Analysis of Xylose Operon from Paenibacillus polymyxa ATCC842 and Development of Tools for Gene Expression
Abstract
1. Introduction
2. Results
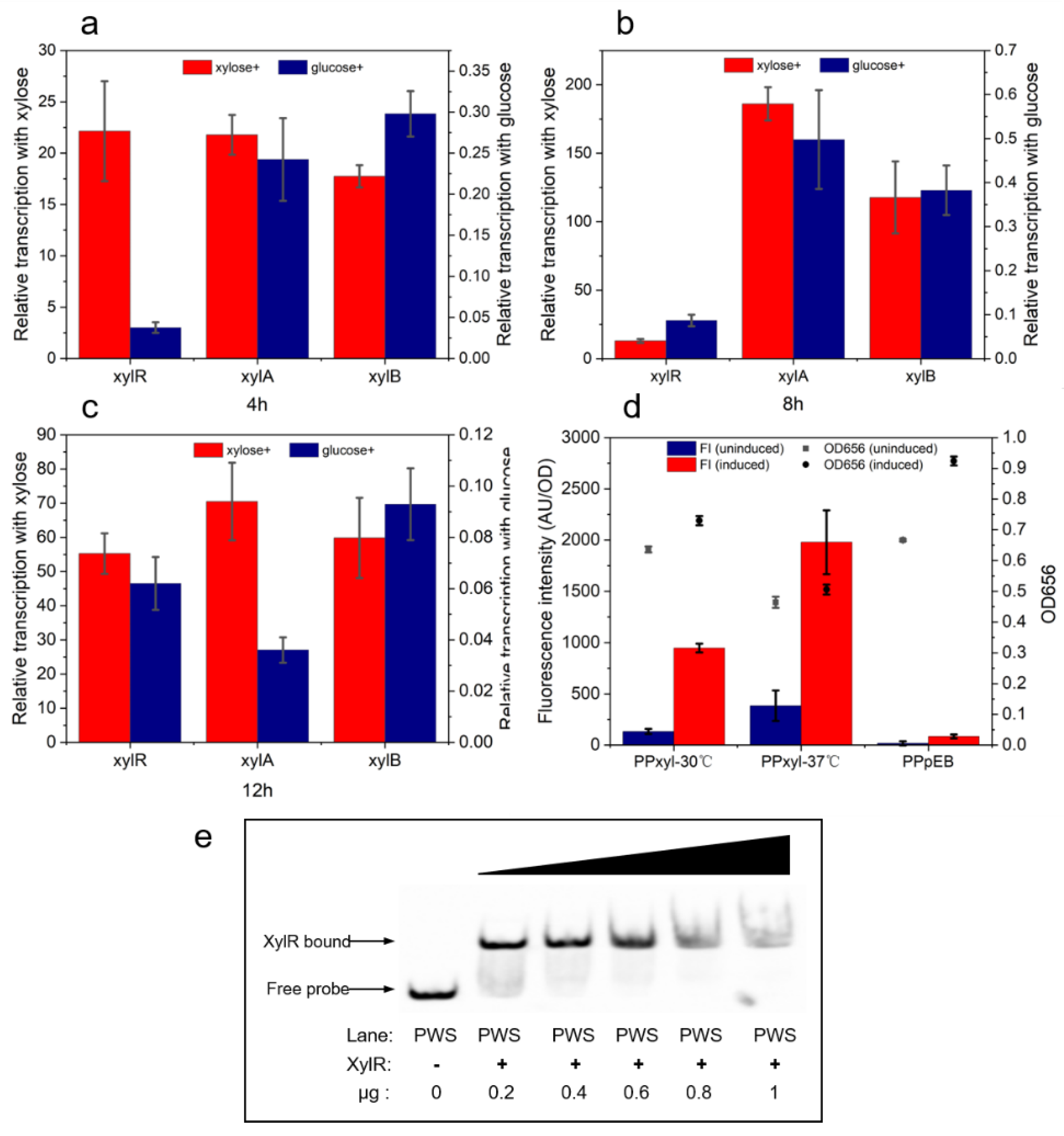
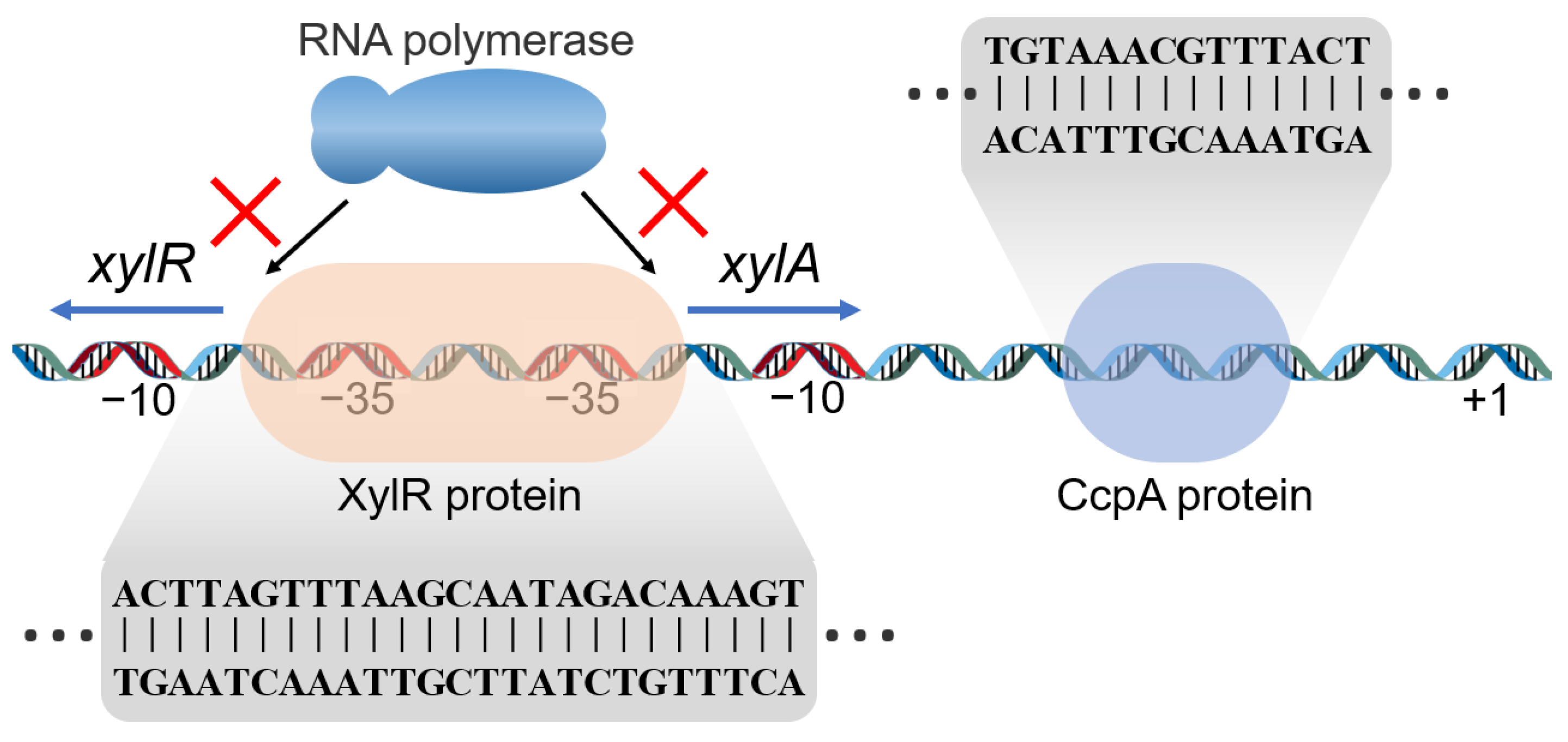
2.1. Analysis of Xylose Operon Elements and Function in P. polymyxa
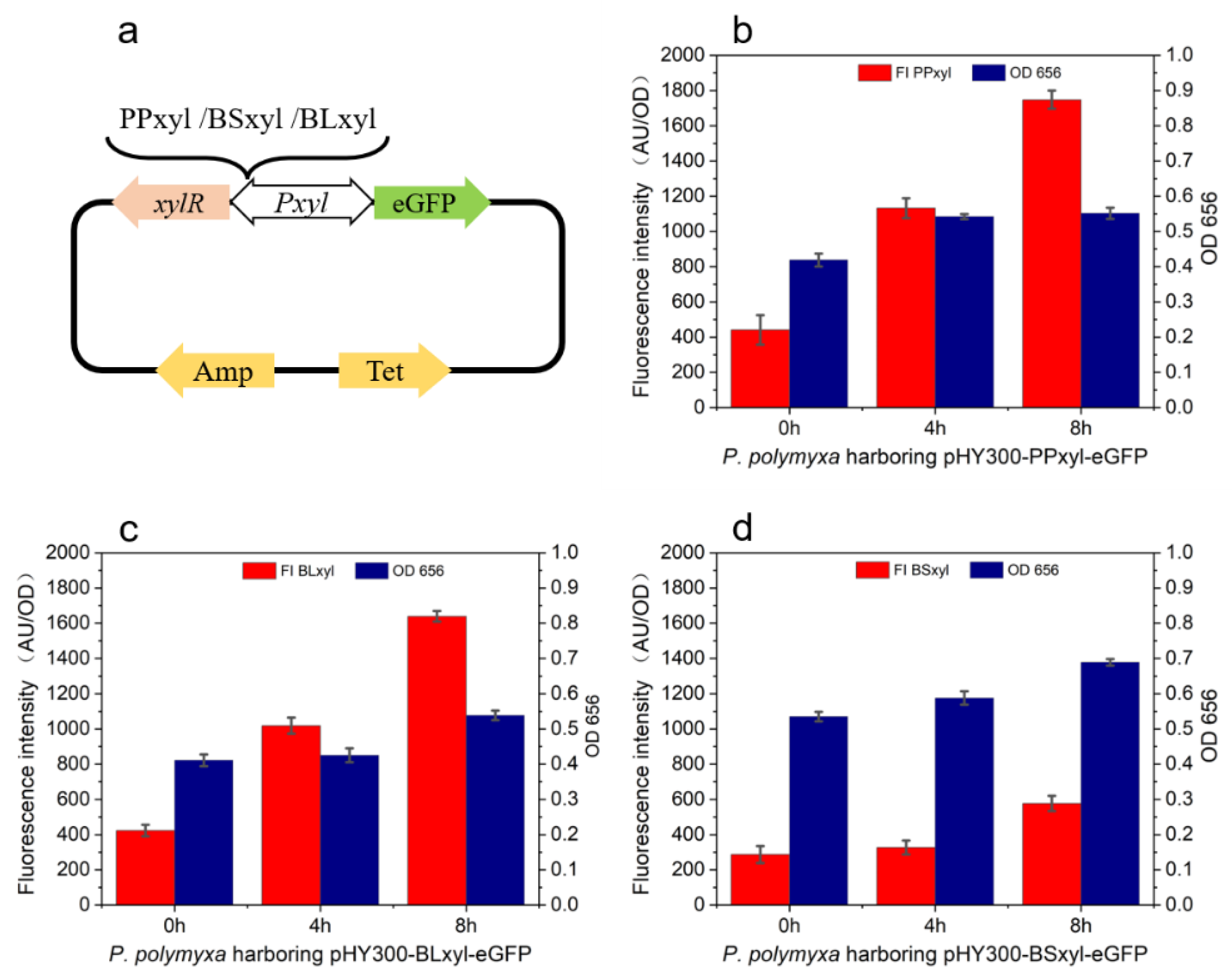
2.2. Efficient Expression of Protein by PPxyl in P. polymyxa
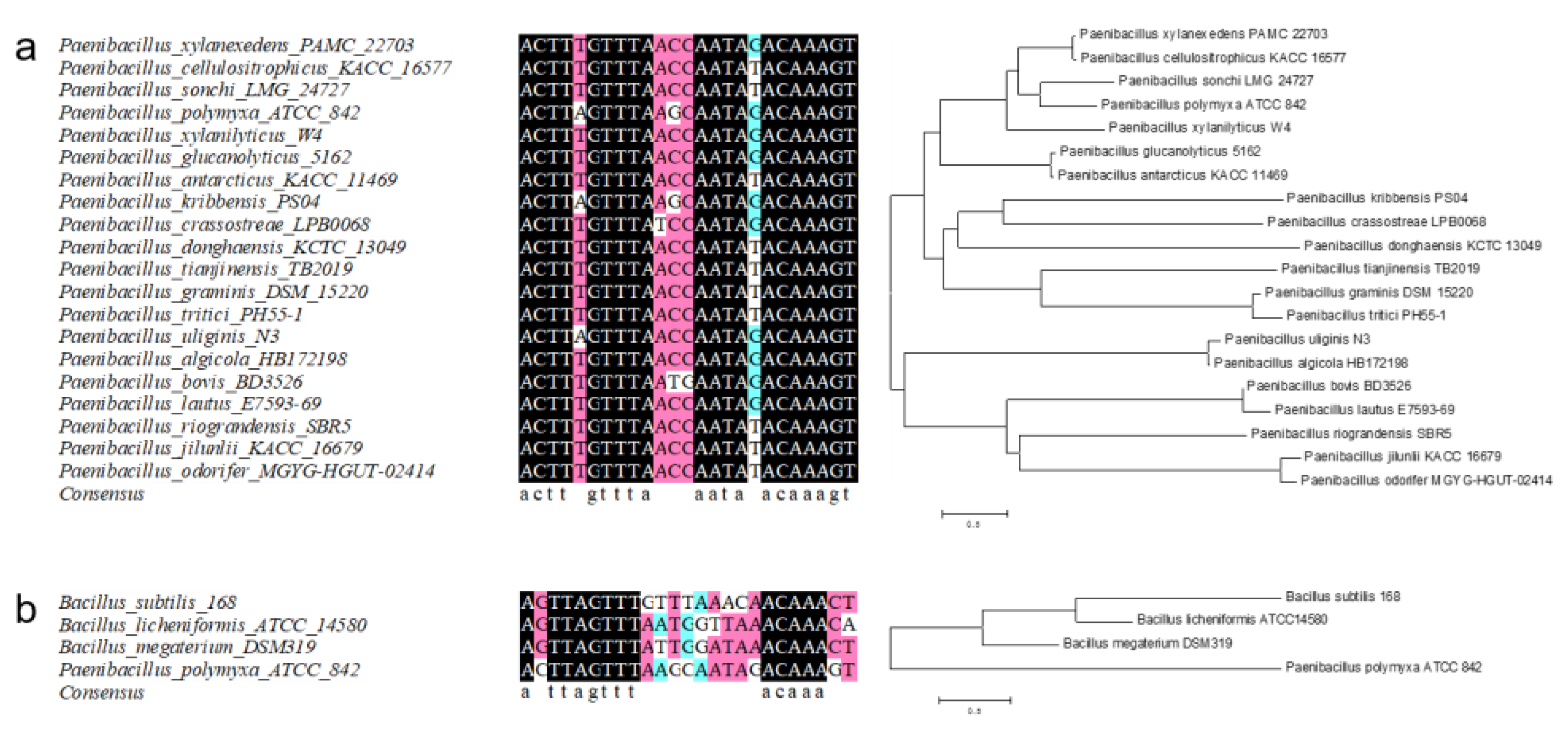
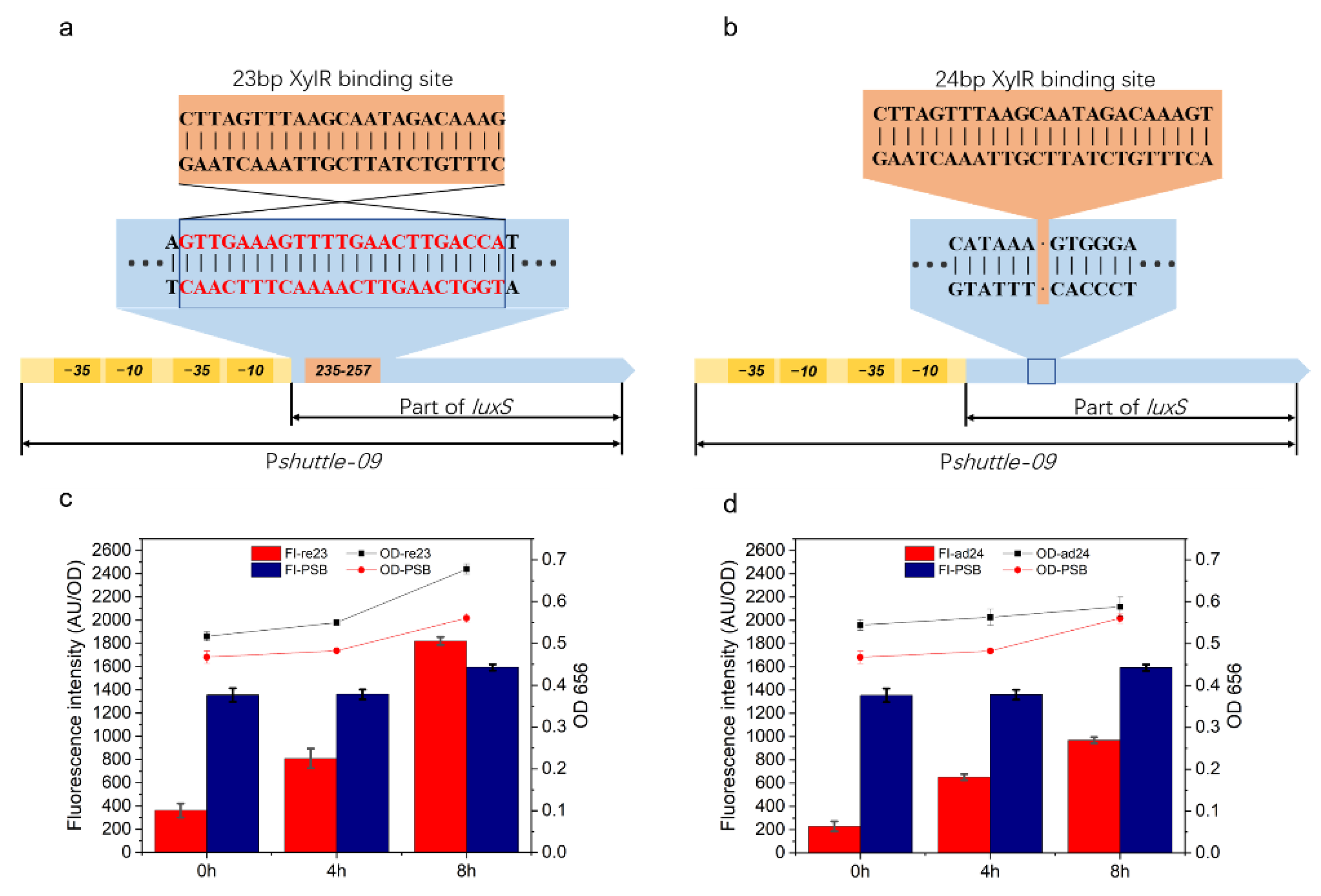
2.3. Similar Xylose Operons from P. polymyxa with Different Binding Sites
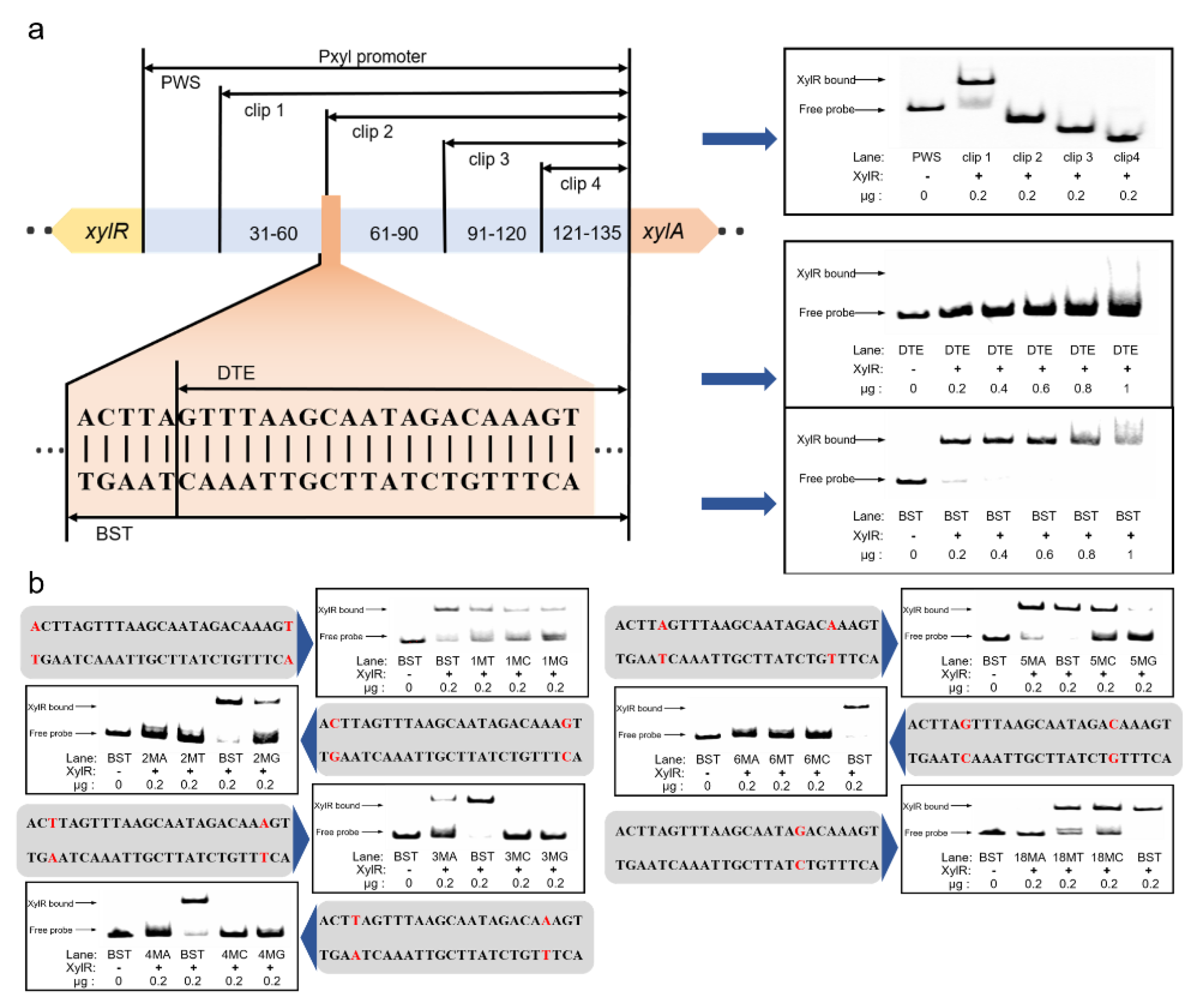
2.4. Promoter Transformation Based on Pxyl Binding Site
3. Discussion
4. Materials and Methods
4.1. Bacterial Strain and Culture Medium
4.2. Promoter-Function-Characterizing Plasmid Construction
4.3. P. polymyxa Competent Cell Preparation
4.4. P. polymyxa Transformation
4.5. eGFP Fluorescence Measurement
4.6. Electrophoretic Mobility Shift Assay
4.7. Reverse Transcription Quantitative PCR (RT-qPCR)
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Choi, S.; Park, S.; Kim, R.; Lee, C.-H.; Kim, J.F.; Park, S.-H. Identification and functional analysis of the fusaricidin biosynthetic gene of Paenibacillus polymyxa E681. Biochem. Biophys. Res. Commun. 2008, 365, 89–95. [Google Scholar] [CrossRef]
- Choi, S.; Park, S.; Kim, R.; Kim, S.-B.; Lee, C.-H.; Kim, J.F.; Park, S.-H. Identification of a Polymyxin Synthetase Gene Cluster of Paenibacillus polymyxa and Heterologous Expression of the Gene in Bacillus subtilis. J. Bacteriol. 2009, 191, 3350–3358. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Zhang, R.; Wang, Y.; Zhang, N.; Shao, J.; Qiu, M.; Shen, B.; Yin, X.; Shen, Q. Promoter analysis and transcription regulation of fus gene cluster responsible for fusaricidin synthesis of Paenibacillus polymyxa SQR-21. Appl. Microbiol. Biotechnol. 2013, 97, 9479–9489. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Wu, X.; Bai, X.; Zhang, H.; Dong, D.; Zhang, T.; Wu, H. Fusaricidins in Paenibacillus polymyxa A21 and their antagonistic activity against Botrytis cinerea on tomato. Front. Agric. Sci. Eng. 2018, 5, 262–270. [Google Scholar] [CrossRef]
- Liu, H.; Wang, J.; Sun, H.; Han, X.; Peng, Y.; Liu, J.; Liu, K.; Ding, Y.; Wang, C.; Du, B. Transcriptome Profiles Reveal the Growth-Promoting Mechanisms of Paenibacillus polymyxa YC0136 on Tobacco (Nicotiana tabacum L.). Front. Microbiol. 2020, 11, 584174. [Google Scholar] [CrossRef] [PubMed]
- Niu, B.; Rückert, C.; Blom, J.; Wang, Q.; Borriss, R. The Genome of the Plant Growth-Promoting Rhizobacterium Paenibacillus polymyxa M-1 Contains Nine Sites Dedicated to Nonribosomal Synthesis of Lipopeptides and Polyketides. J. Bacteriol. 2011, 193, 5862–5863. [Google Scholar] [CrossRef] [PubMed]
- Pu, X.; Chen, F.; Yang, Y.; Qu, X.; Zhang, G.; Luo, Y. Isolation and characterization of Paenibacillus polymyxa LY214, a camptothecin-producing endophytic bacterium from Camptotheca acuminata. J. Ind. Microbiol. Biotechnol. 2015, 42, 1197–1202. [Google Scholar] [CrossRef]
- Rütering, M.; Schmid, J.; Rühmann, B.; Schilling, M.; Sieber, V. Controlled production of polysaccharides–exploiting nutrient supply for levan and heteropolysaccharide formation in Paenibacillus sp. Carbohydr. Polym. 2016, 148, 326–334. [Google Scholar] [CrossRef]
- Yu, Z.; Sun, Z.; Yin, J.; Qiu, J. Enhanced Production of Polymyxin E in Paenibacillus polymyxa by Replacement of Glucose by Starch. BioMed Res. Int. 2018, 2018, 1934309. [Google Scholar] [CrossRef]
- Yu, Z.; Guo, C.; Qiu, J. Precursor Amino Acids Inhibit Polymyxin E Biosynthesis in Paenibacillus polymyxa, Probably by Affecting the Expression of Polymyxin E Biosynthesis-Associated Genes. BioMed Res. Int. 2015, 2015, 690830. [Google Scholar] [CrossRef]
- Zhang, L.; Cao, C.; Jiang, R.; Xu, H.; Xue, F.; Huang, W.; Ni, H.; Gao, J. Production of R,R-2,3-butanediol of ultra-high optical purity from Paenibacillus polymyxa ZJ-9 using homologous recombination. Bioresour. Technol. 2018, 261, 272–278. [Google Scholar] [CrossRef] [PubMed]
- Rütering, M.; Cress, B.F.; Schilling, M.; Rühmann, B.; Koffas, M.A.G.; Sieber, V.; Schmid, J. Tailor-made exopolysaccharides—CRISPR-Cas9 mediated genome editing in Paenibacillus polymyxa. Synth. Biol. 2017, 2, ysx007. [Google Scholar] [CrossRef] [PubMed]
- Brito, L.F.; Irla, M.; Walter, T.; Wendisch, V.F. Magnesium aminoclay-based transformation of Paenibacillus riograndensis and Paenibacillus polymyxa and development of tools for gene expression. Appl. Microbiol. Biotechnol. 2017, 101, 735–747. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.-Z.; Yan, X.; Cui, Z.-L.; Hong, Q.; Li, S.-P. mazF, a novel counter-selectable marker for unmarked chromosomal manipulation in Bacillus subtilis. Nucleic Acids Res. 2006, 34, e71. [Google Scholar] [CrossRef]
- Nijland, R.; Lindner, C.; van Hartskamp, M.; Hamoen, L.W.; Kuipers, O.P. Heterologous production and secretion of Clostridium perfringens β-toxoid in closely related Gram-positive hosts. J. Biotechnol. 2007, 127, 361–372. [Google Scholar] [CrossRef]
- Ming, Y.M.; Wei, Z.W.; Lin, C.Y.; Sheng, G.Y. Development of a Bacillus subtilis expression system using the improved Pglv promoter. Microb. Cell Factories 2010, 9, 55. [Google Scholar] [CrossRef]
- Lee, S.-J.; Pan, J.-G.; Park, S.-H.; Choi, S.-K. Development of a stationary phase-specific autoinducible expression system in Bacillus subtilis. J. Biotechnol. 2010, 149, 16–20. [Google Scholar] [CrossRef]
- Inácio, J.M.; Costa, C.; de Sá-Nogueira, I. Distinct molecular mechanisms involved in carbon catabolite repression of the arabinose regulon in Bacillus subtilis. Microbiology 2003, 149, 2345–2355. [Google Scholar] [CrossRef]
- Sun, T.; Altenbuchner, J. Characterization of a Mannose Utilization System in Bacillus subtilis. J. Bacteriol. 2010, 192, 2128–2139. [Google Scholar] [CrossRef]
- Martin-Verstraete, I.; Dkbarbouilk, M.; Rapoport, G. Levanase operon of Bacillus subtilis includes a fructose-specific phos-photransferase system regulating the expression of the operon. J. Mol. Biol. 1990, 214, 657–671. [Google Scholar] [CrossRef]
- Choi, E.-S.; Sohn, J.-H.; Rhee, S.-K. Optimization of the expression system using galactose-inducible promoter for the production of anticoagulant hirudin in Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 1994, 42, 587–594. [Google Scholar] [CrossRef] [PubMed]
- Rygus, T.; Hillen, W. Catabolite repression of the xyl operon in Bacillus megaterium. J. Bacteriol. 1992, 174, 3049–3055. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hartsock, C.E.; Lewis, J.K.; Leslie, I.; Pope, J.A.; Tsai, L.B.; Sachdev, R.; Meng, S.-Y. Assessing post-induction cellular response in a recombinant E. coli lactose-inducible system by monitoring β-galactosidase levels. Biotechnol. Lett. 1995, 17, 1025–1030. [Google Scholar] [CrossRef]
- Yue, J.; Zhang, D.; Wen, J.; Fu, G. A new maltose-inducible high-performance heterologous expression system in Bacillus subtilis. Biotechnol. Lett. 2017, 39, 1237–1244. [Google Scholar] [CrossRef]
- Liu, X.J.; Prat, S.; Willmitzer, L.; Frommer, W. Cis regulatory elements directing tuber-specific and sucrose-inducible expression of a chimeric class I patatin promoter/GUS-gene fusion. Mol. Genet. Genom. 1990, 223, 401–406. [Google Scholar] [CrossRef]
- Bodelle, P.; Schopp, C.; Sarthy, A.V.; Fonzi, W. Construction of a Sorbitol-Based Vector for Expression of Heterologous Pro-teins in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 1998, 64, 793–794. [Google Scholar]
- Hoffmann, J.; Altenbuchner, J. Functional Characterization of the Mannitol Promoter of Pseudomonas fluorescens DSM 50106 and Its Application for a Mannitol-Inducible Expression System for Pseudomonas putida KT2440. PLoS ONE 2015, 10, e0133248. [Google Scholar] [CrossRef]
- Yurimoto, H.; Komeda, T.; Lim, C.R.; Nakagawa, T.; Kondo, K.; Kato, N.; Sakai, Y. Regulation and evaluation of five methanol-inducible promoters in the methylotrophic yeast Candida boidinii. Biochim. Biophys. Acta 2000, 1493, 56–63. [Google Scholar] [CrossRef]
- Lee, S.; Lee, Y.J.; Choi, S.; Park, S.B.; Tran, Q.G.; Heo, J.; Kim, H.S. Development of an alcohol-inducible gene expression system for recombinant protein expression in Chlamydomonas reinhardtii. J. Appl. Phycol. 2018, 30, 2297–2304. [Google Scholar] [CrossRef]
- Gärtner, D.; Degenkolb, J.; Ripperger, J.A.E.; Allmansberger, R.; Hillen, W. Regulation of the Bacillus subtilis W23 xylose utilization operon: Interaction of the Xyl repressor with the xyl operator and the inducer xylose. Mol. Genet. Genom. 1992, 232, 415–422. [Google Scholar] [CrossRef]
- Scheler, A.; Hlllen, W. Regulation of xylose utilization in Bacillus licheniformis: Xyl repressor-xyl-operator interaction studied by DNA modification protection and interference. Mol. Microbiol. 1994, 13, 505–512. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Z.; Lin, X.; Jiang, T.; Ye, W.; Ouyang, J. Genomic analysis of a xylose operon and characterization of novel xylose isomerase and xylulokinase from Bacillus coagulans NL01. Biotechnol. Lett. 2016, 38, 1331–1339. [Google Scholar] [CrossRef] [PubMed]
- Teo, W.S.; Chang, M.W. Bacterial XylRs and synthetic promoters function as genetically encoded xylose biosensors in Saccharomyces cerevisiae. Biotechnol. J. 2015, 10, 315–322. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.-P.; Shang, Y.; Zhang, P.; Liu, Y.; You, D.; Yin, B.-C.; Ye, B.-C. Engineering Prokaryotic Transcriptional Activator XylR as a Xylose-Inducible Biosensor for Transcription Activation in Yeast. ACS Synth. Biol. 2020, 9, 1022–1029. [Google Scholar] [CrossRef] [PubMed]
- Bréchemier-Baey, D.; Domínguez-Ramírez, L.; Plumbridge, J. The linker sequence, joining the DNA-binding domain of the homologous transcription factors, Mlc and NagC, to the rest of the protein, determines the specificity of their DNA target recognition in Escherichia coli. Mol. Microbiol. 2012, 85, 1007–1019. [Google Scholar] [CrossRef]
- Bréchemier-Baey, D.; Domínguez-Ramírez, L.; Oberto, J.; Plumbridge, J. Operator recognition by the ROK transcription factor family members, NagC and Mlc. Nucleic Acids Res. 2014, 43, 361–372. [Google Scholar] [CrossRef] [PubMed]
- Gösseringer, R.; Küster-Schöck, E.; Galinier, A.; Deutscher, J.; Hillen, W. Cooperative and non-cooperative DNA binding modes of catabolite control protein CcpA from Bacillus megaterium result from sensing two different signals. J. Mol. Biol. 1997, 266, 665–676. [Google Scholar] [CrossRef]
- Qi, L.S.; Larson, M.H.; Gilbert, L.A.; Doudna, J.A.; Weissman, J.S.; Arkin, A.P.; Lim, W.A. Repurposing CRISPR as an RNA-Guided Platform for Sequence-Specific Control of Gene Expression. Cell 2013, 152, 1173–1183. [Google Scholar] [CrossRef]
- Warmbold, B.; Ronzheimer, S.; Freibert, S.-A.; Seubert, A.; Hoffmann, T.; Bremer, E. Two MarR-Type Repressors Balance Precursor Uptake and Glycine Betaine Synthesis in Bacillus subtilis to Provide Cytoprotection Against Sustained Osmotic Stress. Front. Microbiol. 2020, 11, 1700. [Google Scholar] [CrossRef]
- Weickert, M.J.; Chambliss, G.H. Site-directed mutagenesis of a catabolite repression operator sequence in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 1990, 87, 6238–6242. [Google Scholar] [CrossRef]
- Bruder, M.; Moo-Young, M.; Chung, D.A.; Chou, C.P. Elimination of carbon catabolite repression in Clostridium acetobutylicum—A journey toward simultaneous use of xylose and glucose. Appl. Microbiol. Biotechnol. 2015, 99, 7579–7588. [Google Scholar] [CrossRef] [PubMed]
- Kraus, A.; Hueck, C.; Gartner, D.; Hillen, W. Catabolite repression of the Bacillus subtilis xyl operon involves a cis element func-tional in the context of an unrelated sequence, and glucose exerts additional xylR-dependent repression. J. Bacteriol. 1994, 176, 1738–1745. [Google Scholar] [CrossRef]
- Li, Y.; Jin, K.; Zhang, L.; Ding, Z.; Gu, Z.; Shi, G. Development of an Inducible Secretory Expression System in Bacillus licheniformis Based on an Engineered Xylose Operon. J. Agric. Food Chem. 2018, 66, 9456–9464. [Google Scholar] [CrossRef] [PubMed]
- Dahl, M.K.; Degenkolb, J.; Hillen, W. Transcription of the xyl Operon is Controlled in Bacillus subtilis by Tandem Overlapping Operators Spaced by four Base-pairs. J. Mol. Biol. 1994, 243, 413–424. [Google Scholar] [CrossRef] [PubMed]
- Light, S.H.; Cahoon, L.A.; Halavaty, A.S.; Freitag, N.E.; Anderson, W.F. Structure to function of an α-glucan metabolic pathway that promotes Listeria monocytogenes pathogenesis. Nat. Microbiol. 2017, 2, 16202. [Google Scholar] [CrossRef] [PubMed]
- Stephen, C. DNA recognition by proteins with the helix-turn-helix. Annu. Rev. Biochem. 1990, 59, 933–969. [Google Scholar]
- Xiao, F.; Li, Y.; Zhang, Y.; Wang, H.; Zhang, L.; Ding, Z.; Gu, Z.; Xu, S.; Shi, G. Construction of a novel sugar alcohol-inducible expression system in Bacillus licheniformis. Appl. Microbiol. Biotechnol. 2020, 104, 5409–5425. [Google Scholar] [CrossRef]
- Yang, M.; Zhang, W.; Ji, S.; Cao, P.; Chen, Y.; Zhao, X. Generation of an Artificial Double Promoter for Protein Expression in Bacillus subtilis through a Promoter Trap System. PLoS ONE 2013, 8, e56321. [Google Scholar] [CrossRef]
- Xu, X.; Li, X.; Liu, Y.; Zhu, Y.; Li, J.; Du, G.; Chen, J.; Ledesma-Amaro, R.; Liu, L. Pyruvate-responsive genetic circuits for dynamic control of central metabolism. Nat. Chem. Biol. 2020, 16, 1261–1268. [Google Scholar] [CrossRef]
| Primers | Sequences (5’-3’) |
|---|---|
| 16S-f | CCGCACAAGCAGTGGAGT |
| 16S-r | TACCCAACATCTCACGACACGA |
| xA-f | GGAACACGGCGAACATGTTTAG |
| xA-r | CAGCAGTGTTTCGTAGCCTTCAC |
| xB-f | GGAGAAGCTAAATCAGGAGCAG |
| xB-r | GTGATGCTAACGTGCCACTG |
| xR-f | CGATACCGAGCACAAAGC |
| xR-f | CCACCGTGTCCAACCTG |
| xylR-f | CTAGCTAGCATGAATGTCACTGGCGATCAGGCG |
| xylR-r | CCCAAGCTTTAAAGACACGCGTACTCGCCCCA |
| eGFP-f | CCCAAGCTTATGGGTCGCGGATCCATGGTAG |
| eGFP-r | TCCCCCGGGTCACACGTGGTGGTGGTGGTG |
| BSPxyl-f | ATGGAAAAACGCTTTGCCCATTACATTGTAATCATGTCCAGAAAATGATC |
| BSPxyl-r | ACCATGGATCCGCGACCCATGTGATTTCCCCCTTAAAAATAAAT |
| BLPxyl-f | ATGGAAAAACGCTTTGCCCATTAAAATCTCTCGTTCATAAACCGTTCCA |
| BLPxyl-r | ACCATGGATCCGCGACCCAT TCCGATCTCCCCCTTCAC |
| PPPxyl-f | ATGGAAAAACGCTTTGCCCATTATAAAGACACGCGTACTCGC |
| PPPxyl-r | ACCATGGATCCGCGACCCATTATAAGTTCCTCCTTTGTAGTAAACG |
| PE-f | TTATAAAGACACGCGTACTCGC |
| PE-r | TATAAGTTCCTCCTTTGTAGTAAACGTTTACA |
| PxylABf(5′-biotin) | GCGCGGATCTTCCAGAGAT |
| PWS-f | GGTTATTTTCACTTCCTGTTGATGTAAT |
| Clip 1-f | TGATATTATATCATAAAAACAAACTTAGTTTAAGCAATAGACAAAG |
| Clip 2-f | TTAAGCAATAGACAAAGTTTCTTGGC |
| Clip 3-f | CTAGAATCGTTTTTGTAAACGTTTACTACA |
| Clip 4-f | AAGGAGGAACTTATAATGAATCTCTGG |
| BST-f | ACTTAGTTTAAGCAATAGACAAAGTCTTTTGG |
| DTE-f | GTTTAAGCAATAGACAAAGTCTTTTGGC |
| 1T-f | TCTTAGTTTAAGCAATAGACAAAGATTCTTGGCT |
| 1C-f | CCTTAGTTTAAGCAATAGACAAAGGTTCTTGGCT |
| 1G-f | GCTTAGTTTAAGCAATAGACAAAGCTTCTTGGCT |
| 2T-f | ATTTAGTTTAAGCAATAGACAAAATTTCTTGGCT |
| 2A-f | AATTAGTTTAAGCAATAGACAAATTTTCTTGGCT |
| 2G-f | AGTTAGTTTAAGCAATAGACAAACTTTCTTGGCT |
| 3C-f | ACCTAGTTTAAGCAATAGACAAGGTTTCTTGGCT |
| 3G-f | ACGTAGTTTAAGCAATAGACAACGTTTCTTGGCT |
| 3A-f | ACATAGTTTAAGCAATAGACAATGTTTCTTGGCT |
| 4C-f | ACTCAGTTTAAGCAATAGACAGAGTTTCTTGGCT |
| 4G-f | ACTGAGTTTAAGCAATAGACACAGTTTCTTGGCT |
| 4A-f | ACTAAGTTTAAGCAATAGACATAGTTTCTTGGCT |
| 5C-f | ACTTCGTTTAAGCAATAGACGAAGTTTCTTGGCT |
| 5G-f | ACTTGGTTTAAGCAATAGACCAAGTTTCTTGGCT |
| 5A-f | ACTTTGTTTAAGCAATAGACTAAGTTTCTTGGCT |
| 6T-f | ACTTATTTTAAGCAATAGAAAAAGTTTCTTGGCT |
| 6A-f | ACTTAATTTAAGCAATAGATAAAGTTTCTTGGCT |
| 6C-f | ACTTACTTTAAGCAATAGAGAAAGTTTCTTGGCT |
| 17C-f | ACTTAGTTTAAGCAATACACAAAGTTTCTTGGCT |
| 17A-f | ACTTAGTTTAAGCAATAAACAAAGTTTCTTGGCT |
| 17T-f | ACTTAGTTTAAGCAATATACAAAGTTTCTTGGCT |
| -1T-f | TACTTAGTTTAAGCAATAGACAAAGTATCTTGGCT |
| -1C-f | CACTTAGTTTAAGCAATAGACAAAGTGTCTTGGCT |
| -1G-f | GACTTAGTTTAAGCAATAGACAAAGTCTCTTGGCT |
| -2T-f | TAACTTAGTTTAAGCAATAGACAAAGTTACTTGGC |
| -2C-f | CAACTTAGTTTAAGCAATAGACAAAGTTGCTTGGC |
| -2G-f | GAACTTAGTTTAAGCAATAGACAAAGTTCCTTGGC |
| HapaII-f | GATGGCGCATTGTGACGATCAAGCTTTTTTGAGTGATCTTCTCAAAAAATACTACC |
| HapaII-r | CATATGTAAATCGCTCCTTTTTAGGTGGC |
| P09-f | GATCGTCACAATGCGCCATCA |
| P09-r | ACCATGGATCCGCGACCCATGGATCCCACTTTATGGACGCCG |
| 09xylR-f | GGAGCGATTTACATATGAATGTTACTGGCGATCAGGC |
| 09xylR-r | TATGGAAAAACGCTTTGCCCTTACAAAGATACACGTACACGCCCGAGA |
| Psr23-f | CTTAGTTTAAGCAATAGACAAAGTAATGCAGTAAAAGCGCCTTACGTCAGACAC |
| Psr23-r | CTTTGTCTATTGCTTAAACTAAGTGAAGGCATGTTTCCTCTCTCCCCT |
| Psa24-f | CTTAGTTTAAGCAATAGACAAAGTGTGGGATCCATGGGTCGCGGA |
| Psa24-r | ACTTTGTCTATTGCTTAAACTAAGTTTATGGACGCCGCAGTGTCTGACGTAAG |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Z.; Fang, Y.; Shi, Y.; Xin, Y.; Gu, Z.; Yang, T.; Li, Y.; Ding, Z.; Shi, G.; Zhang, L. Analysis of Xylose Operon from Paenibacillus polymyxa ATCC842 and Development of Tools for Gene Expression. Int. J. Mol. Sci. 2022, 23, 5024. https://doi.org/10.3390/ijms23095024
Wang Z, Fang Y, Shi Y, Xin Y, Gu Z, Yang T, Li Y, Ding Z, Shi G, Zhang L. Analysis of Xylose Operon from Paenibacillus polymyxa ATCC842 and Development of Tools for Gene Expression. International Journal of Molecular Sciences. 2022; 23(9):5024. https://doi.org/10.3390/ijms23095024
Chicago/Turabian StyleWang, Zilong, Yakun Fang, Yi Shi, Yu Xin, ZhengHua Gu, Ting Yang, Youran Li, Zhongyang Ding, Guiyang Shi, and Liang Zhang. 2022. "Analysis of Xylose Operon from Paenibacillus polymyxa ATCC842 and Development of Tools for Gene Expression" International Journal of Molecular Sciences 23, no. 9: 5024. https://doi.org/10.3390/ijms23095024
APA StyleWang, Z., Fang, Y., Shi, Y., Xin, Y., Gu, Z., Yang, T., Li, Y., Ding, Z., Shi, G., & Zhang, L. (2022). Analysis of Xylose Operon from Paenibacillus polymyxa ATCC842 and Development of Tools for Gene Expression. International Journal of Molecular Sciences, 23(9), 5024. https://doi.org/10.3390/ijms23095024







