Solvent Vibrations as a Proxy of the Telomere G-Quadruplex Rearrangements across Thermal Unfolding
Abstract
1. Introduction
2. Results
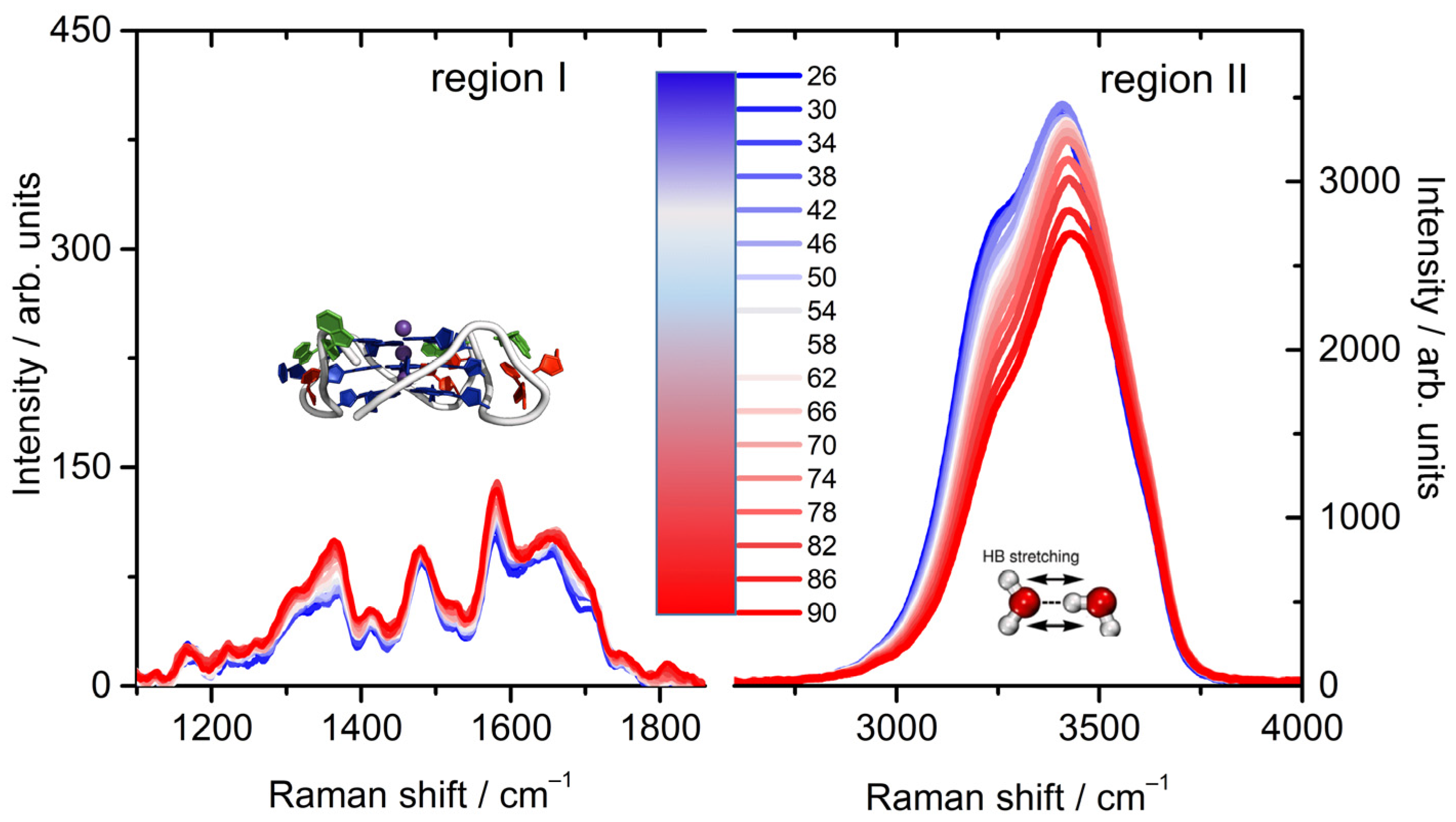
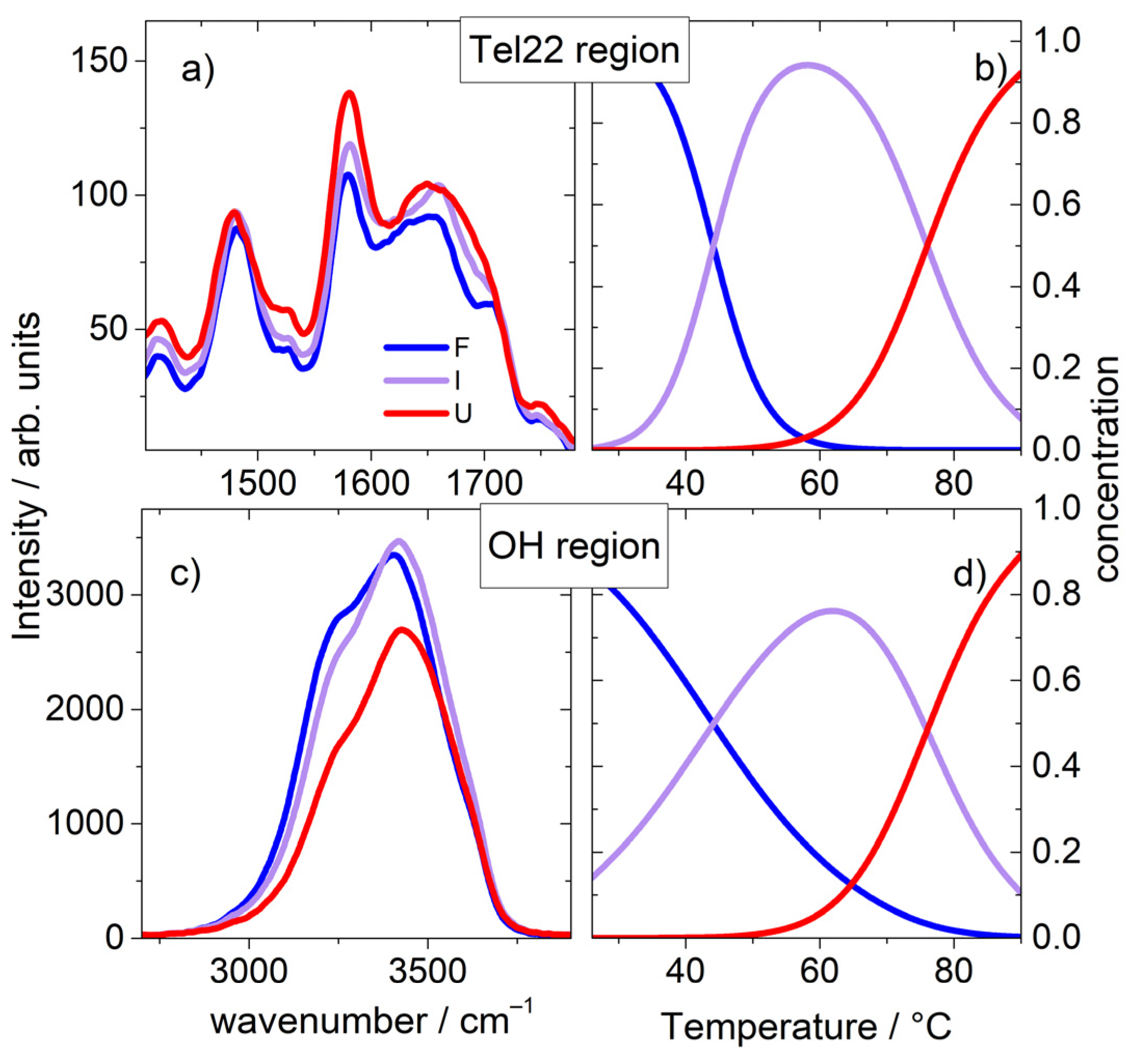
2.1. A Broad Band UVRR Data Treatment
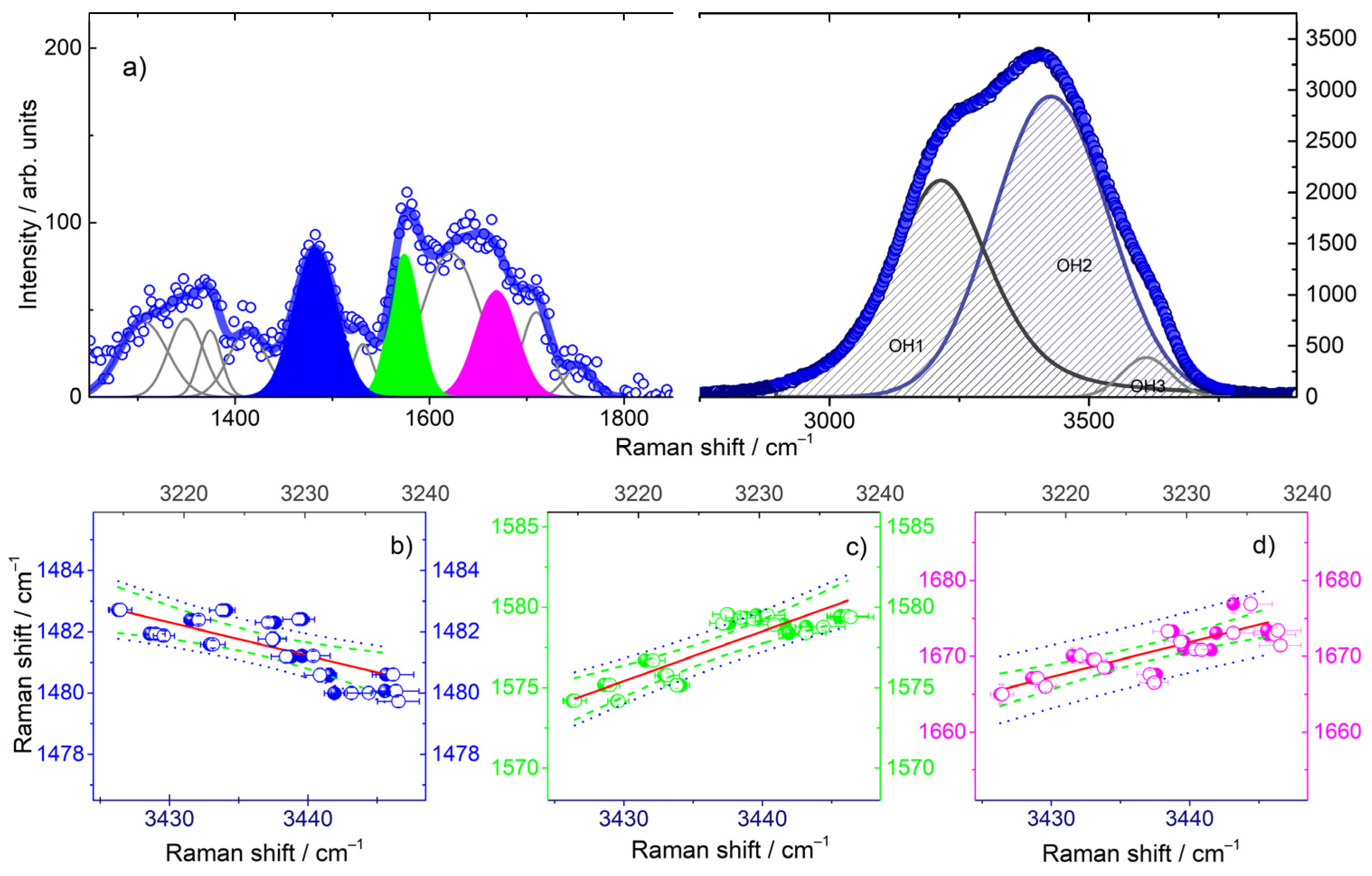
2.2. The Fingerprint Region of Tel22 Cromophores
2.3. The OH Stretching Band
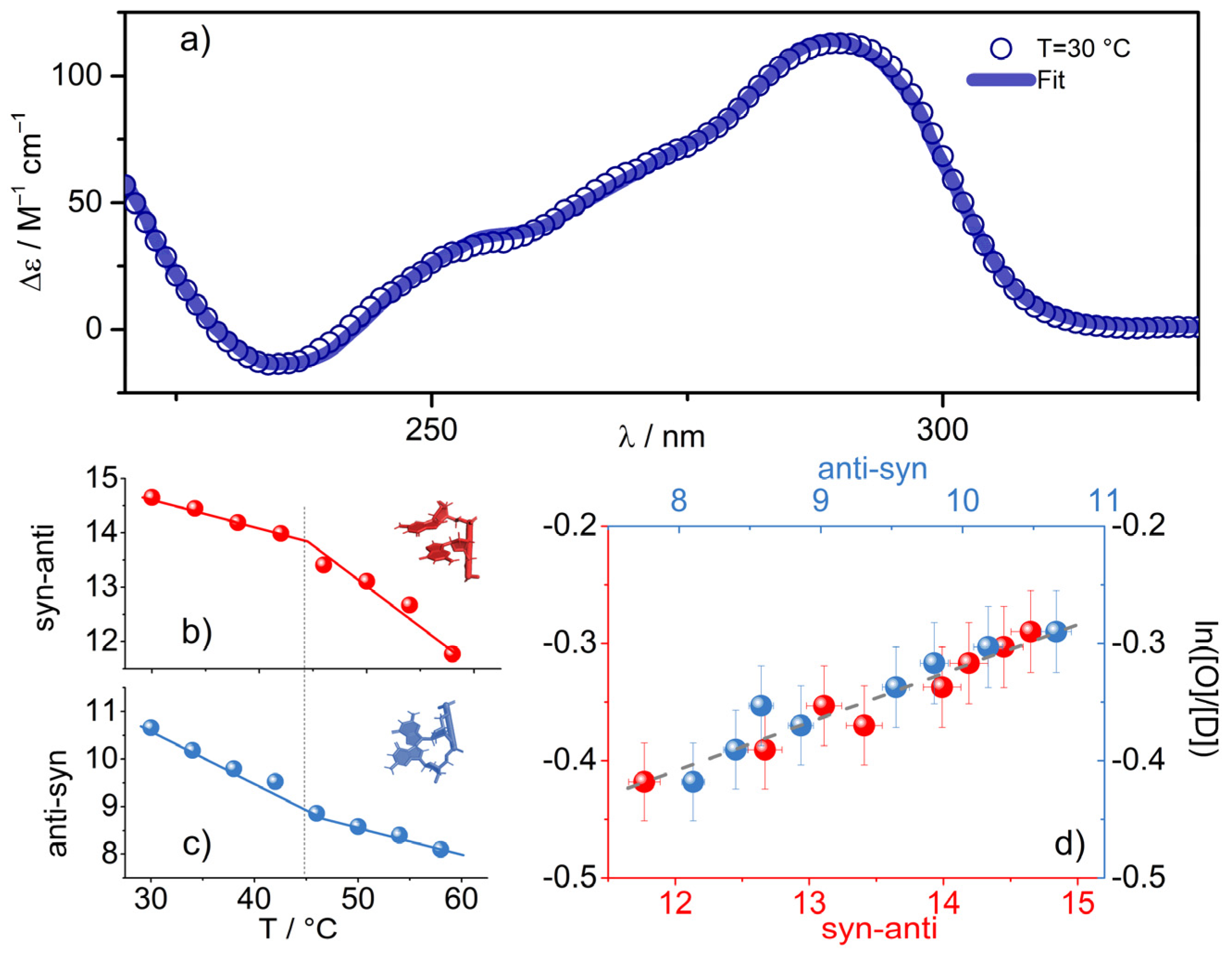
2.4. The Link with the Secondary Structure
3. Discussion
4. Materials and Methods
4.1. UVRR Experiments
4.2. CD Experiments
4.3. SVD Details
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sen, D.; Gilbert, W. Formation of Parallel Four-Stranded Complexes by Guanine-Rich Motifs in DNA and Its Implications for Meiosis. Nature 1988, 334, 364–366. [Google Scholar] [CrossRef] [PubMed]
- Neidle, S.; Parkinson, G. Telomere Maintenance as a Target for Anticancer Drug Discovery. Nat. Rev. Drug Discov. 2002, 1, 383–393. [Google Scholar] [CrossRef] [PubMed]
- Biffi, G.; Tannahill, D.; McCafferty, J.; Balasubramanian, S. Quantitative Visualization of DNA G-Quadruplex Structures in Human Cells. Nat. Chem. 2013, 5, 182–186. [Google Scholar] [CrossRef] [PubMed]
- Henderson, A.; Wu, Y.; Huang, Y.C.; Chavez, E.A.; Platt, J.; Johnson, F.B.; Brosh, R.M.; Sen, D.; Lansdorp, P.M. Detection of G-Quadruplex DNA in Mammalian Cells. Nucleic Acids Res. 2014, 42, 860–869. [Google Scholar] [CrossRef]
- Williamson, J.R.; Raghuraman, M.K.; Cech, T.R. Monovalent Cation-Induced Structure of Telomeric DNA: The G-Quartet Model. Cell 1989, 59, 871–880. [Google Scholar] [CrossRef]
- Neidle, S.; Parkinson, G.N. The Structure of Telomeric DNA. Curr. Opin. Struct. Biol. 2003, 13, 275–283. [Google Scholar] [CrossRef]
- Largy, E.; Mergny, J.-L.; Gabelica, V. Role of Alkali Metal Ions in G-Quadruplex Nucleic Acid Structure and Stability. In The Alkali Metal Ions: Their Role for Life; Springer: Cham, Switzerland, 2016; pp. 203–258. [Google Scholar]
- Da Silva, M.W. Geometric Formalism for DNA Quadruplex Folding. Chem. Eur. J. 2007, 13, 9738–9745. [Google Scholar] [CrossRef]
- Lim, K.W.; Ng, V.C.M.; Martín-Pintado, N.; Heddi, B.; Phan, A.T. Structure of the Human Telomere in Na Solution: An Antiparallel (2 2) G-Quadruplex Scaffold Reveals Additional Diversity. Nucleic Acids Res. 2013, 41, 10556–10562. [Google Scholar] [CrossRef]
- Wang, Y.; Patel, D.J. Solution Structure of a Parallel-Stranded G-Quadruplex DNA. J. Mol. Biol. 1993, 234, 1171–1183. [Google Scholar] [CrossRef]
- Gray, R.D.; Trent, J.O.; Arumugam, S.; Chaires, J.B. Folding Landscape of a Parallel G-Quadruplex. J. Phys. Chem. Lett. 2019, 10, 1146–1151. [Google Scholar] [CrossRef]
- Luu, K.N.; Phan, A.T.; Kuryavyi, V.; Lacroix, L.; Patel, D.J. Structure of the Human Telomere in K Solution: An Intramolecular (3 1) G-Quadruplex Scaffold. J. Am. Chem. Soc. 2006, 128, 9963–9970. [Google Scholar] [CrossRef] [PubMed]
- Dvorkin, S.A.; Karsisiotis, A.I.; Webba da Silva, M. Encoding Canonical DNA Quadruplex Structure. Sci. Adv. 2018, 4, eaat3007. [Google Scholar] [CrossRef] [PubMed]
- Del Villar-Guerra, R.; Trent, J.O.; Chaires, J.B. G-Quadruplex Secondary Structure Obtained From Circular Dichroism Spectroscopy. Angew. Chem. Int. Ed. 2018, 57, 7171–7175. [Google Scholar] [CrossRef] [PubMed]
- Lech, C.J.; Heddi, B.; Phan, A.T. Guanine Base Stacking in G-Quadruplex Nucleic Acids. Nucleic Acids Res. 2013, 41, 2034–2046. [Google Scholar] [CrossRef] [PubMed]
- Orecchini, A.; Paciaroni, A.; De Francesco, A.; Petrillo, C.; Sacchetti, F. Collective Dynamics of Protein Hydration Water by Brillouin Neutron Spectroscopy. J. Am. Chem. Soc. 2009, 131, 4664–4669. [Google Scholar] [CrossRef] [PubMed]
- Comez, L.; Lupi, L.; Morresi, A.; Paolantoni, M.; Sassi, P.; Fioretto, D. More Is Different: Experimental Results on the Effect of Biomolecules on the Dynamics of Hydration Water. J. Phys. Chem. Lett. 2013, 4, 1188–1192. [Google Scholar] [CrossRef]
- Nibali, V.C.; D’Angelo, G.; Paciaroni, A.; Tobias, D.J.; Tarek, M. On the Coupling between the Collective Dynamics of Proteins and Their Hydration Water. J. Phys. Chem. Lett. 2014, 5, 1181–1186. [Google Scholar] [CrossRef]
- Schirò, G.; Fichou, Y.; Gallat, F.-X.; Wood, K.; Gabel, F.; Moulin, M.; Härtlein, M.; Heyden, M.; Colletier, J.-P.; Orecchini, A.; et al. Translational Diffusion of Hydration Water Correlates with Functional Motions in Folded and Intrinsically Disordered Proteins. Nat. Commun. 2015, 6, 6490. [Google Scholar] [CrossRef]
- Laage, D.; Elsaesser, T.; Hynes, J.T. Water Dynamics in the Hydration Shells of Biomolecules. Chem. Rev. 2017, 117, 10694–10725. [Google Scholar] [CrossRef]
- Liu, Y.; Guchhait, B.; Siebert, T.; Fingerhut, B.P.; Elsaesser, T. Molecular Couplings and Energy Exchange between DNA and Water Mapped by Femtosecond Infrared Spectroscopy of Backbone Vibrations. Struct. Dyn. 2017, 4, 044015. [Google Scholar] [CrossRef]
- Franklin, R.E.; Gosling, R.G. Molecular Configuration in Sodium Thymonucleate. Nature 1953, 171, 740–741. [Google Scholar] [CrossRef] [PubMed]
- Miyoshi, D.; Nakamura, K.; Tateishi-Karimata, H.; Ohmichi, T.; Sugimoto, N. Hydration of Watson−Crick Base Pairs and Dehydration of Hoogsteen Base Pairs Inducing Structural Polymorphism under Molecular Crowding Conditions. J. Am. Chem. Soc. 2009, 131, 3522–3531. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, B.; Neidle, S.; David Wilson, W. A Role for Water Molecules in DNA—Ligand Minor Groove Recognition. Acc. Chem. Res. 2009, 42, 11–21. [Google Scholar] [CrossRef] [PubMed]
- Paciaroni, A.; Orecchini, A.; Goracci, G.; Cornicchi, E.; Petrillo, C.; Sacchetti, F. Glassy Character of DNA Hydration Water. J. Phys. Chem. B 2013, 117, 2026–2031. [Google Scholar] [CrossRef] [PubMed]
- Berman, H.M. Hydration of DNA: Take 2. Curr. Opin. Struct. Biol. 1994, 4, 345–350. [Google Scholar] [CrossRef]
- Halle, B.; Denisov, V.P. Water and Monovalent Ions in the Minor Groove of B-DNA Oligonucleotides as Seen by NMR. Biopolymers 1998, 48, 210–233. [Google Scholar] [CrossRef]
- Feig, M.; Montgomery Pettitt, B. A Molecular Simulation Picture of DNA Hydration around A- and B-DNA. Biopolymers 1998, 48, 199. [Google Scholar] [CrossRef]
- Paciaroni, A.; Comez, L.; Longo, M.; Sebastiani, F.; Bianchi, F.; Orecchini, A.; Zanatta, M.; Verbeni, R.; Bosak, A.; Sacchetti, F.; et al. Terahertz Collective Dynamics of DNA as Affected by Hydration and Counterions. J. Mol. Liq. 2020, 318, 113956. [Google Scholar] [CrossRef]
- Li, K.; Yatsunyk, L.; Neidle, S. Water Spines and Networks in G-Quadruplex Structures. Nucleic Acids Res. 2021, 49, 519–528. [Google Scholar] [CrossRef]
- Lane, A.N.; Chaires, J.B.; Gray, R.D.; Trent, J.O. Stability and Kinetics of G-Quadruplex Structures. Nucleic Acids Res. 2008, 36, 5482–5515. [Google Scholar] [CrossRef]
- Olsen, C.M.; Gmeiner, W.H.; Marky, L.A. Unfolding of G-Quadruplexes: Energetic, and Ion and Water Contributions of G-Quartet Stacking. J. Phys. Chem. B 2006, 110, 6962–6969. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Gaerig, V.; Cui, Y.; Kang, H.; Gokhale, V.; Zhao, Y.; Hurley, L.H.; Mao, H. Tertiary DNA Structure in the Single-Stranded hTERT Promoter Fragment Unfolds and Refolds by Parallel Pathways via Cooperative or Sequential Events. J. Am. Chem. Soc. 2012, 134, 5157–5164. [Google Scholar] [CrossRef] [PubMed]
- Petraccone, L.; Pagano, B.; Giancola, C. Studying the Effect of Crowding and Dehydration on DNA G-Quadruplexes. Methods 2012, 57, 76–83. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Gu, X.; Nakano, S.-I.; Miyoshi, D.; Sugimoto, N. Beads-on-a-String Structure of Long Telomeric DNAs under Molecular Crowding Conditions. J. Am. Chem. Soc. 2012, 134, 20060–20069. [Google Scholar] [CrossRef]
- Spink, C.H.; Chaires, J.B. Effects of Hydration, Ion Release, and Excluded Volume on the Melting of Triplex and Duplex DNA. Biochemistry 1999, 38, 496–508. [Google Scholar] [CrossRef] [PubMed]
- Miyoshi, D.; Karimata, H.; Sugimoto, N. Hydration Regulates Thermodynamics of G-Quadruplex Formation under Molecular Crowding Conditions. J Am. Chem. Soc. 2006, 128, 7957–7963. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.C.; Clarke Miller, M.; Buscaglia, R.; Chaires, J.B.; Lane, A.N.; Trent, J.O. Hydration Is a Major Determinant of the G-Quadruplex Stability and Conformation of the Human Telomere 3′ Sequence of d(AG3(TTAG3)3). J. Am. Chem. Soc. 2010, 132, 17105–17107. [Google Scholar] [CrossRef] [PubMed]
- Miyoshi, D.; Karimata, H.; Sugimoto, N. Hydration Regulates The Thermodynamic Stability Of Dna Structures Under Molecular Crowding Conditions. Nucleosides Nucleotides Nucleic Acids 2007, 26, 589–595. [Google Scholar] [CrossRef]
- Yan, Y.-Y.; Tan, J.-H.; Lu, Y.-J.; Yan, S.-C.; Wong, K.-Y.; Li, D.; Gu, L.-Q.; Huang, Z.-S. G-Quadruplex Conformational Change Driven by pH Variation with Potential Application as a Nanoswitch. Biochim. Biophys. Acta (BBA) Gen. Subj. 2013, 1830, 4935–4942. [Google Scholar] [CrossRef]
- Takahashi, S.; Sugimoto, N. Effect of Pressure on the Stability of G-Quadruplex DNA: Thermodynamics under Crowding Conditions. Angew. Chem. Int. Ed. 2013, 52, 13774–13778. [Google Scholar] [CrossRef]
- Gill, D.; Kilponen, R.G.; Rimai, L. Resonance Raman Scattering of Laser Radiation by Vibrational Modes of Carotenoid Pigment Molecules in Intact Plant Tissues. Nature 1970, 227, 743–744. [Google Scholar] [CrossRef] [PubMed]
- Asher, S.A. UV Resonance Raman Spectroscopy for Analytical, Physical, and Biophysical Chemistry. Anal. Chem. 1993, 65, 59A–66A. [Google Scholar] [CrossRef]
- Oladepo, S.A.; Xiong, K.; Hong, Z.; Asher, S.A.; Handen, J.; Lednev, I.K. UV Resonance Raman Investigations of Peptide and Protein Structure and Dynamics. Chem. Rev. 2012, 112, 2604–2628. [Google Scholar] [CrossRef] [PubMed]
- Fodor, S.P.A.; Spiro, T.G. Ultraviolet Resonance Raman Spectroscopy of DNA with 200-266-Nm Laser Excitation. J. Am. Chem. Soc. 1986, 108, 3198–3205. [Google Scholar] [CrossRef]
- Chan, S.S.; Austin, R.H.; Mukerji, I.; Spiro, T.G. Temperature-Dependent Ultraviolet Resonance Raman Spectroscopy of the Premelting State of dA.dT DNA. Biophys. J. 1997, 72, 1512–1520. [Google Scholar] [CrossRef]
- Ambrus, A.; Chen, D.; Dai, J.; Bialis, T.; Jones, R.A.; Yang, D. Human Telomeric Sequence Forms a Hybrid-Type Intramolecular G-Quadruplex Structure with Mixed Parallel/antiparallel Strands in Potassium Solution. Nucleic Acids Res. 2006, 34, 2723–2735. [Google Scholar] [CrossRef]
- Phan, A.T.; Luu, K.N.; Patel, D.J. Different Loop Arrangements of Intramolecular Human Telomeric (3 1) G-Quadruplexes in K Solution. Nucleic Acids Res. 2006, 34, 5715–5719. [Google Scholar] [CrossRef]
- Renčiuk, D.; Kejnovská, I.; Školáková, P.; Bednářová, K.; Motlová, J.; Vorlíčková, M. Arrangements of Human Telomere DNA Quadruplex in Physiologically Relevant K Solutions. Nucleic Acids Res. 2009, 37, 6625–6634. [Google Scholar] [CrossRef] [PubMed]
- Palacký, J.; Vorlíčková, M.; Kejnovská, I.; Mojzeš, P. Polymorphism of Human Telomeric Quadruplex Structure Controlled by DNA Concentration: A Raman Study. Nucleic Acids Res. 2013, 41, 1005–1016. [Google Scholar] [CrossRef]
- Bianchi, F.; Comez, L.; Biehl, R.; D’Amico, F.; Gessini, A.; Longo, M.; Masciovecchio, C.; Petrillo, C.; Radulescu, A.; Rossi, B.; et al. Structure of Human Telomere G-Quadruplex in the Presence of a Model Drug along the Thermal Unfolding Pathway. Nucleic Acids Res. 2018, 46, 11927–11938. [Google Scholar] [CrossRef]
- Marchand, A.; Rosu, F.; Zenobi, R.; Gabelica, V. Thermal Denaturation of DNA G-Quadruplexes and Their Complexes with Ligands: Thermodynamic Analysis of the Multiple States Revealed by Mass Spectrometry. J. Am. Chem. Soc. 2018, 140, 12553–12565. [Google Scholar] [CrossRef] [PubMed]
- Comez, L.; Bianchi, F.; Libera, V.; Longo, M.; Petrillo, C.; Sacchetti, F.; Sebastiani, F.; D’Amico, F.; Rossi, B.; Gessini, A.; et al. Polymorphism of Human Telomeric Quadruplexes with Drugs: A Multi-Technique Biophysical Study. Phys. Chem. Chem. Phys. 2020, 22, 11583–11592. [Google Scholar] [CrossRef] [PubMed]
- Chaires, J.B. Human Telomeric G-Quadruplex: Thermodynamic and Kinetic Studies of Telomeric Quadruplex Stability. FEBS J. 2010, 277, 1098–1106. [Google Scholar] [CrossRef] [PubMed]
- Gray, R.D.; Chaires, J.B. Analysis of Multidimensional G-Quadruplex Melting Curves. Curr. Protoc. Nucleic Acid Chem. 2011, Chapter 17, Unit17.4. [Google Scholar] [CrossRef]
- Gray, R.D.; Buscaglia, R.; Chaires, J.B. Populated Intermediates in the Thermal Unfolding of the Human Telomeric Quadruplex. J. Am. Chem. Soc. 2012, 134, 16834–16844. [Google Scholar] [CrossRef] [PubMed]
- Benevides, J.M.; Overman, S.A.; Thomas, G.J. Raman, Polarized Raman and Ultraviolet Resonance Raman Spectroscopy of Nucleic Acids and Their Complexes. J. Raman Spectrosc. 2005, 36, 279–299. [Google Scholar] [CrossRef]
- Fodor, S.P.A.; Rava, R.P.; Hays, T.R.; Spiro, T.G. Ultraviolet resonance Raman spectroscopy of nucleotides with 266-,240-,218-, and 200- nm pulsed laser excitation. Chem. Inf. 1985, 107, 1520–1529. [Google Scholar]
- Krafft, C. Secondary Structure Polymorphism in Oxytricha Nova Telomeric DNA. Nucleic Acids Res. 2002, 30, 3981–3991. [Google Scholar] [CrossRef][Green Version]
- Di Fonzo, S.; Bottari, C.; Brady, J.W.; Tavagnacco, L.; Caterino, M.; Petraccone, L.; Amato, J.; Giancola, C.; Cesàro, A. Crowding and Conformation Interplay on Human DNA G-Quadruplex by Ultraviolet Resonant Raman Scattering. Phys. Chem. Chem. Phys. 2019, 21, 2093–2101. [Google Scholar] [CrossRef]
- Amato, J.; Iaccarino, N.; DAria, F.; Randazzo, A.; Giancola, C.; Cesàro, A.; Di Fonzo, S.; Pagano, B. Conformational plasticity of DNA secondary structures: Probing the conversion between i-motif and hairpin species by circular dichroism and ultraviolet resonance Raman spectroscopies. Phys. Chem. Chem. Phys. 2022, 24, 7028–7044. [Google Scholar] [CrossRef]
- Palacký, J.; Mojzeš, P.; Kejnovská, I.; Vorlíčková, M. Does Raman Spectroscopy Recognize Different G-quadruplex Arrangements? J. Raman Spectrosc. 2020, 51, 301–312. [Google Scholar] [CrossRef]
- Bottari, C.; Comez, L.; Paolantoni, M.; Corezzi, S.; D’Amico, F.; Gessini, A.; Masciovecchio, C.; Rossi, B. Hydration Properties and Water Structure in Aqueous Solutions of Native and Modified Cyclodextrins by UV Raman and Brillouin Scattering. J. Raman Spectrosc. 2018, 49, 1076–1085. [Google Scholar] [CrossRef]
- Green, J.L.; Lacey, A.R.; Sceats, M.G. Spectroscopic Evidence for Spatial Correlations of Hydrogen Bonds in Liquid Water. J. Phys. Chem. 1986, 90, 3958–3964. [Google Scholar] [CrossRef]
- Walrafen, G.E.; Chu, Y.C. Linearity between Structural Correlation Length and Correlated-Proton Raman Intensity from Amorphous Ice and Supercooled Water up to Dense Supercritical Steam. J. Phys. Chem. 1995, 99, 11225–11229. [Google Scholar] [CrossRef]
- Walrafen, G.E. Effects of Equilibrium H-Bond Distance and Angle Changes on Raman Intensities from Water. J. Chem. Phys. 2004, 120, 4868–4876. [Google Scholar] [CrossRef] [PubMed]
- Eaves, J.D.; Loparo, J.J.; Fecko, C.J.; Roberts, S.T.; Tokmakoff, A.; Geissler, P.L. Hydrogen Bonds in Liquid Water Are Broken Only Fleetingly. Proc. Natl. Acad. Sci. USA 2005, 102, 13019–13022. [Google Scholar] [CrossRef] [PubMed]
- Paolantoni, M.; Faginas Lago, N.; Albertí, M.; Laganà, A. Tetrahedral Ordering in Water: Raman Profiles and Their Temperature Dependence. J. Phys. Chem. A 2009, 113, 15100–15105. [Google Scholar] [CrossRef] [PubMed]
- Libera, V.; Andreeva, E.A.; Martel, A.; Thureau, A.; Longo, M.; Petrillo, C.; Paciaroni, A.; Schirò, G.; Comez, L. Porphyrin Binding and Irradiation Promote G-Quadruplex DNA Dimeric Structure. J. Phys. Chem. Lett. 2021, 12, 8096–8102. [Google Scholar] [CrossRef]
- Bao, H.-L.; Liu, H.-s.; Xu, Y. Hybrid-type and two-tetrad antiparallel telomere DNA G-quadruplex structures in living human cells. Nucleic Acids Res. 2019, 47, 4940–4947. [Google Scholar] [CrossRef]
- Bao, H.-L.; Ishizuka, T.; Yamashita, A.; Furukoji, E.; Asada, Y.; Xu, Y. Improving Thermodynamic Stability and Anticoagulant Activity of a Thrombin Binding Aptamer by Incorporation of 8-trifluoromethyl-2′-deoxyguanosine. J. Med. Chem. 2021, 64, 711–718. [Google Scholar] [CrossRef]
- Neidle, S. Structured Waters Mediate Small Molecule Binding to G-Quadruplex Nucleic Acids. Pharmaceuticals 2021, 15, 7. [Google Scholar] [CrossRef] [PubMed]
- Del Villar-Guerra, R.; Gray, R.D.; Chaires, J.B. Characterization of Quadruplex DNA Structure by Circular Dichroism. Curr. Protoc. Nucleic Acid Chem. 2017, 68, 17.8.1–17.8.16. [Google Scholar] [CrossRef] [PubMed]
- Rossi, B.; Bottari, C.; Catalini, S.; D’Amico, F.; Gessini, A.; Masciovecchio, C. Synchrotron-Based Ultraviolet Resonance Raman Scattering for Material Science. In Molecular and Laser Spectroscopy—Advances and Applications; Elsevier: Amsterdam, The Netherlands, 2020; Volume 2, pp. 447–482. [Google Scholar]
- Privalov, P.L.; Khechinashvili, N.N. A Thermodynamic Approach to the Problem of Stabilization of Globular Protein Structure: A Calorimetric Study. J. Mol. Biol. 1974, 86, 665–684. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Libera, V.; Bianchi, F.; Rossi, B.; D’Amico, F.; Masciovecchio, C.; Petrillo, C.; Sacchetti, F.; Paciaroni, A.; Comez, L. Solvent Vibrations as a Proxy of the Telomere G-Quadruplex Rearrangements across Thermal Unfolding. Int. J. Mol. Sci. 2022, 23, 5123. https://doi.org/10.3390/ijms23095123
Libera V, Bianchi F, Rossi B, D’Amico F, Masciovecchio C, Petrillo C, Sacchetti F, Paciaroni A, Comez L. Solvent Vibrations as a Proxy of the Telomere G-Quadruplex Rearrangements across Thermal Unfolding. International Journal of Molecular Sciences. 2022; 23(9):5123. https://doi.org/10.3390/ijms23095123
Chicago/Turabian StyleLibera, Valeria, Federico Bianchi, Barbara Rossi, Francesco D’Amico, Claudio Masciovecchio, Caterina Petrillo, Francesco Sacchetti, Alessandro Paciaroni, and Lucia Comez. 2022. "Solvent Vibrations as a Proxy of the Telomere G-Quadruplex Rearrangements across Thermal Unfolding" International Journal of Molecular Sciences 23, no. 9: 5123. https://doi.org/10.3390/ijms23095123
APA StyleLibera, V., Bianchi, F., Rossi, B., D’Amico, F., Masciovecchio, C., Petrillo, C., Sacchetti, F., Paciaroni, A., & Comez, L. (2022). Solvent Vibrations as a Proxy of the Telomere G-Quadruplex Rearrangements across Thermal Unfolding. International Journal of Molecular Sciences, 23(9), 5123. https://doi.org/10.3390/ijms23095123







