A Blueprint of Microstructures and Stage-Specific Transcriptome Dynamics of Cuticle Formation in Bombyx mori
Abstract
:1. Introduction
2. Results
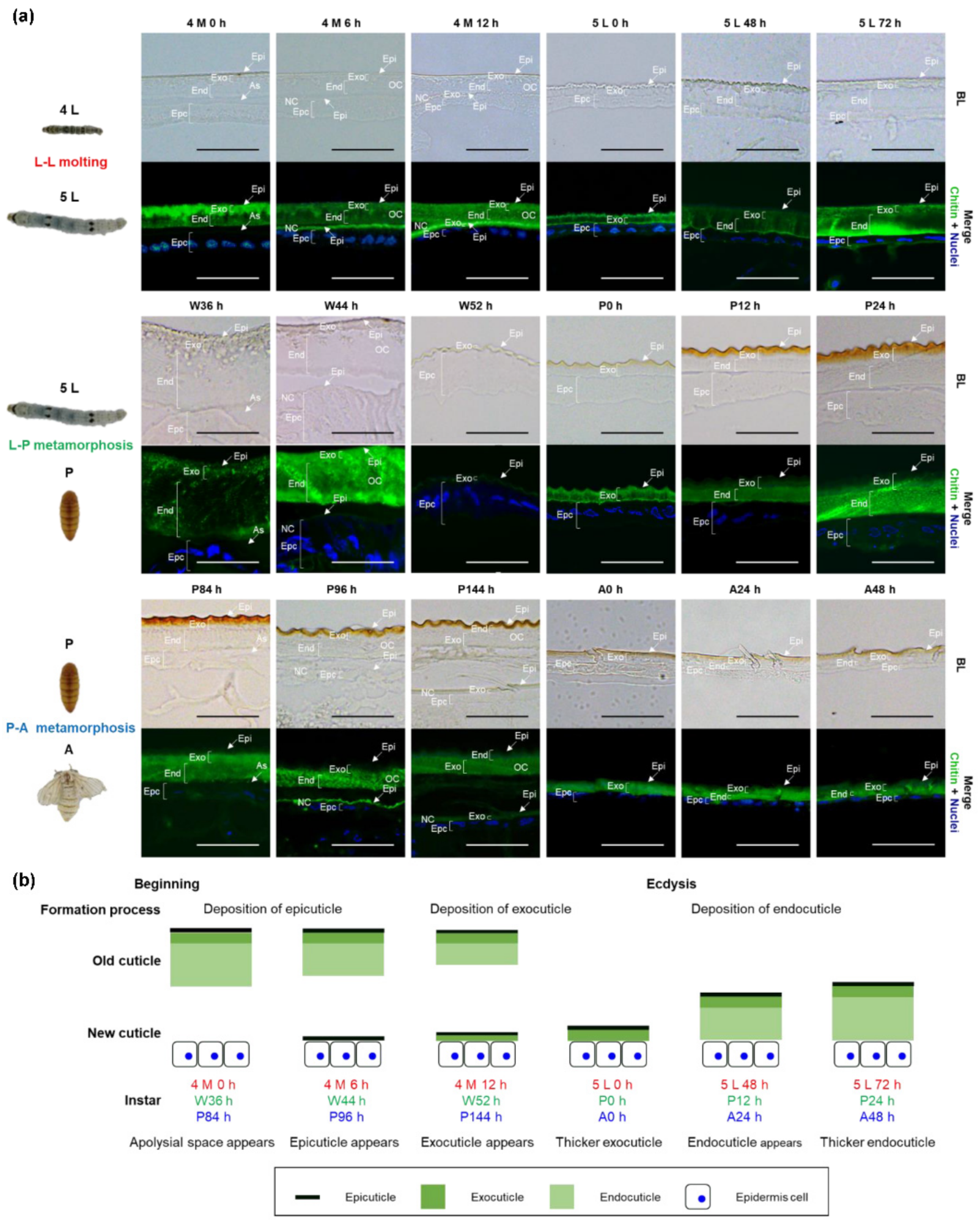
2.1. Microstructures and Formation Processes of Larval, Pupal, and Adult Cuticles in B. mori
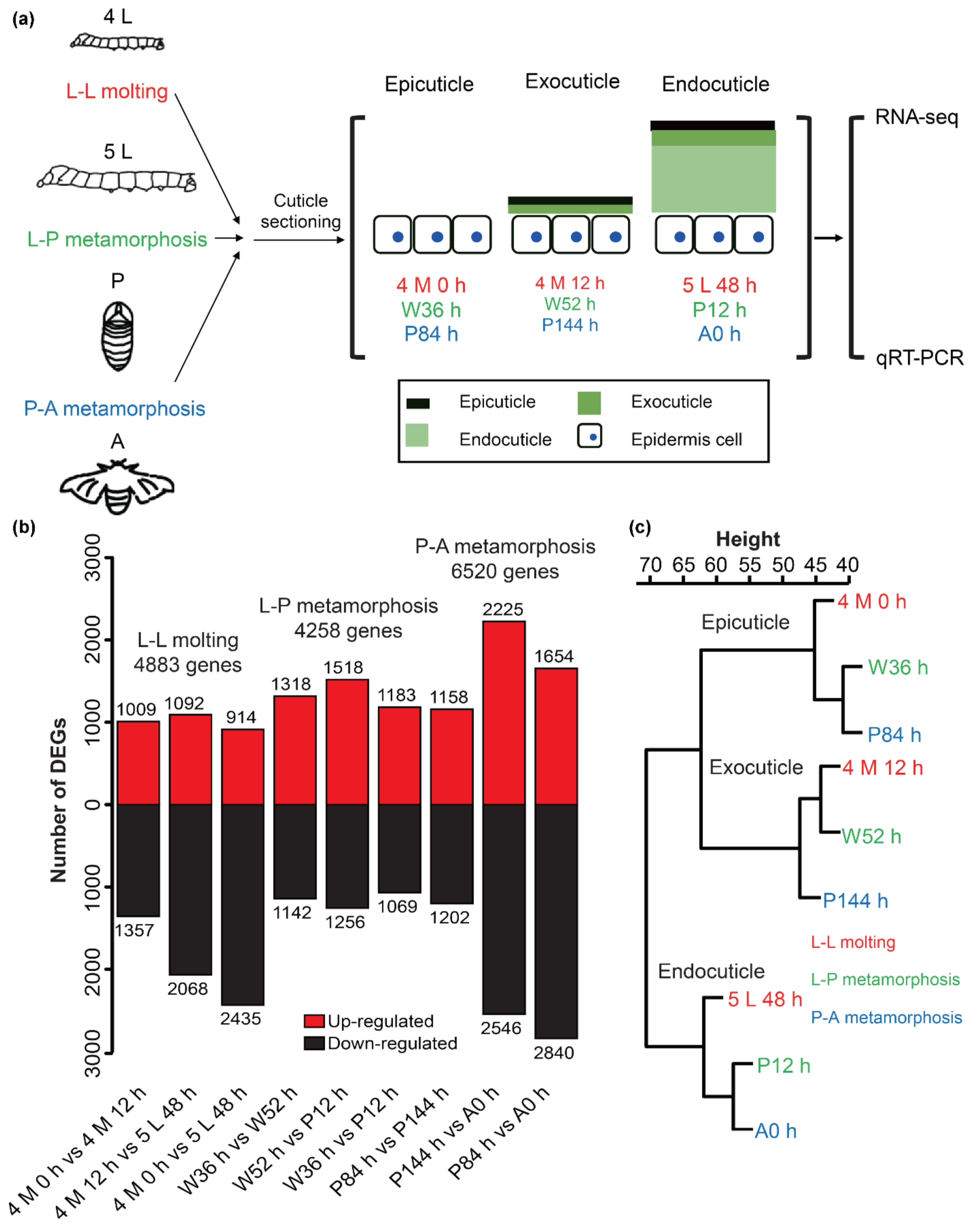
2.2. Transcriptome Analysis during Larval, Pupal, and Adult Cuticle Formations in B. mori
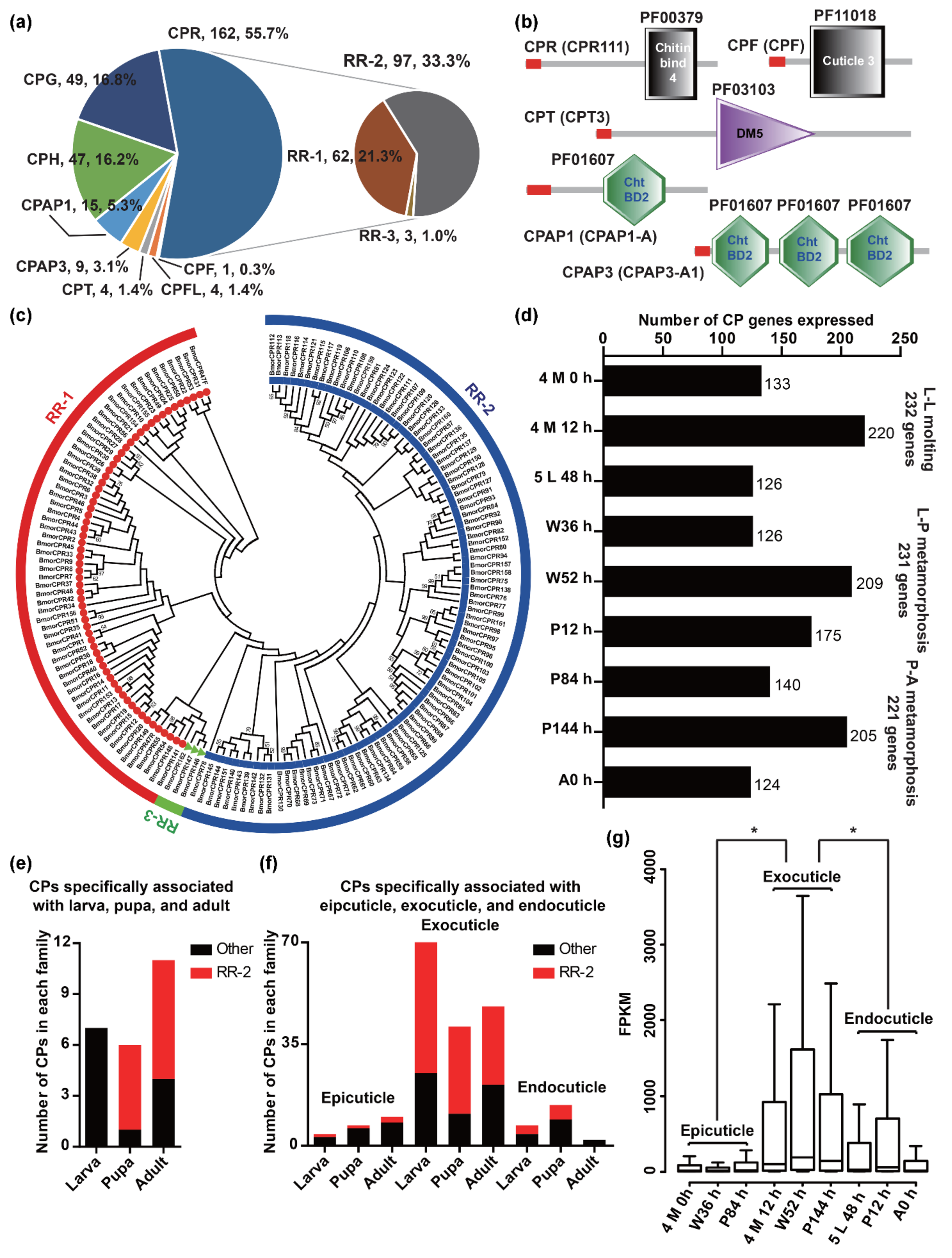
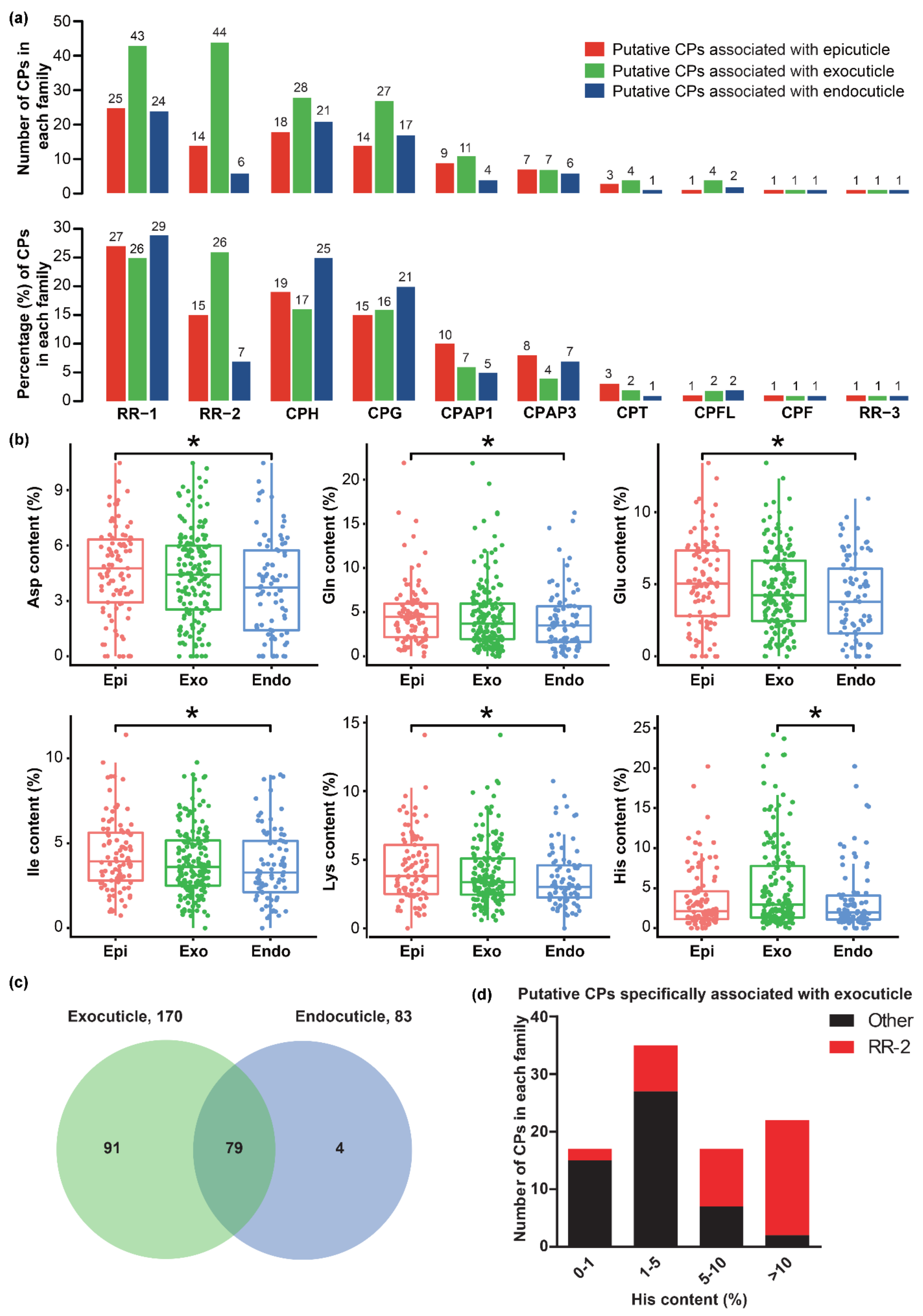
2.3. CPs associated with Cuticle Formation in B. mori
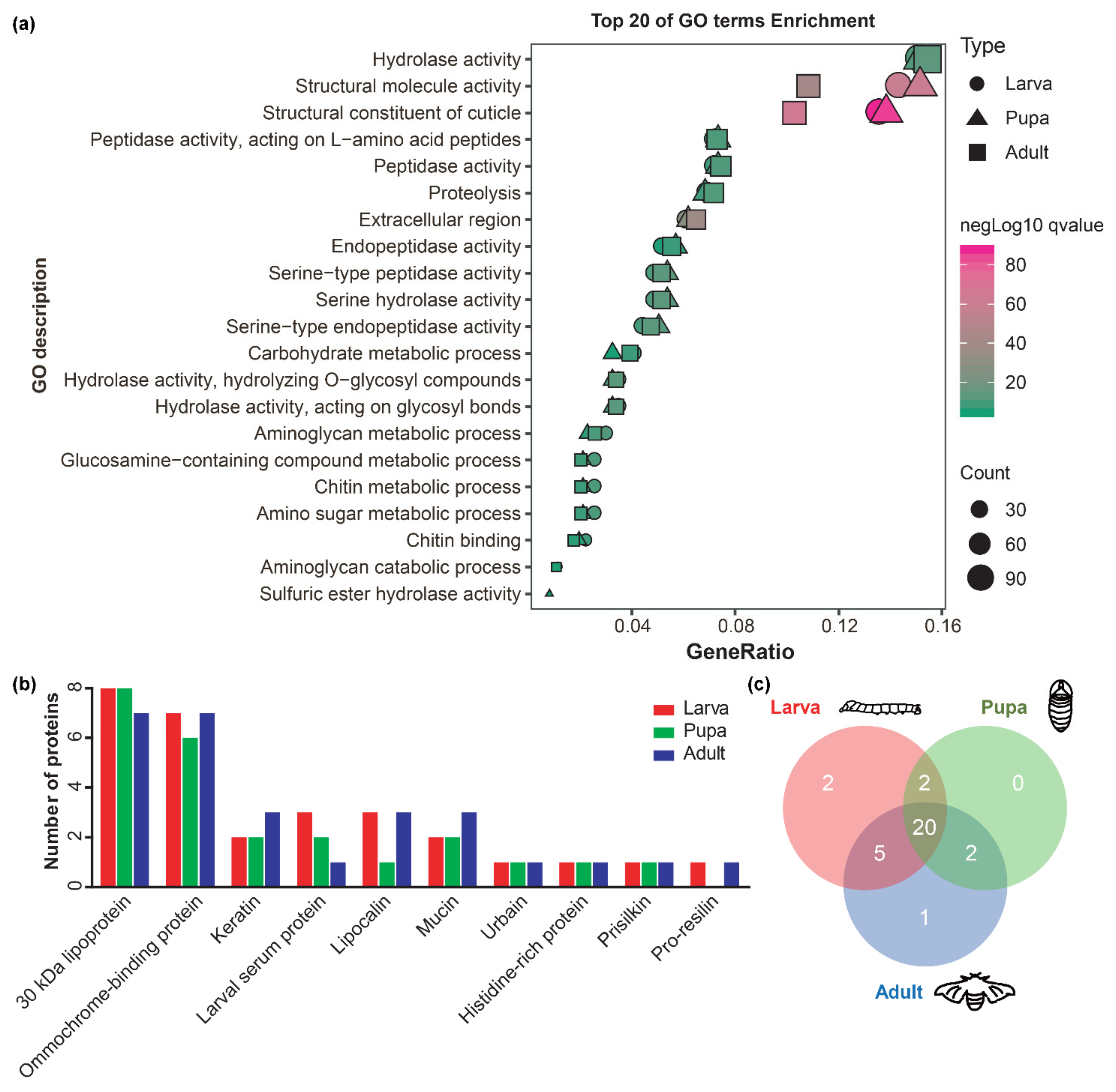
2.4. Other protein Components Involved in Cuticle Formation in B. mori
2.5. BmorCPAP1-H Is Involved in Cuticle Formation in B. mori
3. Discussion
4. Materials and Methods
4.1. Insects
4.2. Sectioning, Staining, and Cuticular Structure Analysis
4.3. Sample Collection, Library Preparation, Sequencing, and Analysis of Transcriptome
4.4. qRT-PCR Confirmation of Transcriptome Data
4.5. Identification, Classification, Analysis of CPs in B. mori
4.6. Identification of Other Protein Components Associated with Cuticle Formation
4.7. Spatiotemporal Expression Pattern
4.8. CRISPR/Cas9-Mediated Gene Knockout
4.9. Other Analysis Tools
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cohen, E.; Moussian, B. Extracellular Composite Matrices in Arthropods, 1st ed.; Springer: Cham, Switzerland, 2016; pp. 1–22. [Google Scholar]
- Xiong, G.; Tong, X.; Yan, Z.; Hu, H.; Duan, X.; Li, C.; Han, M.; Lu, C.; Dai, F. Cuticular protein defective bamboo mutant of Bombyx mori is sensitive to environmental stresses. Pestic. Biochem. Phys. 2018, 148, 111–115. [Google Scholar] [CrossRef] [PubMed]
- Nation, J.L. Integument: Structure and function. In Encyclopedia of Entomology, 1st ed.; Capinera, J.L., Ed.; Springer: Dordrecht, The Netherlands, 2004; pp. 1200–1203. [Google Scholar]
- Qiao, L.; Xiong, G.; Wang, R.-X.; He, S.-Z.; Chen, J.; Tong, X.-L.; Hu, H.; Li, C.-L.; Gai, T.-T.; Xin, Y.-Q.; et al. Mutation of a cuticular protein, BmorCPR2, alters larval body shape and adaptability in silkworm, Bombyx mori. Genetics 2014, 196, 1103–1115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ascough, J.C.; Fathelrahman, E.M.; McMaster, G.S. Insect pest models and insecticide application. In Encyclopedia of Ecology, 1st ed.; Jorgensen, S.E., Ed.; Elsevier: Fort Collins, CO, USA, 2008; pp. 1978–1985. [Google Scholar]
- Locke, M. Pore canals and related structures in insect cuticle. J. Biophys. Biochem. Cytol. 1961, 10, 589–618. [Google Scholar] [CrossRef] [PubMed]
- Andersen, S.O. Characteristic properties of proteins from pre-ecdysial cuticle of larvae and pupae of the mealworm Tenebrio molitor. Insect. Biochem. Mol. Biol. 2002, 32, 1077–1087. [Google Scholar] [CrossRef]
- Charles, J.P. The regulation of expression of insect cuticle protein genes. Insect. Biochem. Mol. Biol. 2010, 40, 205–213. [Google Scholar] [CrossRef]
- Shahin, R.; Iwanaga, M.; Kawasaki, H. Expression profiles of cuticular protein genes in wing tissues during pupal to adult stages and the deduced adult cuticular structure of Bombyx mori. Gene 2018, 646, 181–194. [Google Scholar] [CrossRef]
- Xiong, G.; Tong, X.; Gai, T.; Li, C.; Qiao, L.; Monteiro, A.; Hu, H.; Han, M.-J.; Ding, X.; Wu, S.; et al. Body shape and coloration of silkworm larvae are influenced by a novel cuticular protein. Genetics 2017, 207, 1053–1066. [Google Scholar] [CrossRef] [Green Version]
- Vannini, L.; Willis, J.H. Localization of RR-1 and RR-2 cuticular proteins within the cuticle of Anopheles gambiae. Arthropod. Struct. Dev. 2017, 46, 13–29. [Google Scholar] [CrossRef] [Green Version]
- Yu, R.R.; Zhang, R.; Liu, W.M.; Zhao, X.M.; Zhu, K.Y.; Moussian, B.; Zhang, J.Z. The DOMON domain protein LmKnk contributes to correct chitin content, pore canal formation and lipid deposition in the cuticle of Locusta migratoria during moulting. Insect. Mol. Biol. 2022, 31, 127–138. [Google Scholar] [CrossRef]
- Pan, P.-L.; Ye, Y.-X.; Lou, Y.-H.; Lu, J.-B.; Cheng, C.; Shen, Y.; Moussian, B.; Zhang, C.-X. A comprehensive omics analysis and functional survey of cuticular proteins in the brown planthopper. Proc. Natl. Acad. Sci. USA 2018, 115, 5175–5180. [Google Scholar] [CrossRef] [Green Version]
- Jasrapuria, S.; Specht, C.A.; Kramer, K.J.; Beeman, R.W.; Muthukrishnan, S. Gene families of cuticular proteins analogous to peritrophins (CPAPs) in Tribolium castaneum have diverse functions. PLoS ONE 2012, 7, e49844. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kiguchi, K.; Agui, N. Ecdysteroid levels and developmental events during larval moulting in the silkworm, Bombyx mori. J. Insect. Physiol. 1981, 27, 805–812. [Google Scholar] [CrossRef]
- Moussian, B.; Seifarth, C.; Müller, U.; Berger, J.; Schwarz, H. Cuticle differentiation during Drosophila embryogenesis. Arthropod. Struct. Dev. 2006, 35, 137–152. [Google Scholar] [CrossRef] [PubMed]
- Sobala, L.F.; Adler, P.N. The gene expression program for the formation of wing cuticle in Drosophila. PLoS Genet. 2016, 12, e1006100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Noh, M.Y.; Muthukrishnan, S.; Kramer, K.J.; Arakane, Y. Development and ultrastructure of the rigid dorsal and flexible ventral cuticles of the elytron of the red flour beetle, Tribolium castaneum. Insect. Biochem. Mol. Biol. 2017, 91, 21–33. [Google Scholar] [CrossRef]
- Locke, M. The structure and formation of the cuticulin layer in the epicuticle of an insect, Calpodes ethlius (Lepidoptera, Hesperiidae). J. Morphol. 1966, 118, 461–494. [Google Scholar] [CrossRef]
- Leopold, R.A.; Newman, S.M.; Helgeson, G. A Comparison of cuticle dposition during the preeclosion and posteclosion stages of the adult weevil, Anthonomus Grandis Boheman (Coleoptera, Curculionidae). Int. J. Insect. Morphol. 1992, 21, 37–62. [Google Scholar] [CrossRef]
- Futahashi, R.; Okamoto, S.; Kawasaki, H.; Zhong, Y.-S.; Iwanaga, M.; Mita, K.; Fujiwara, H. Genome-wide identification of cuticular protein genes in the silkworm, Bombyx mori. Insect. Biochem. Mol. Biol. 2008, 38, 1138–1146. [Google Scholar] [CrossRef]
- Liu, J.; Chen, Z.; Xiao, Y.; Asano, T.; Li, S.; Peng, L.; Chen, E.; Zhang, J.; Li, W.; Zhang, Y.; et al. Lepidopteran wing scales contain abundant cross-linked film-forming histidine-rich cuticular proteins. Commun. Biol. 2021, 4, 491. [Google Scholar] [CrossRef]
- Liang, J.B.; Wang, T.; Xiang, Z.H.; He, N.J. Tweedle cuticular protein BmCPT1 is involved in innate immunity by participating in recognition of Escherichia coli. Insect. Biochem. Mol. Biol. 2015, 58, 76–88. [Google Scholar] [CrossRef]
- Fristrom, D.; Doctor, J.; Fristrom, J.W. Procuticle proteins and chitin-like material in the inner epicuticle of the Drosophila pupal cuticle. Tissue Cell 1986, 18, 531–543. [Google Scholar] [CrossRef]
- Iconomidou, V.A.; Willis, J.H.; Hamodrakas, S.J. Unique features of the structural model of ’hard’ cuticle proteins: Implications for chitin–protein interactions and cross-linking in cuticle. Insect. Biochem. Mol. Biol. 2005, 35, 553–560. [Google Scholar] [CrossRef] [PubMed]
- Koeppe, J.K.; Gilbert, L.I. Immunochemical evidence for the transport of haemolymph protein into the cuticle of Manduca sexta. J. Insect. Physiol. 1973, 19, 615–621. [Google Scholar] [CrossRef]
- Knig, M.; Agrawal, O.P.; Schenkel, H.; Scheller, K. Incorporation of calliphorin into the cuticle of the developing blowfly, Calliphora vicina. Rouxs. Arch. Dev. Biol. 1986, 195, 296–301. [Google Scholar] [CrossRef]
- Kong, Y.; Jing, G.; Yan, Z.; Li, C.; Gong, N.; Zhu, F.; Li, D.; Zhang, Y.; Zheng, G.; Wang, H.; et al. Cloning and characterization of Prisilkin-39, a novel matrix protein serving a dual role in the prismatic layer formation from the oyster Pinctada fucata. J. Biol. Chem. 2009, 284, 10841–10854. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.F.; Zhou, Y.Q.; Chen, Y.R.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef] [Green Version]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Yan, Z.W.; He, Z.B.; Yan, Z.T.; Si, F.L.; Zhou, Y.; Chen, B. Genome-wide and expression-profiling analyses suggest the main cytochrome P450 genes related to pyrethroid resistance in the malaria vector, Anopheles sinensis (Diptera Culicidae). Pest. Manag. Sci. 2018, 74, 1810–1820. [Google Scholar] [CrossRef]
- Dai, Z.; Ren, J.; Tong, X.; Hu, H.; Lu, K.; Dai, F.; Han, M.-J. The landscapes of full-length transcripts and splice isoforms as well as transposons exonization in the lepidopteran model system, Bombyx mori. Front. Genet. 2021, 12, 704162. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.-W.; Wang, L.-L.; Tang, X.; Dong, Z.; Guo, P.-C.; Zhao, D.-C.; Xia, Q.-Y.; Zhao, P. Proteomic analysis of Bombyx mori molting fluid: Insights into the molting process. J. Proteom. 2018, 173, 115–125. [Google Scholar] [CrossRef] [PubMed]
- Quevillon, E.; Silventoinen, V.; Pillai, S.; Harte, N.; Mulder, N.; Apweiler, R.; Lopez, R. InterProScan: Protein domains identifier. Nucleic Acids Res. 2005, 33, W116–W120. [Google Scholar] [CrossRef] [PubMed] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yan, Z.; Tong, X.; Xiong, G.; Yang, W.; Lu, K.; Yuan, Y.; Han, M.; Hu, H.; Wei, W.; Dai, F. A Blueprint of Microstructures and Stage-Specific Transcriptome Dynamics of Cuticle Formation in Bombyx mori. Int. J. Mol. Sci. 2022, 23, 5155. https://doi.org/10.3390/ijms23095155
Yan Z, Tong X, Xiong G, Yang W, Lu K, Yuan Y, Han M, Hu H, Wei W, Dai F. A Blueprint of Microstructures and Stage-Specific Transcriptome Dynamics of Cuticle Formation in Bombyx mori. International Journal of Molecular Sciences. 2022; 23(9):5155. https://doi.org/10.3390/ijms23095155
Chicago/Turabian StyleYan, Zhengwen, Xiaoling Tong, Gao Xiong, Weike Yang, Kunpeng Lu, Yajie Yuan, Minjin Han, Hai Hu, Wei Wei, and Fangyin Dai. 2022. "A Blueprint of Microstructures and Stage-Specific Transcriptome Dynamics of Cuticle Formation in Bombyx mori" International Journal of Molecular Sciences 23, no. 9: 5155. https://doi.org/10.3390/ijms23095155
APA StyleYan, Z., Tong, X., Xiong, G., Yang, W., Lu, K., Yuan, Y., Han, M., Hu, H., Wei, W., & Dai, F. (2022). A Blueprint of Microstructures and Stage-Specific Transcriptome Dynamics of Cuticle Formation in Bombyx mori. International Journal of Molecular Sciences, 23(9), 5155. https://doi.org/10.3390/ijms23095155





