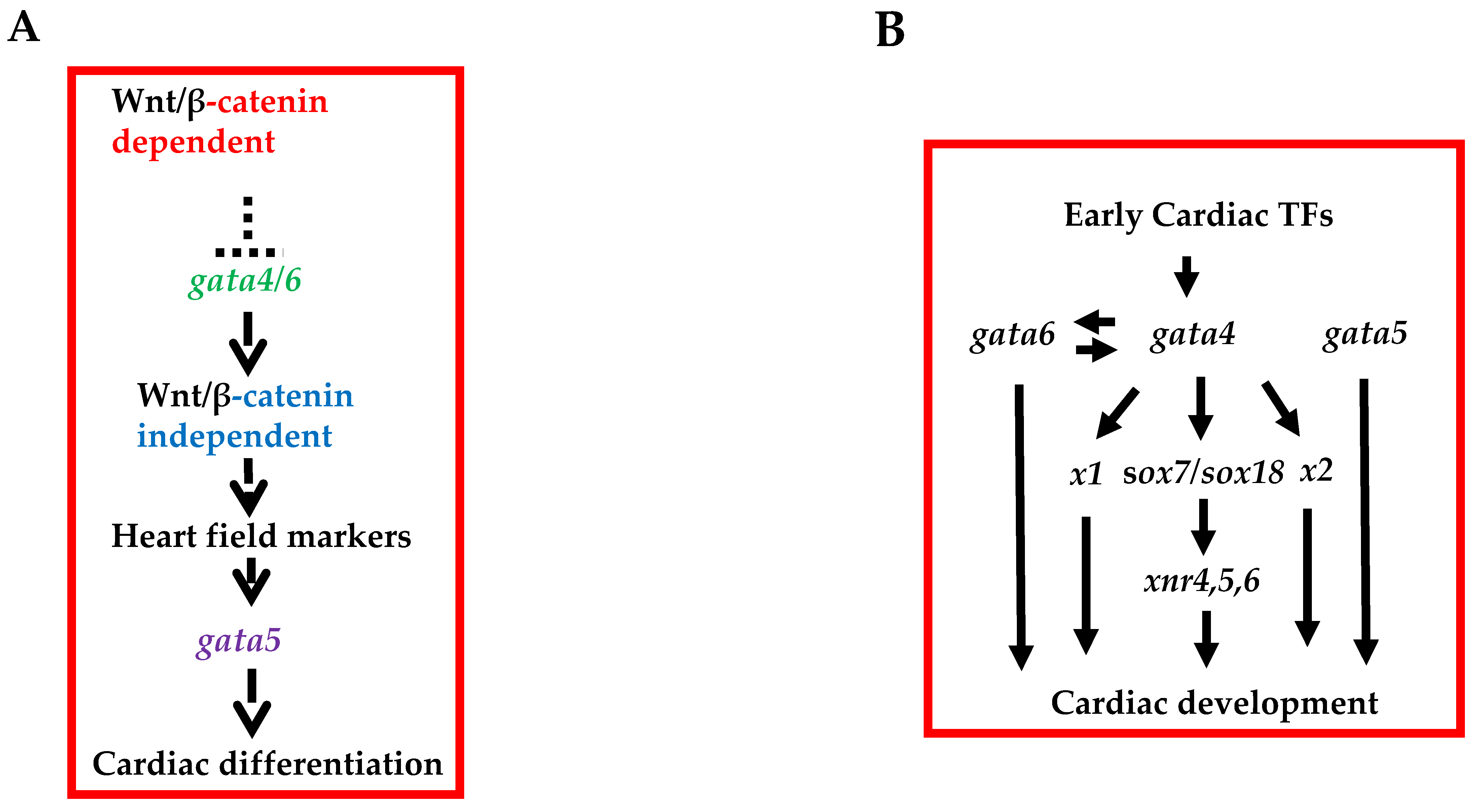
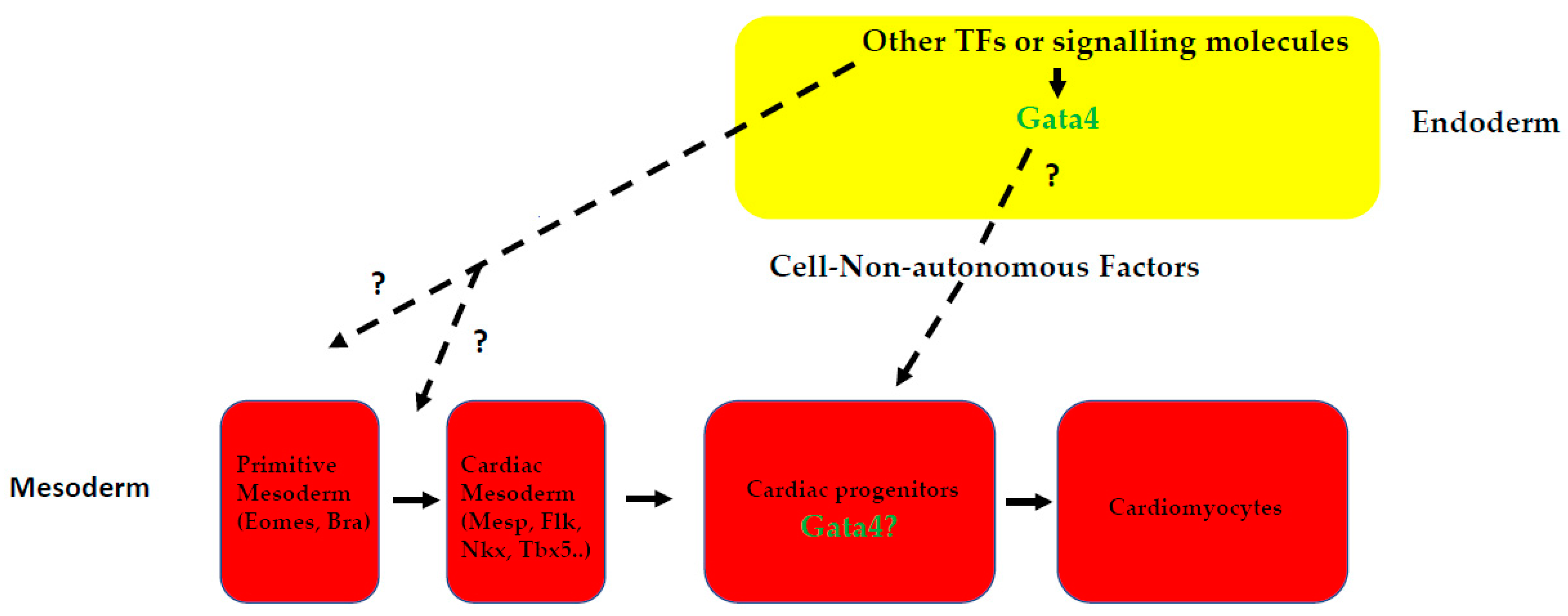
6. Direct and Indirect Roles of GATA4 in Cardiac Development: Is GATA4 the Boss?
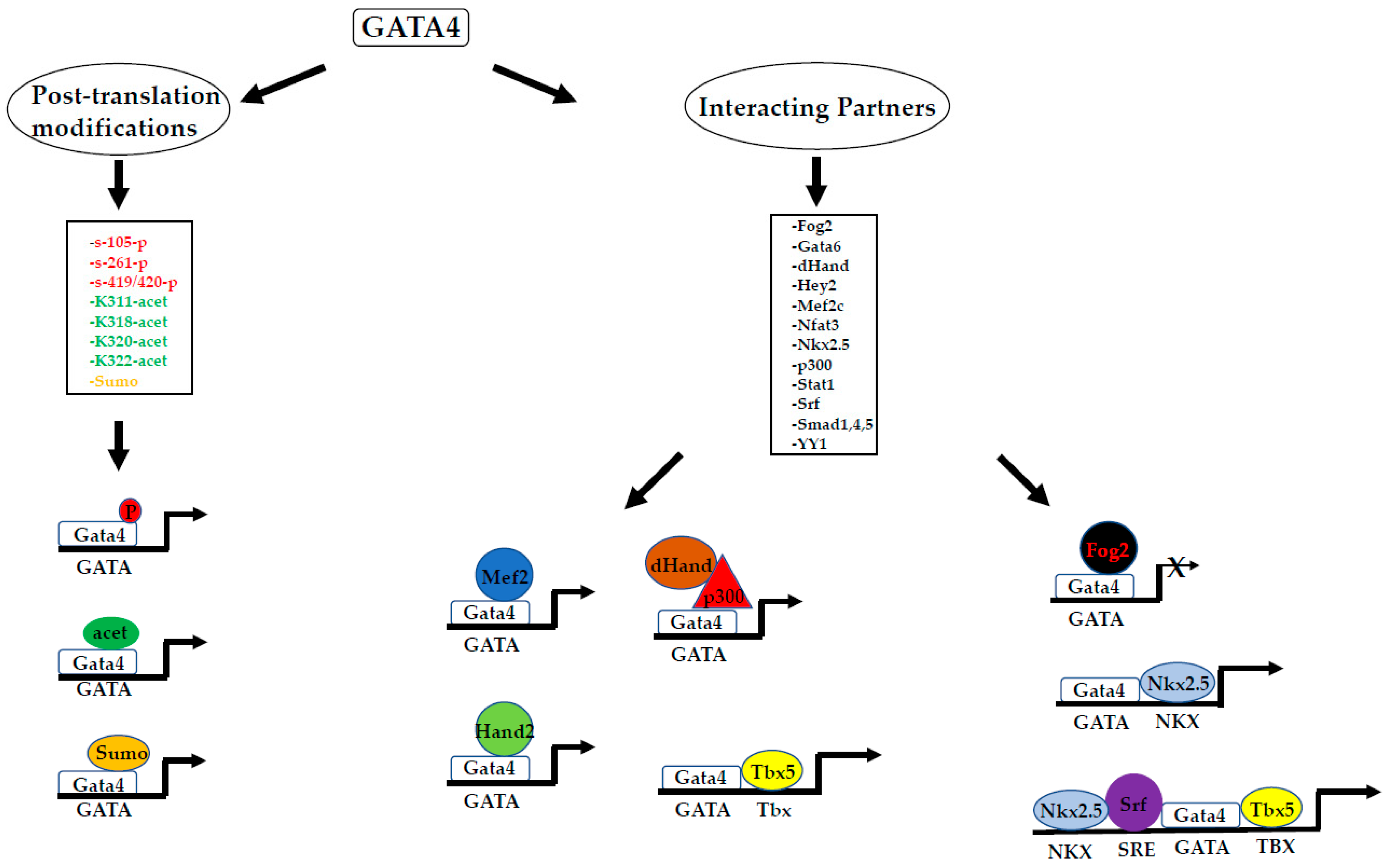
6.1. Co-Operative Activity with Gata4 in Cardiogenesis: With a Little Help from My Friends
6.2. Gata4 Possesses Intrinsic Cardiogenic Activities
6.3. Context-Dependent Activities of GATA4
7. Conclusions and Further Perspectives
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- McFadden, D.G.; Olson, E.N. Heart development: Learning from mistakes. Curr. Opin. Genet. Dev. 2002, 12, 328–335. [Google Scholar] [CrossRef]
- Fahed, A.C.; Gelb, B.D.; Seidman, J.G.; Seidman, C.E. Genetics of congenital heart disease: The glass half empty. Circ. Res. 2013, 112, 707–720. [Google Scholar] [CrossRef] [PubMed]
- Patient, R.K.; McGhee, J.D. The GATA family (vertebrates and invertebrates). Curr. Opin. Genet. Dev. 2002, 12, 416–422. [Google Scholar] [CrossRef]
- Zon, L.I.; Mather, C.; Burgess, S.; Bolce, M.E.; Harland, R.M.; Orkin, S.H. Expression of GATA-binding proteins during embryonic development in Xenopus laevis. Proc. Natl. Acad. Sci. USA 1991, 88, 10642–10646. [Google Scholar] [CrossRef]
- Molkentin, J.D. The zinc finger-containing transcription factors GATA-4, -5, and -6. Ubiquitously expressed regulators of tissue-specific gene expression. J. Biol. Chem. 2000, 275, 38949–38952. [Google Scholar] [CrossRef]
- Jiang, Y.; Evans, T. The Xenopus GATA-4/5/6 genes are associated with cardiac specification and can regulate cardiac-specific transcription during embryogenesis. Dev. Biol. 1996, 174, 258–270. [Google Scholar] [CrossRef][Green Version]
- Grepin, C.; Nemer, G.; Nemer, M. Enhanced cardiogenesis in embryonic stem cells overexpressing the GATA-4 transcription factor. Development 1997, 124, 2387–2395. [Google Scholar] [CrossRef]
- Latinkic, B.V.; Kotecha, S.; Mohun, T.J. Induction of cardiomyocytes by GATA4 in Xenopus ectodermal explants. Development 2003, 130, 3865–3876. [Google Scholar] [CrossRef]
- Afouda, B.A.; Martin, J.; Liu, F.; Ciau-Uitz, A.; Patient, R.; Hoppler, S. GATA transcription factors integrate Wnt signalling during heart development. Development 2008, 135, 3185–3190. [Google Scholar] [CrossRef]
- Afouda, B.A.; Hoppler, S. Xenopus explants as an experimental model system for studying heart development. Trends. Cardiovasc. Med. 2009, 19, 220–226. [Google Scholar] [CrossRef]
- Afouda, B.A.; Hoppler, S. Different requirements for GATA factors in cardiogenesis are mediated by non-canonical Wnt signaling. Dev. Dyn. 2011, 240, 649–662. [Google Scholar] [CrossRef] [PubMed]
- Turbendian, H.K.; Gordillo, M.; Tsai, S.Y.; Lu, J.; Kang, G.; Liu, T.C.; Tang, A.; Liu, S.; Fishman, G.I.; Evans, T. GATA factors efficiently direct cardiac fate from embryonic stem cells. Development 2013, 140, 1639–1644. [Google Scholar] [CrossRef] [PubMed]
- Liang, Q.; De Windt, L.J.; Witt, S.A.; Kimball, T.R.; Markham, B.E.; Molkentin, J.D. The transcription factors GATA4 and GATA6 regulate cardiomyocyte hypertrophy in vitro and in vivo. J. Biol. Chem. 2001, 276, 30245–30253. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Tarzami, S.; Burch, J.B.; Evans, T. Common role for each of the cGATA-4/5/6 genes in the regulation of cardiac morphogenesis. Dev. Genet. 1998, 22, 263–277. [Google Scholar] [CrossRef]
- Laforest, B.; Nemer, M. GATA5 interacts with GATA4 and GATA6 in outflow tract development. Dev. Biol. 2011, 358, 368–378. [Google Scholar] [CrossRef]
- Gharibeh, L.; Komati, H.; Bosse, Y.; Boodhwani, M.; Heydarpour, M.; Fortier, M.; Hassanzadeh, R.; Ngu, J.; Mathieu, P.; Body, S.; et al. GATA6 Regulates Aortic Valve Remodeling, and Its Haploinsufficiency Leads to Right-Left Type Bicuspid Aortic Valve. Circulation 2018, 138, 1025–1038. [Google Scholar] [CrossRef]
- Xu, Y.J.; Di, R.M.; Qiao, Q.; Li, X.M.; Huang, R.T.; Xue, S.; Liu, X.Y.; Wang, J.; Yang, Y.Q. GATA6 loss-of-function mutation contributes to congenital bicuspid aortic valve. Gene 2018, 663, 115–120. [Google Scholar] [CrossRef]
- Gharibeh, L.; Yamak, A.; Whitcomb, J.; Lu, A.; Joyal, M.; Komati, H.; Liang, W.; Fiset, C.; Nemer, M. GATA6 is a regulator of sinus node development and heart rhythm. Proc. Natl. Acad. Sci. USA 2021, 118. [Google Scholar] [CrossRef]
- Nemer, G.; Nemer, M. Cooperative interaction between GATA5 and NF-ATc regulates endothelial-endocardial differentiation of cardiogenic cells. Development 2002, 129, 4045–4055. [Google Scholar] [CrossRef]
- Messaoudi, S.; He, Y.; Gutsol, A.; Wight, A.; Hebert, R.L.; Vilmundarson, R.O.; Makrigiannis, A.P.; Chalmers, J.; Hamet, P.; Tremblay, J.; et al. Endothelial Gata5 transcription factor regulates blood pressure. Nat. Commun. 2015, 6, 8835. [Google Scholar] [CrossRef]
- Kelley, C.; Blumberg, H.; Zon, L.I.; Evans, T. GATA-4 is a novel transcription factor expressed in endocardium of the developing heart. Development 1993, 118, 817–827. [Google Scholar] [CrossRef] [PubMed]
- Molkentin, J.D.; Lin, Q.; Duncan, S.A.; Olson, E.N. Requirement of the transcription factor GATA4 for heart tube formation and ventral morphogenesis. Genes Dev. 1997, 11, 1061–1072. [Google Scholar] [CrossRef] [PubMed]
- Kuo, C.T.; Morrisey, E.E.; Anandappa, R.; Sigrist, K.; Lu, M.M.; Parmacek, M.S.; Soudais, C.; Leiden, J.M. GATA4 transcription factor is required for ventral morphogenesis and heart tube formation. Genes Dev. 1997, 11, 1048–1060. [Google Scholar] [CrossRef] [PubMed]
- Afouda, B.A.; Lynch, A.T.; de Paiva Alves, E.; Hoppler, S. Genome-wide transcriptomics analysis identifies sox7 and sox18 as specifically regulated by gata4 in cardiomyogenesis. Dev. Biol. 2018, 434, 108–120. [Google Scholar] [CrossRef]
- Afouda, B.A.; Lynch, A.T.; de Paiva Alves, E.; Hoppler, S. Genome-wide transcriptomics analysis of genes regulated by GATA4, 5 and 6 during cardiomyogenesis in Xenopus laevis. Data Brief. 2018, 17, 559–563. [Google Scholar] [CrossRef]
- Peterkin, T.; Gibson, A.; Patient, R. Redundancy and evolution of GATA factor requirements in development of the myocardium. Dev. Biol. 2007, 311, 623–635. [Google Scholar] [CrossRef]
- Sam, J.; Mercer, E.J.; Torregroza, I.; Banks, K.M.; Evans, T. Specificity, redundancy and dosage thresholds among gata4/5/6 genes during zebrafish cardiogenesis. Biol. Open 2020, 9. [Google Scholar] [CrossRef]
- Liu, J.; Cheng, H.; Xiang, M.; Zhou, L.; Wu, B.; Moskowitz, I.P.; Zhang, K.; Xie, L. Gata4 regulates hedgehog signaling and Gata6 expression for outflow tract development. PLoS Genet. 2019, 15, e1007711. [Google Scholar] [CrossRef]
- Nemer, G.; Nemer, M. Transcriptional activation of BMP-4 and regulation of mammalian organogenesis by GATA-4 and -6. Dev. Biol. 2003, 254, 131–148. [Google Scholar] [CrossRef]
- Peterkin, T.; Gibson, A.; Patient, R. GATA-6 maintains BMP-4 and Nkx2 expression during cardiomyocyte precursor maturation. EMBO J. 2003, 22, 4260–4273. [Google Scholar] [CrossRef]
- Holtzinger, A.; Rosenfeld, G.E.; Evans, T. Gata4 directs development of cardiac-inducing endoderm from ES cells. Dev. Biol. 2010, 337, 63–73. [Google Scholar] [CrossRef] [PubMed]
- Martin, J.; Afouda, B.A.; Hoppler, S. Wnt/beta-catenin signalling regulates cardiomyogenesis via GATA transcription factors. J. Anat. 2010, 216, 92–107. [Google Scholar] [CrossRef] [PubMed]
- Novikov, N.; Evans, T. Tmem88a mediates GATA-dependent specification of cardiomyocyte progenitors by restricting WNT signaling. Development 2013, 140, 3787–3798. [Google Scholar] [CrossRef] [PubMed]
- Yu, W.; Huang, X.; Tian, X.; Zhang, H.; He, L.; Wang, Y.; Nie, Y.; Hu, S.; Lin, Z.; Zhou, B.; et al. GATA4 regulates Fgf16 to promote heart repair after injury. Development 2016, 143, 936–949. [Google Scholar] [CrossRef] [PubMed]
- Morrisey, E.E.; Tang, Z.; Sigrist, K.; Lu, M.M.; Jiang, F.; Ip, H.S.; Parmacek, M.S. GATA6 regulates HNF4 and is required for differentiation of visceral endoderm in the mouse embryo. Genes Dev. 1998, 12, 3579–3590. [Google Scholar] [CrossRef]
- Koutsourakis, M.; Langeveld, A.; Patient, R.; Beddington, R.; Grosveld, F. The transcription factor GATA6 is essential for early extraembryonic development. Development 1999, 126, 723–732. [Google Scholar] [CrossRef]
- Gove, C.; Walmsley, M.; Nijjar, S.; Bertwistle, D.; Guille, M.; Partington, G.; Bomford, A.; Patient, R. Over-expression of GATA-6 in Xenopus embryos blocks differentiation of heart precursors. EMBO J. 1997, 16, 355–368. [Google Scholar] [CrossRef]
- Narita, N.; Heikinheimo, M.; Bielinska, M.; White, R.A.; Wilson, D.B. The gene for transcription factor GATA-6 resides on mouse chromosome 18 and is expressed in myocardium and vascular smooth muscle. Genomics 1996, 36, 345–348. [Google Scholar] [CrossRef]
- Charron, F.; Paradis, P.; Bronchain, O.; Nemer, G.; Nemer, M. Cooperative interaction between GATA-4 and GATA-6 regulates myocardial gene expression. Mol. Cell Biol. 1999, 19, 4355–4365. [Google Scholar] [CrossRef]
- Kodo, K.; Nishizawa, T.; Furutani, M.; Arai, S.; Yamamura, E.; Joo, K.; Takahashi, T.; Matsuoka, R.; Yamagishi, H. GATA6 mutations cause human cardiac outflow tract defects by disrupting semaphorin-plexin signaling. Proc. Natl. Acad. Sci. USA 2009, 106, 13933–13938. [Google Scholar] [CrossRef]
- van Berlo, J.H.; Elrod, J.W.; van den Hoogenhof, M.M.; York, A.J.; Aronow, B.J.; Duncan, S.A.; Molkentin, J.D. The transcription factor GATA-6 regulates pathological cardiac hypertrophy. Circ. Res. 2010, 107, 1032–1040. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Yuan, L.; Goss, A.M.; Wang, T.; Yang, J.; Lepore, J.J.; Zhou, D.; Schwartz, R.J.; Patel, V.; Cohen, E.D.; et al. Characterization and in vivo pharmacological rescue of a Wnt2-Gata6 pathway required for cardiac inflow tract development. Dev. Cell 2010, 18, 275–287. [Google Scholar] [CrossRef] [PubMed]
- Reiter, J.F.; Alexander, J.; Rodaway, A.; Yelon, D.; Patient, R.; Holder, N.; Stainier, D.Y. Gata5 is required for the development of the heart and endoderm in zebrafish. Genes Dev. 1999, 13, 2983–2995. [Google Scholar] [CrossRef] [PubMed]
- Laforest, B.; Andelfinger, G.; Nemer, M. Loss of Gata5 in mice leads to bicuspid aortic valve. J. Clin. Investig. 2011, 121, 2876–2887. [Google Scholar] [CrossRef]
- Wei, D.; Bao, H.; Liu, X.Y.; Zhou, N.; Wang, Q.; Li, R.G.; Xu, Y.J.; Yang, Y.Q. GATA5 loss-of-function mutations underlie tetralogy of fallot. Int. J. Med. Sci. 2013, 10, 34–42. [Google Scholar] [CrossRef]
- Shi, L.M.; Tao, J.W.; Qiu, X.B.; Wang, J.; Yuan, F.; Xu, L.; Liu, H.; Li, R.G.; Xu, Y.J.; Wang, Q.; et al. GATA5 loss-of-function mutations associated with congenital bicuspid aortic valve. Int. J. Mol. Med. 2014, 33, 1219–1226. [Google Scholar] [CrossRef]
- Singh, M.K.; Li, Y.; Li, S.; Cobb, R.M.; Zhou, D.; Lu, M.M.; Epstein, J.A.; Morrisey, E.E.; Gruber, P.J. Gata4 and Gata5 cooperatively regulate cardiac myocyte proliferation in mice. J. Biol. Chem. 2010, 285, 1765–1772. [Google Scholar] [CrossRef]
- Garg, V.; Kathiriya, I.S.; Barnes, R.; Schluterman, M.K.; King, I.N.; Butler, C.A.; Rothrock, C.R.; Eapen, R.S.; Hirayama-Yamada, K.; Joo, K.; et al. GATA4 mutations cause human congenital heart defects and reveal an interaction with TBX5. Nature 2003, 424, 443–447. [Google Scholar] [CrossRef]
- Rajagopal, S.K.; Ma, Q.; Obler, D.; Shen, J.; Manichaikul, A.; Tomita-Mitchell, A.; Boardman, K.; Briggs, C.; Garg, V.; Srivastava, D.; et al. Spectrum of heart disease associated with murine and human GATA4 mutation. J. Mol. Cell Cardiol. 2007, 43, 677–685. [Google Scholar] [CrossRef]
- Moskowitz, I.P.; Wang, J.; Peterson, M.A.; Pu, W.T.; Mackinnon, A.C.; Oxburgh, L.; Chu, G.C.; Sarkar, M.; Berul, C.; Smoot, L.; et al. Transcription factor genes Smad4 and Gata4 cooperatively regulate cardiac valve development. [corrected]. Proc. Natl. Acad. Sci. USA 2011, 108, 4006–4011. [Google Scholar] [CrossRef]
- Ounzain, S.; Kobayashi, S.; Peterson, R.E.; He, A.; Motterle, A.; Samani, N.J.; Menick, D.R.; Pu, W.T.; Liang, Q.; Chong, N.W. Cardiac expression of ms1/STARS, a novel gene involved in cardiac development and disease, is regulated by GATA4. Mol. Cell Biol. 2012, 32, 1830–1843. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Misra, C.; Sachan, N.; McNally, C.R.; Koenig, S.N.; Nichols, H.A.; Guggilam, A.; Lucchesi, P.A.; Pu, W.T.; Srivastava, D.; Garg, V. Congenital heart disease-causing Gata4 mutation displays functional deficits in vivo. PLoS Genet. 2012, 8, e1002690. [Google Scholar] [CrossRef] [PubMed]
- Wang, E.; Sun, S.; Qiao, B.; Duan, W.; Huang, G.; An, Y.; Xu, S.; Zheng, Y.; Su, Z.; Gu, X.; et al. Identification of functional mutations in GATA4 in patients with congenital heart disease. PLoS ONE 2013, 8, e62138. [Google Scholar] [CrossRef] [PubMed]
- Misra, C.; Chang, S.W.; Basu, M.; Huang, N.; Garg, V. Disruption of myocardial Gata4 and Tbx5 results in defects in cardiomyocyte proliferation and atrioventricular septation. Hum. Mol. Genet. 2014, 23, 5025–5035. [Google Scholar] [CrossRef]
- Han, H.; Chen, Y.; Liu, G.; Han, Z.; Zhao, Z.; Tang, Y. GATA4 transgenic mice as an in vivo model of congenital heart disease. Int. J. Mol. Med. 2015, 35, 1545–1553. [Google Scholar] [CrossRef][Green Version]
- Li, R.G.; Xu, Y.J.; Wang, J.; Liu, X.Y.; Yuan, F.; Huang, R.T.; Xue, S.; Li, L.; Liu, H.; Li, Y.J.; et al. GATA4 Loss-of-Function Mutation and the Congenitally Bicuspid Aortic Valve. Am. J. Cardiol. 2018, 121, 469–474. [Google Scholar] [CrossRef]
- Dixit, R.; Narasimhan, C.; Balekundri, V.I.; Agrawal, D.; Kumar, A.; Mohapatra, B. Functionally significant, novel GATA4 variants are frequently associated with Tetralogy of Fallot. Hum. Mutat. 2018, 39, 1957–1972. [Google Scholar] [CrossRef]
- LaHaye, S.; Majumdar, U.; Yasuhara, J.; Koenig, S.N.; Matos-Nieves, A.; Kumar, R.; Garg, V. Developmental origins for semilunar valve stenosis identified in mice harboring congenital heart disease-associated GATA4 mutation. Dis. Model. Mech. 2019, 12. [Google Scholar] [CrossRef]
- Li, B.; Li, Z.; Yang, J.; Hong, N.; Jin, L.; Xu, Y.; Fu, Q.; Sun, K.; Yu, Y.; Lu, Y.; et al. Predisposition to atrioventricular septal defects may be caused by SOX7 variants that impair interaction with GATA4. Mol. Genet. Genom. 2022. [Google Scholar] [CrossRef]
- Holtzinger, A.; Evans, T. Gata4 regulates the formation of multiple organs. Development 2005, 132, 4005–4014. [Google Scholar] [CrossRef]
- Watt, A.J.; Battle, M.A.; Li, J.; Duncan, S.A. GATA4 is essential for formation of the proepicardium and regulates cardiogenesis. Proc. Natl. Acad. Sci. USA 2004, 101, 12573–12578. [Google Scholar] [CrossRef] [PubMed]
- Zhao, R.; Watt, A.J.; Battle, M.A.; Li, J.; Bondow, B.J.; Duncan, S.A. Loss of both GATA4 and GATA6 blocks cardiac myocyte differentiation and results in acardia in mice. Dev. Biol. 2008, 317, 614–619. [Google Scholar] [CrossRef] [PubMed]
- Rivera-Feliciano, J.; Lee, K.H.; Kong, S.W.; Rajagopal, S.; Ma, Q.; Springer, Z.; Izumo, S.; Tabin, C.J.; Pu, W.T. Development of heart valves requires Gata4 expression in endothelial-derived cells. Development 2006, 133, 3607–3618. [Google Scholar] [CrossRef] [PubMed]
- Stefanovic, S.; Barnett, P.; van Duijvenboden, K.; Weber, D.; Gessler, M.; Christoffels, V.M. GATA-dependent regulatory switches establish atrioventricular canal specificity during heart development. Nat. Commun. 2014, 5, 3680. [Google Scholar] [CrossRef]
- Si, L.; Shi, J.; Gao, W.; Zheng, M.; Liu, L.; Zhu, J.; Tian, J. Smad4 mediated BMP2 signal is essential for the regulation of GATA4 and Nkx2.5 by affecting the histone H3 acetylation in H9c2 cells. Biochem. Biophys. Res. Commun. 2014, 450, 81–86. [Google Scholar] [CrossRef]
- Stefanovic, S.; Christoffels, V.M. GATA-dependent transcriptional and epigenetic control of cardiac lineage specification and differentiation. Cell Mol. Life Sci. 2015, 72, 3871–3881. [Google Scholar] [CrossRef]
- Iyer, L.M.; Nagarajan, S.; Woelfer, M.; Schoger, E.; Khadjeh, S.; Zafiriou, M.P.; Kari, V.; Herting, J.; Pang, S.T.; Weber, T.; et al. A context-specific cardiac beta-catenin and GATA4 interaction influences TCF7L2 occupancy and remodels chromatin driving disease progression in the adult heart. Nucleic Acids Res. 2018, 46, 2850–2867. [Google Scholar] [CrossRef]
- Stone, N.R.; Gifford, C.A.; Thomas, R.; Pratt, K.J.B.; Samse-Knapp, K.; Mohamed, T.M.A.; Radzinsky, E.M.; Schricker, A.; Ye, L.; Yu, P.; et al. Context-Specific Transcription Factor Functions Regulate Epigenomic and Transcriptional Dynamics during Cardiac Reprogramming. Cell Stem Cell 2019, 25, 87–102.e109. [Google Scholar] [CrossRef]
- Nemer, M. Genetic insights into normal and abnormal heart development. Cardiovasc. Pathol. Off. J. Soc. Cardiovasc. Pathol. 2008, 17, 48–54. [Google Scholar] [CrossRef]
- Meganathan, K.; Sotiriadou, I.; Natarajan, K.; Hescheler, J.; Sachinidis, A. Signaling molecules, transcription growth factors and other regulators revealed from in-vivo and in-vitro models for the regulation of cardiac development. Int. J. Cardiol. 2015, 183, 117–128. [Google Scholar] [CrossRef]
- Whitcomb, J.; Gharibeh, L.; Nemer, M. From embryogenesis to adulthood: Critical role for GATA factors in heart development and function. IUBMB Life 2020, 72, 53–67. [Google Scholar] [CrossRef] [PubMed]
- Crispino, J.D.; Lodish, M.B.; Thurberg, B.L.; Litovsky, S.H.; Collins, T.; Molkentin, J.D.; Orkin, S.H. Proper coronary vascular development and heart morphogenesis depend on interaction of GATA-4 with FOG cofactors. Genes Dev. 2001, 15, 839–844. [Google Scholar] [CrossRef] [PubMed]
- Padang, R.; Bagnall, R.D.; Richmond, D.R.; Bannon, P.G.; Semsarian, C. Rare non-synonymous variations in the transcriptional activation domains of GATA5 in bicuspid aortic valve disease. J. Mol. Cell Cardiol. 2012, 53, 277–281. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.Q.; Wang, X.H.; Tan, H.W.; Jiang, W.F.; Fang, W.Y.; Liu, X. Prevalence and spectrum of GATA6 mutations associated with familial atrial fibrillation. Int. J. Cardiol. 2012, 155, 494–496. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.X.; Krane, M.; Deutsch, M.A.; Wang, L.; Rav-Acha, M.; Gregoire, S.; Engels, M.C.; Rajarajan, K.; Karra, R.; Abel, E.D.; et al. Inefficient reprogramming of fibroblasts into cardiomyocytes using Gata4, Mef2c, and Tbx5. Circ. Res. 2012, 111, 50–55. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.Q.; Li, R.G.; Wang, J.; Liu, X.Y.; Xu, Y.J.; Fang, W.Y.; Chen, X.Z.; Zhang, W.; Wang, X.Z.; Yang, Y.Q. Prevalence and spectrum of GATA5 mutations associated with congenital heart disease. Int. J. Cardiol. 2013, 165, 570–573. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.L.; Dai, N.; Tang, K.; Chen, Y.Q.; Chen, W.; Wang, J.; Zhao, C.M.; Yuan, F.; Qiu, X.B.; Qu, X.K.; et al. GATA5 loss-of-function mutation in familial dilated cardiomyopathy. Int. J. Mol. Med. 2015, 35, 763–770. [Google Scholar] [CrossRef]
- Prendiville, T.W.; Guo, H.; Lin, Z.; Zhou, P.; Stevens, S.M.; He, A.; VanDusen, N.; Chen, J.; Zhong, L.; Wang, D.Z.; et al. Novel Roles of GATA4/6 in the Postnatal Heart Identified through Temporally Controlled, Cardiomyocyte-Specific Gene Inactivation by Adeno-Associated Virus Delivery of Cre Recombinase. PLoS ONE 2015, 10, e0128105. [Google Scholar] [CrossRef]
- Narita, N.; Bielinska, M.; Wilson, D.B. Cardiomyocyte differentiation by GATA-4-deficient embryonic stem cells. Development 1997, 124, 3755–3764. [Google Scholar] [CrossRef]
- Xin, M.; Davis, C.A.; Molkentin, J.D.; Lien, C.L.; Duncan, S.A.; Richardson, J.A.; Olson, E.N. A threshold of GATA4 and GATA6 expression is required for cardiovascular development. Proc. Natl. Acad. Sci. USA 2006, 103, 11189–11194. [Google Scholar] [CrossRef]
- Maitra, M.; Schluterman, M.K.; Nichols, H.A.; Richardson, J.A.; Lo, C.W.; Srivastava, D.; Garg, V. Interaction of Gata4 and Gata6 with Tbx5 is critical for normal cardiac development. Dev. Biol. 2009, 326, 368–377. [Google Scholar] [CrossRef]
- Borok, M.J.; Papaioannou, V.E.; Sussel, L. Unique functions of Gata4 in mouse liver induction and heart development. Dev. Biol. 2016, 410, 213–222. [Google Scholar] [CrossRef] [PubMed]
- Merika, M.; Orkin, S.H. DNA-binding specificity of GATA family transcription factors. Mol. Cell Biol. 1993, 13, 3999–4010. [Google Scholar] [CrossRef] [PubMed]
- Ko, L.J.; Engel, J.D. DNA-binding specificities of the GATA transcription factor family. Mol. Cell Biol. 1993, 13, 4011–4022. [Google Scholar] [CrossRef] [PubMed]
- Sorrentino, R.P.; Gajewski, K.M.; Schulz, R.A. GATA factors in Drosophila heart and blood cell development. Semin. Cell Dev. Biol. 2005, 16, 107–116. [Google Scholar] [CrossRef] [PubMed]
- Davidson, E.H.; Erwin, D.H. Gene regulatory networks and the evolution of animal body plans. Science 2006, 311, 796–800. [Google Scholar] [CrossRef]
- Maitra, M.; Koenig, S.N.; Srivastava, D.; Garg, V. Identification of GATA6 sequence variants in patients with congenital heart defects. Pediatr. Res. 2010, 68, 281–285. [Google Scholar] [CrossRef]
- Nam, Y.S.; Kim, Y.; Joung, H.; Kwon, D.H.; Choe, N.; Min, H.K.; Kim, Y.S.; Kim, H.S.; Kim, D.K.; Cho, Y.K.; et al. Small heterodimer partner blocks cardiac hypertrophy by interfering with GATA6 signaling. Circ. Res. 2014, 115, 493–503. [Google Scholar] [CrossRef]
- Sharma, A.; Wasson, L.K.; Willcox, J.A.; Morton, S.U.; Gorham, J.M.; DeLaughter, D.M.; Neyazi, M.; Schmid, M.; Agarwal, R.; Jang, M.Y.; et al. GATA6 mutations in hiPSCs inform mechanisms for maldevelopment of the heart, pancreas, and diaphragm. Elife 2020, 9. [Google Scholar] [CrossRef]
- van Berlo, J.H.; Aronow, B.J.; Molkentin, J.D. Parsing the roles of the transcription factors GATA-4 and GATA-6 in the adult cardiac hypertrophic response. PLoS ONE 2013, 8, e84591. [Google Scholar] [CrossRef]
- Wada, H.; Hasegawa, K.; Morimoto, T.; Kakita, T.; Yanazume, T.; Sasayama, S. A p300 protein as a coactivator of GATA-6 in the transcription of the smooth muscle-myosin heavy chain gene. J. Biol. Chem. 2000, 275, 25330–25335. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Jin, Y.; Merenick, B.L.; Ding, M.; Fetalvero, K.M.; Wagner, R.J.; Mai, A.; Gleim, S.; Tucker, D.F.; Birnbaum, M.J.; et al. Phosphorylation of GATA-6 is required for vascular smooth muscle cell differentiation after mTORC1 inhibition. Sci. Signal. 2015, 8, ra44. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Sun, W.; Zhu, J.; Yuan, H.; Chu, M.; Wen, B. Modification of cardiac transcription factor Gata6 by SUMO. Biochimie 2020, 170, 212–218. [Google Scholar] [CrossRef] [PubMed]
- Kume, T. The cooperative roles of Foxc1 and Foxc2 in cardiovascular development. Adv. Exp. Med. Biol. 2009, 665, 63–77. [Google Scholar] [CrossRef]
- Jha, R.; Li, D.; Wu, Q.; Ferguson, K.E.; Forghani, P.; Gibson, G.C.; Xu, C. A long non-coding RNA GATA6-AS1 adjacent to GATA6 is required for cardiomyocyte differentiation from human pluripotent stem cells. FASEB J. 2020, 34, 14336–14352. [Google Scholar] [CrossRef]
- Nemer, G.; Qureshi, S.T.; Malo, D.; Nemer, M. Functional analysis and chromosomal mapping of Gata5, a gene encoding a zinc finger DNA-binding protein. Mamm. Genome 1999, 10, 993–999. [Google Scholar] [CrossRef]
- Brewer, A.; Gove, C.; Davies, A.; McNulty, C.; Barrow, D.; Koutsourakis, M.; Farzaneh, F.; Pizzey, J.; Bomford, A.; Patient, R. The human and mouse GATA-6 genes utilize two promoters and two initiation codons. J. Biol. Chem. 1999, 274, 38004–38016. [Google Scholar] [CrossRef]
- MacNeill, C.; Ayres, B.; Laverriere, A.C.; Burch, J.B. Transcripts for functionally distinct isoforms of chicken GATA-5 are differentially expressed from alternative first exons. J. Biol. Chem. 1997, 272, 8396–8401. [Google Scholar] [CrossRef]
- Lou, X.; Deshwar, A.R.; Crump, J.G.; Scott, I.C. Smarcd3b and Gata5 promote a cardiac progenitor fate in the zebrafish embryo. Development 2011, 138, 3113–3123. [Google Scholar] [CrossRef]
- Wen, B.; Yuan, H.; Liu, X.; Wang, H.; Chen, S.; Chen, Z.; de The, H.; Zhou, J.; Zhu, J. GATA5 SUMOylation is indispensable for zebrafish cardiac development. Biochim. Biophys. Acta. Gen. Subj. 2017, 1861, 1691–1701. [Google Scholar] [CrossRef]
- Raid, R.; Krinka, D.; Bakhoff, L.; Abdelwahid, E.; Jokinen, E.; Karner, M.; Malva, M.; Meier, R.; Pelliniemi, L.J.; Ploom, M.; et al. Lack of Gata3 results in conotruncal heart anomalies in mouse. Mech. Dev. 2009, 126, 80–89. [Google Scholar] [CrossRef] [PubMed]
- Epstein, J.A.; Buck, C.A. Transcriptional regulation of cardiac development: Implications for congenital heart disease and DiGeorge syndrome. Pediatr. Res. 2000, 48, 717–724. [Google Scholar] [CrossRef] [PubMed]
- Wilson, D.I.; Britton, S.B.; McKeown, C.; Kelly, D.; Cross, I.E.; Strobel, S.; Scambler, P.J. Noonan’s and DiGeorge syndromes with monosomy 22q11. Arch. Dis. Child. 1993, 68, 187–189. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Wang, Z.; Wu, J.; Liu, M.; Li, M.; Sun, Y.; Huang, W.; Li, Y.; Zhang, Y.; Tang, W.; et al. Endothelial SIRT6 Is Vital to Prevent Hypertension and Associated Cardiorenal Injury Through Targeting Nkx3.2-GATA5 Signaling. Circ. Res. 2019, 124, 1448–1461. [Google Scholar] [CrossRef]
- Haworth, K.E.; Kotecha, S.; Mohun, T.J.; Latinkic, B.V. GATA4 and GATA5 are essential for heart and liver development in Xenopus embryos. BMC Dev. Biol. 2008, 8, 74. [Google Scholar] [CrossRef]
- Grepin, C.; Robitaille, L.; Antakly, T.; Nemer, M. Inhibition of transcription factor GATA-4 expression blocks in vitro cardiac muscle differentiation. Mol. Cell Biol. 1995, 15, 4095–4102. [Google Scholar] [CrossRef]
- Narita, N.; Bielinska, M.; Wilson, D.B. Wild-type endoderm abrogates the ventral developmental defects associated with GATA-4 deficiency in the mouse. Dev. Biol. 1997, 189, 270–274. [Google Scholar] [CrossRef]
- Hu, D.L.; Chen, F.K.; Liu, Y.Q.; Sheng, Y.H.; Yang, R.; Kong, X.Q.; Cao, K.J.; Gu, H.T.; Qian, L.M. GATA-4 promotes the differentiation of P19 cells into cardiac myocytes. Int. J. Mol. Med. 2010, 26, 365–372. [Google Scholar]
- Lough, J.; Sugi, Y. Endoderm and heart development. Dev. Dyn. 2000, 217, 327–342. [Google Scholar] [CrossRef]
- Foley, A.C.; Mercola, M. Heart induction by Wnt antagonists depends on the homeodomain transcription factor Hex. Genes Dev. 2005, 19, 387–396. [Google Scholar] [CrossRef]
- Foley, A.C.; Gupta, R.W.; Guzzo, R.M.; Korol, O.; Mercola, M. Embryonic heart induction. Ann. N. Y. Acad. Sci. 2006, 1080, 85–96. [Google Scholar] [CrossRef] [PubMed]
- Fujikura, J.; Yamato, E.; Yonemura, S.; Hosoda, K.; Masui, S.; Nakao, K.; Miyazaki Ji, J.; Niwa, H. Differentiation of embryonic stem cells is induced by GATA factors. Genes Dev. 2002, 16, 784–789. [Google Scholar] [CrossRef] [PubMed]
- Afouda, B.A.; Ciau-Uitz, A.; Patient, R. GATA4, 5 and 6 mediate TGFbeta maintenance of endodermal gene expression in Xenopus embryos. Development 2005, 132, 763–774. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Liu, Y.; Asakura, M.; Inoue, H.; Nakamura, T.; Sano, M.; Niu, Z.; Chen, M.; Schwartz, R.J.; Schneider, M.D. Sox17 is essential for the specification of cardiac mesoderm in embryonic stem cells. Proc. Natl. Acad. Sci. USA 2007, 104, 3859–3864. [Google Scholar] [CrossRef] [PubMed]
- Foley, A.C.; Korol, O.; Timmer, A.M.; Mercola, M. Multiple functions of Cerberus cooperate to induce heart downstream of Nodal. Dev. Biol. 2007, 303, 57–65. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Takeuchi, J.K.; Bruneau, B.G. Directed transdifferentiation of mouse mesoderm to heart tissue by defined factors. Nature 2009, 459, 708–711. [Google Scholar] [CrossRef]
- Zhou, L.; Liu, Y.; Lu, L.; Lu, X.; Dixon, R.A. Cardiac gene activation analysis in mammalian non-myoblasic cells by Nkx2-5, Tbx5, Gata4 and Myocd. PLoS ONE 2012, 7, e48028. [Google Scholar] [CrossRef]
- Christoforou, N.; Chellappan, M.; Adler, A.F.; Kirkton, R.D.; Wu, T.; Addis, R.C.; Bursac, N.; Leong, K.W. Transcription factors MYOCD, SRF, Mesp1 and SMARCD3 enhance the cardio-inducing effect of GATA4, TBX5, and MEF2C during direct cellular reprogramming. PLoS ONE 2013, 8, e63577. [Google Scholar] [CrossRef]
- Fu, J.D.; Stone, N.R.; Liu, L.; Spencer, C.I.; Qian, L.; Hayashi, Y.; Delgado-Olguin, P.; Ding, S.; Bruneau, B.G.; Srivastava, D. Direct reprogramming of human fibroblasts toward a cardiomyocyte-like state. Stem Cell Rep. 2013, 1, 235–247. [Google Scholar] [CrossRef]
- Wang, L.; Liu, Z.; Yin, C.; Asfour, H.; Chen, O.; Li, Y.; Bursac, N.; Liu, J.; Qian, L. Stoichiometry of Gata4, Mef2c, and Tbx5 influences the efficiency and quality of induced cardiac myocyte reprogramming. Circ. Res. 2015, 116, 237–244. [Google Scholar] [CrossRef]
- Ieda, M.; Fu, J.D.; Delgado-Olguin, P.; Vedantham, V.; Hayashi, Y.; Bruneau, B.G.; Srivastava, D. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell 2010, 142, 375–386. [Google Scholar] [CrossRef] [PubMed]
- Song, K.; Nam, Y.J.; Luo, X.; Qi, X.; Tan, W.; Huang, G.N.; Acharya, A.; Smith, C.L.; Tallquist, M.D.; Neilson, E.G.; et al. Heart repair by reprogramming non-myocytes with cardiac transcription factors. Nature 2012, 485, 599–604. [Google Scholar] [CrossRef] [PubMed]
- Jayawardena, T.M.; Egemnazarov, B.; Finch, E.A.; Zhang, L.; Payne, J.A.; Pandya, K.; Zhang, Z.; Rosenberg, P.; Mirotsou, M.; Dzau, V.J. MicroRNA-mediated in vitro and in vivo direct reprogramming of cardiac fibroblasts to cardiomyocytes. Circ. Res. 2012, 110, 1465–1473. [Google Scholar] [CrossRef] [PubMed]
- Protze, S.; Khattak, S.; Poulet, C.; Lindemann, D.; Tanaka, E.M.; Ravens, U. A new approach to transcription factor screening for reprogramming of fibroblasts to cardiomyocyte-like cells. J. Mol. Cell Cardiol. 2012, 53, 323–332. [Google Scholar] [CrossRef]
- Srivastava, D.; Ieda, M. Critical factors for cardiac reprogramming. Circ. Res. 2012, 111, 5–8. [Google Scholar] [CrossRef] [PubMed]
- Wada, R.; Muraoka, N.; Inagawa, K.; Yamakawa, H.; Miyamoto, K.; Sadahiro, T.; Umei, T.; Kaneda, R.; Suzuki, T.; Kamiya, K.; et al. Induction of human cardiomyocyte-like cells from fibroblasts by defined factors. Proc. Natl. Acad. Sci. USA 2013, 110, 12667–12672. [Google Scholar] [CrossRef]
- Smagulova, F.O.; Manuylov, N.L.; Leach, L.L.; Tevosian, S.G. GATA4/FOG2 transcriptional complex regulates Lhx9 gene expression in murine heart development. BMC Dev. Biol. 2008, 8, 67. [Google Scholar] [CrossRef]
- Zhou, B.; Ma, Q.; Kong, S.W.; Hu, Y.; Campbell, P.H.; McGowan, F.X.; Ackerman, K.G.; Wu, B.; Zhou, B.; Tevosian, S.G.; et al. Fog2 is critical for cardiac function and maintenance of coronary vasculature in the adult mouse heart. J. Clin. Investig. 2009, 119, 1462–1476. [Google Scholar] [CrossRef]
- Garnatz, A.S.; Gao, Z.; Broman, M.; Martens, S.; Earley, J.U.; Svensson, E.C. FOG-2 mediated recruitment of the NuRD complex regulates cardiomyocyte proliferation during heart development. Dev. Biol. 2014, 395, 50–61. [Google Scholar] [CrossRef]
- Zhang, W.; Shen, L.; Deng, Z.; Ding, Y.; Mo, X.; Xu, Z.; Gao, Q.; Yi, L. Novel missense variants of ZFPM2/FOG2 identified in conotruncal heart defect patients do not impair interaction with GATA4. PLoS ONE 2014, 9, e102379. [Google Scholar] [CrossRef]
- Carter, D.R.; Buckle, A.D.; Tanaka, K.; Perdomo, J.; Chong, B.H. Art27 interacts with GATA4, FOG2 and NKX2.5 and is a novel co-repressor of cardiac genes. PLoS ONE 2014, 9, e95253. [Google Scholar] [CrossRef] [PubMed]
- Gallagher, J.M.; Komati, H.; Roy, E.; Nemer, M.; Latinkic, B.V. Dissociation of cardiogenic and postnatal myocardial activities of GATA4. Mol. Cell. Biol. 2012, 32, 2214–2223. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Liang, Q.; Wiese, R.J.; Bueno, O.F.; Dai, Y.S.; Markham, B.E.; Molkentin, J.D. The transcription factor GATA4 is activated by extracellular signal-regulated kinase 1- and 2-mediated phosphorylation of serine 105 in cardiomyocytes. Mol. Cell. Biol. 2001, 21, 7460–7469. [Google Scholar] [CrossRef] [PubMed]
- Gallagher, J.M.; Yamak, A.; Kirilenko, P.; Black, S.; Bochtler, M.; Lefebvre, C.; Nemer, M.; Latinkic, B.V. Carboxy terminus of GATA4 transcription factor is required for its cardiogenic activity and interaction with CDK4. Mech. Dev. 2014, 134, 31–41. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Rojas, A.; Kong, S.W.; Agarwal, P.; Gilliss, B.; Pu, W.T.; Black, B.L. GATA4 is a direct transcriptional activator of cyclin D2 and Cdk4 and is required for cardiomyocyte proliferation in anterior heart field-derived myocardium. Mol. Cell. Biol. 2008, 28, 5420–5431. [Google Scholar] [CrossRef]
- Ma, C.X.; Song, Y.L.; Xiao, L.; Xue, L.X.; Li, W.J.; Laforest, B.; Komati, H.; Wang, W.P.; Jia, Z.Q.; Zhou, C.Y.; et al. EGF is required for cardiac differentiation of P19CL6 cells through interaction with GATA-4 in a time- and dose-dependent manner. Cell. Mol. Life Sci. 2015, 72, 2005–2022. [Google Scholar] [CrossRef]
- van Berlo, J.H.; Elrod, J.W.; Aronow, B.J.; Pu, W.T.; Molkentin, J.D. Serine 105 phosphorylation of transcription factor GATA4 is necessary for stress-induced cardiac hypertrophy in vivo. Proc. Natl. Acad. Sci. USA 2011, 108, 12331–12336. [Google Scholar] [CrossRef]
- Yamak, A.; Temsah, R.; Maharsy, W.; Caron, S.; Paradis, P.; Aries, A.; Nemer, M. Cyclin D2 rescues size and function of GATA4 haplo-insufficient hearts. Am. J. Physiol. Heart Circ. Physiol. 2012, 303, H1057–H1066. [Google Scholar] [CrossRef][Green Version]
- Katanasaka, Y.; Suzuki, H.; Sunagawa, Y.; Hasegawa, K.; Morimoto, T. Regulation of Cardiac Transcription Factor GATA4 by Post-Translational Modification in Cardiomyocyte Hypertrophy and Heart Failure. Int. Heart J. 2016, 57, 672–675. [Google Scholar] [CrossRef]
- Xu, Y.; Liang, C.; Luo, Y.; Xing, W.; Zhang, T. Possible mechanism of GATA4 inhibiting myocardin activity during cardiac hypertrophy. J. Cell Biochem. 2019, 120, 9047–9055. [Google Scholar] [CrossRef]
- Zhou, P.; He, A.; Pu, W.T. Regulation of GATA4 transcriptional activity in cardiovascular development and disease. Curr. Top. Dev. Biol. 2012, 100, 143–169. [Google Scholar] [CrossRef] [PubMed]
- Walsh, E.P.; Cecchin, F. Arrhythmias in adult patients with congenital heart disease. Circulation 2007, 115, 534–545. [Google Scholar] [CrossRef] [PubMed]
- Schlesinger, J.; Schueler, M.; Grunert, M.; Fischer, J.J.; Zhang, Q.; Krueger, T.; Lange, M.; Tonjes, M.; Dunkel, I.; Sperling, S.R. The cardiac transcription network modulated by Gata4, Mef2a, Nkx2.5, Srf, histone modifications, and microRNAs. PLoS Genet. 2011, 7, e1001313. [Google Scholar] [CrossRef] [PubMed]
- He, A.; Gu, F.; Hu, Y.; Ma, Q.; Ye, L.Y.; Akiyama, J.A.; Visel, A.; Pennacchio, L.A.; Pu, W.T. Dynamic GATA4 enhancers shape the chromatin landscape central to heart development and disease. Nat. Commun. 2014, 5, 4907. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Basta, T.; Klymkowsky, M.W. SOX7 and SOX18 are essential for cardiogenesis in Xenopus. Dev. Dyn. 2005, 234, 878–891. [Google Scholar] [CrossRef] [PubMed]
| GATA Factors | Cardiac Defects Associated with Human Mutations | Mouse Phenotypes | Cardiac Expression | Biological Functions |
|---|---|---|---|---|
| GATA6/Gata6 | -Atrial septal defects -Atrioventricular septal defects -Bicuspid aortic valves -Dilated cardiomyopathy -Double outlet left ventricle -Double outlet right ventricle -Tetralogy of Fallot -Ventricular septal defects | Embryonic lethal at E5.5–E7 | Yes | -Cardiac hypertrophy [13] -Cardiac morphogenesis [14] -Outflow tract development [15,16,17,18] |
| GATA5/Gata5 | -Atrial septal defects -Atrioventricular septal defects -Bicuspid aortic valves -Dilated cardiomyopathy -Double outlet right ventricle -Hypertrophic cardiomyopathy -Tetralogy of Fallot -Ventricular septal defects | Viable and fertile | Yes | -Endocardial cell development [19] -Vascular endothelial homeostasis [20] |
| GATA4/Gata4 | -Atrial septal defects -Atrioventricular septal defects -Bicuspid aortic valves -Dilated cardiomyopathy -Double inlet left ventricle -Double outlet right ventricle -Hypertrophic cardiomyopathy -Tetralogy of Fallot -Ventricular septal defects | Embryonic lethal at E9.5 | Yes | -Cardiomyocyte proliferation, differentiation and hypertrophy [13] -Endocardial cell proliferation [21] -Heart tube formation and ventral morphogenesis [22,23] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Afouda, B.A. Towards Understanding the Gene-Specific Roles of GATA Factors in Heart Development: Does GATA4 Lead the Way? Int. J. Mol. Sci. 2022, 23, 5255. https://doi.org/10.3390/ijms23095255
Afouda BA. Towards Understanding the Gene-Specific Roles of GATA Factors in Heart Development: Does GATA4 Lead the Way? International Journal of Molecular Sciences. 2022; 23(9):5255. https://doi.org/10.3390/ijms23095255
Chicago/Turabian StyleAfouda, Boni A. 2022. "Towards Understanding the Gene-Specific Roles of GATA Factors in Heart Development: Does GATA4 Lead the Way?" International Journal of Molecular Sciences 23, no. 9: 5255. https://doi.org/10.3390/ijms23095255
APA StyleAfouda, B. A. (2022). Towards Understanding the Gene-Specific Roles of GATA Factors in Heart Development: Does GATA4 Lead the Way? International Journal of Molecular Sciences, 23(9), 5255. https://doi.org/10.3390/ijms23095255






