Molecular Regulation of Cotton Fiber Development: A Review
Abstract
1. Introduction
2. Transcriptional Regulation of Cotton Fiber Development
3. Genetic Control of Cotton Fiber Quality and Yield
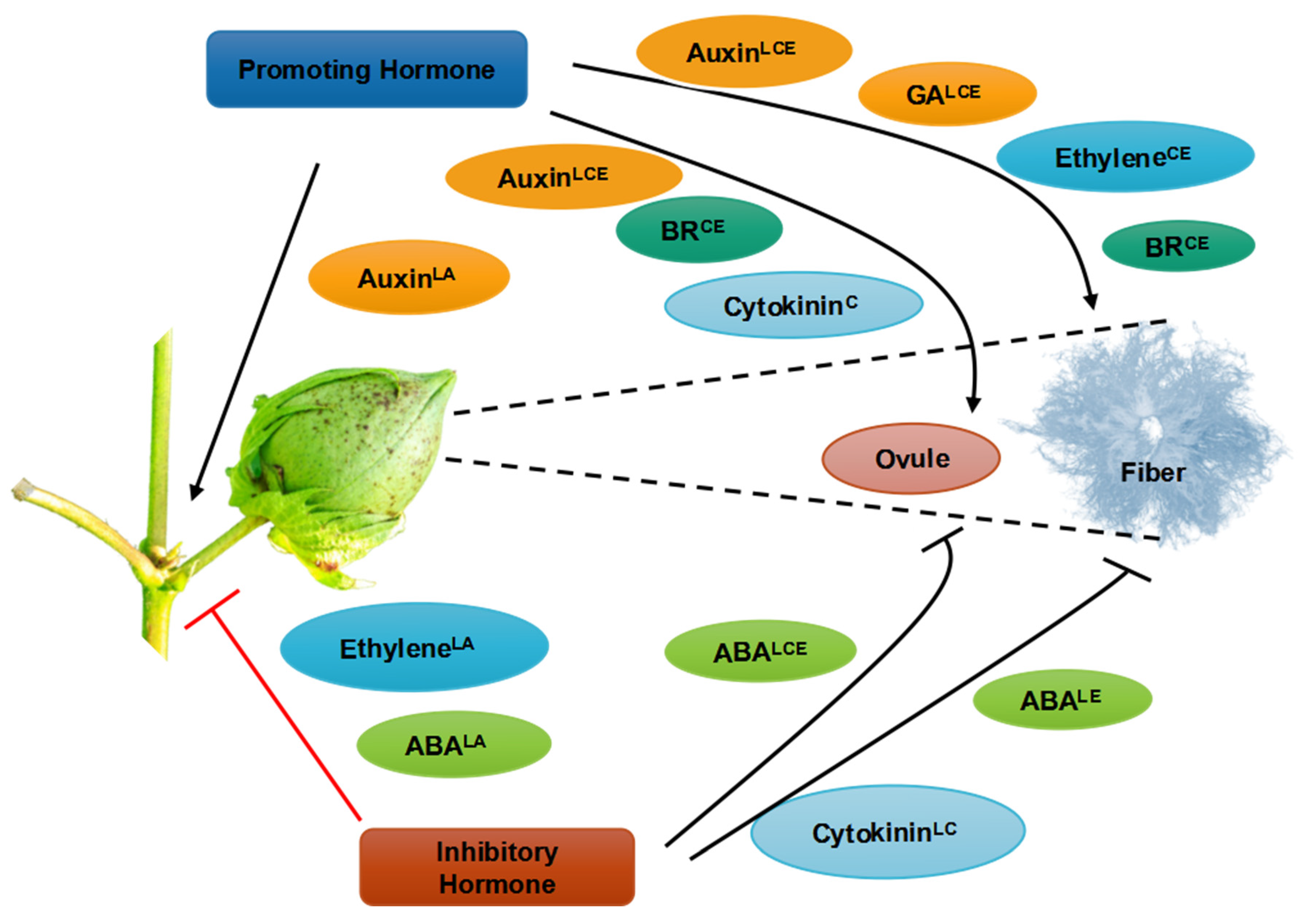
4. Role of Plant Growth Regulators in Cotton Growth and Development
5. Genes Involved in Fiber Development
6. Interactions of Phytohormones with Fiber Development
6.1. Gibberellic Acid
6.2. Jasmonic Acid
6.3. Brassinosteroids
6.4. Auxins
6.5. Ethylene
6.6. Cytokinin and Abscisic Acid
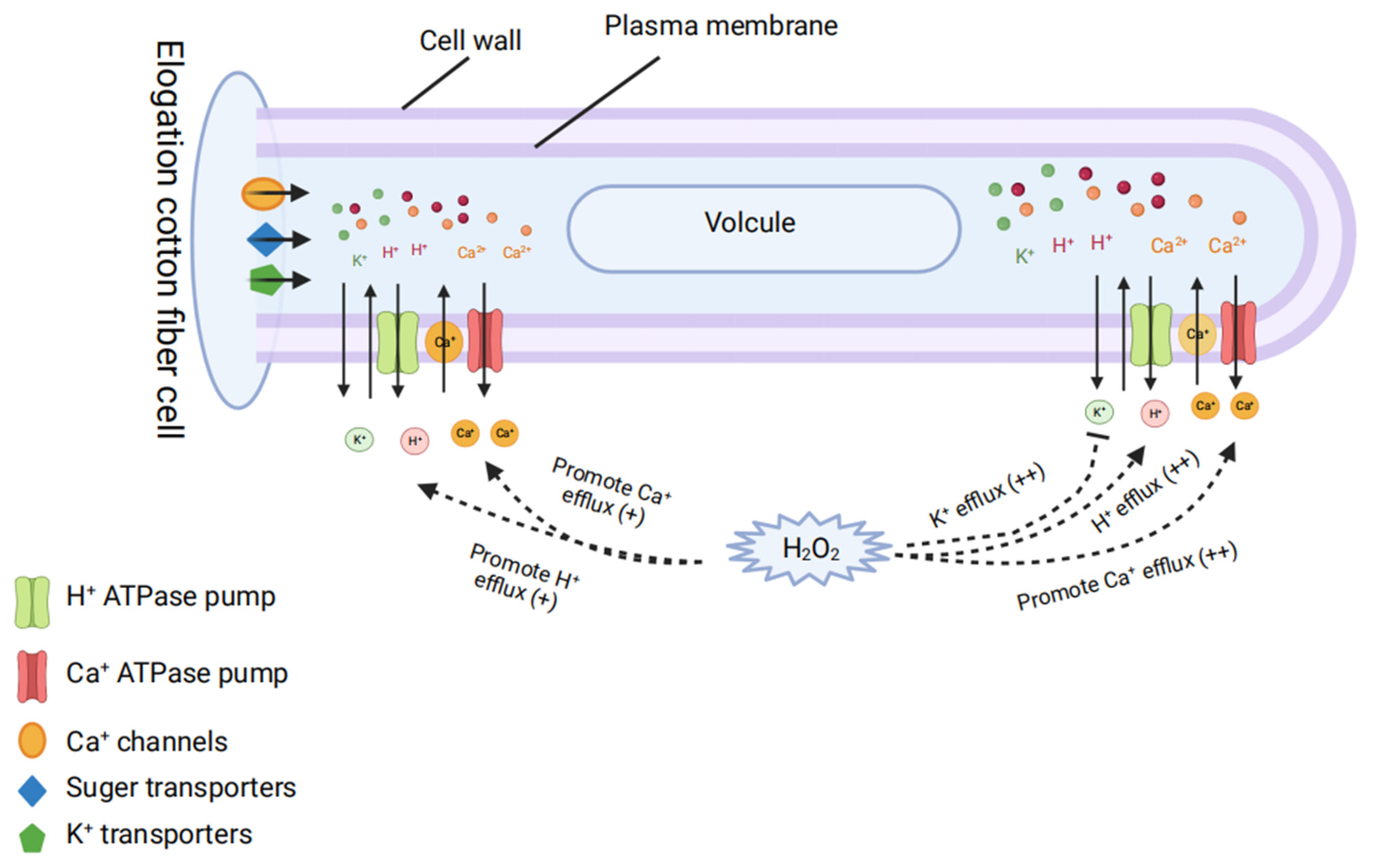
7. Role of Other Factors in Fiber Development
8. Challenges, Conclusions, and Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gao, X.; Guo, H.; Zhang, Q.; Guo, H.; Zhang, L.; Zhang, C.; Gou, Z.; Liu, Y.; Wei, J.; Chen, A.; et al. Arbuscular mycorrhizal fungi (AMF) enhanced the growth, yield, fiber quality and phosphorus regulation in upland cotton (Gossypium hirsutum L.). Sci. Rep. 2020, 10, 2084. [Google Scholar] [CrossRef] [PubMed]
- Leal, A.J.F.; Piati, G.L.; da Costa Leite, R.; Zanella, M.S.; de Souza Osório, C.R.W.; Lima, S.F. Nitrogen and mepiquat chloride can affect fiber quality and cotton yield. Rev. Bras. Eng. Agric. Ambient. 2020, 24, 238–243. [Google Scholar] [CrossRef]
- Zhang, M.; Zheng, X.; Song, S.; Zeng, Q.; Hou, L.; Li, D.; Zhao, J.; Wei, Y.; Li, X.; Luo, M.; et al. Spatiotemporal manipulation of auxin biosynthesis in cotton ovule epidermal cells enhances fiber yield and quality. Nat. Biotechnol. 2011, 29, 453–458. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Gong, J.; Zhang, Z.; Gong, W.; Li, J.; Shi, Y.; Liu, A.; Ge, Q.; Pan, J.; Fan, S.; et al. Identification and analysis of oil candidate genes reveals the molecular basis of cottonseed oil accumulation in Gossypium hirsutum L. Theor. Appl. Genet. 2022, 135, 449–460. [Google Scholar] [CrossRef] [PubMed]
- Abdelrahman, M.; Jogaiah, S.; Burritt, D.J.; Tran, L.-S.P. Legume genetic resources and transcriptome dynamics under abiotic stress conditions. Plant. Cell Environ. 2018, 41, 1972–1983. [Google Scholar] [CrossRef] [PubMed]
- Hinchliffe, D.J.; Meredith, W.R.; Yeater, K.M.; Kim, H.J.; Woodward, A.W.; Chen, Z.J.; Triplett, B.A. Near-isogenic cotton germplasm lines that differ in fiber-bundle strength have temporal differences in fiber gene expression patterns as revealed by comparative high-throughput profiling. Theor. Appl. Genet. 2010, 120, 1347–1366. [Google Scholar] [CrossRef]
- Salih, H.; Leng, X.; He, S.-P.; Jia, Y.; Gong, W.; Du, X.-M. Characterization of the early fiber development gene, Ligon-lintless 1 (Li1), using microarray. Plant Gene 2016, 6, 59–66. [Google Scholar] [CrossRef]
- Pesch, M.; Schultheiß, I.; Digiuni, S.; Uhrig, J.F.; Hülskamp, M. Mutual control of intracellular localisation of the patterning proteins AtMYC1, GL1 and TRY/CPC in Arabidopsis. Development 2013, 140, 3456–3467. [Google Scholar] [CrossRef][Green Version]
- Han, X.; Xu, X.; Fang, D.D.; Zhang, T.; Guo, W. Cloning and expression analysis of novel Aux/IAA family genes in Gossypium hirsutum. Gene 2012, 503, 83–91. [Google Scholar] [CrossRef]
- Niu, X.; Fu, D. The Roles of BLH Transcription Factors in Plant Development and Environmental Response. Int. J. Mol. Sci. 2022, 23, 3731. [Google Scholar] [CrossRef]
- Zang, Y.; Hu, Y.; Dai, F.; Zhang, T. Comparative transcriptome analysis reveals the regulation network for fiber strength in cotton. Biotechnol. Lett. 2022, 44, 547–560. [Google Scholar] [CrossRef] [PubMed]
- Walford, S.-A.; Wu, Y.; Llewellyn, D.J.; Dennis, E.S. GhMYB25-like: A key factor in early cotton fibre development. Plant J. 2011, 65, 785–797. [Google Scholar] [CrossRef] [PubMed]
- Davière, J.-M.; Achard, P. A Pivotal Role of DELLAs in Regulating Multiple Hormone Signals. Mol. Plant 2016, 9, 10–20. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Sun, Y.; Tian, Z.; Fu, G.; Pei, X.; Pan, Z.; Nazir, M.F.; Song, S.; Li, H.; Wang, X.; et al. GhGASA10–1 promotes the cell elongation in fiber development through the phytohormones IAA-induced. BMC Plant Biol. 2021, 21, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Mishra, B.S.; Sharma, M.; Laxmi, A. Role of sugar and auxin crosstalk in plant growth and development. Physiol. Plant. 2022, 174, e13546. [Google Scholar] [CrossRef] [PubMed]
- Beasley, C.A.; Birnbaum, E.H.; Dugger, W.M.; Ting, I.P. A quantitative procedure for estimating cotton fiber growth. Stain Technol. 1974, 49, 85–92. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; He, X.; Tu, L.; Zhu, L.; Zhu, S.; Ge, Z.; Zhang, X. GhJAZ2 negatively regulates cotton fiber initiation by interacting with the R2R3-MYB transcription factor GhMYB25-like. Plant J. 2016, 88, 921–935. [Google Scholar] [CrossRef]
- Manghwar, H.; Hussain, A.; Ali, Q.; Liu, F. Brassinosteroids (BRs) Role in Plant Development and Coping with Different Stresses. Int. J. Mol. Sci. 2022, 23, 1012. [Google Scholar] [CrossRef]
- Xiao, G.; He, P.; Zhao, P.; Liu, H.; Zhang, L.; Pang, C.; Yu, J. Genome-wide identification of the GhARF gene family reveals that GhARF2 and GhARF18 are involved in cotton fibre cell initiation. J. Exp. Bot. 2018, 69, 4323–4337. [Google Scholar] [CrossRef]
- Hande, A.S.; Katageri, I.S.; Jadhav, M.P.; Adiger, S.; Gamanagatti, S.; Padmalatha, K.V.; Dhandapani, G.; Kanakachari, M.; Kumar, P.A.; Reddy, V.S. Transcript profiling of genes expressed during fibre development in diploid cotton (Gossypium arboreum L.). BMC Genomics 2017, 18, 675. [Google Scholar] [CrossRef]
- Wang, M.; Sun, R.; Li, C.; Wang, Q.; Zhang, B. MicroRNA expression profiles during cotton (Gossypium hirsutum L.) fiber early development. Sci. Rep. 2017, 7, 44454. [Google Scholar] [CrossRef] [PubMed]
- Ioannidi, E.; Rigas, S.; Tsitsekian, D.; Daras, G.; Alatzas, A.; Makris, A.; Tanou, G.; Argiriou, A.; Alexandrou, D.; Poethig, S.; et al. Trichome patterning control involves TTG1 interaction with SPL transcription factors. Plant Mol. Biol. 2016, 92, 675–687. [Google Scholar] [CrossRef] [PubMed]
- McKhann, H.I.; Frugier, F.; Petrovics, G.; Coba de la Peña, T.; Jurkevitch, E.; Brown, S.; Kondorosi, E.; Kondorosi, A.; Crespi, M. Cloning of a WD-repeat-containing gene from alfalfa (Medicago sativa): A role in hormone-mediated cell division? Plant Mol. Biol. 1997, 34, 771–780. [Google Scholar] [CrossRef] [PubMed]
- Naoumkina, M.; Thyssen, G.N.; Fang, D.D. RNA-seq analysis of short fiber mutants Ligon-lintless-1 (Li1) and—2 (Li2) revealed important role of aquaporins in cotton (Gossypium hirsutum L.) fiber elongation. BMC Plant Biol. 2015, 15, 65. [Google Scholar] [CrossRef]
- Shi, Y.-H.; Zhu, S.-W.; Mao, X.-Z.; Feng, J.-X.; Qin, Y.-M.; Zhang, L.; Cheng, J.; Wei, L.-P.; Wang, Z.-Y.; Zhu, Y.-X. Transcriptome profiling, molecular biological, and physiological studies reveal a major role for ethylene in cotton fiber cell elongation. Plant Cell 2006, 18, 651–664. [Google Scholar] [CrossRef]
- Qin, Y.-M.; Hu, C.-Y.; Pang, Y.; Kastaniotis, A.J.; Hiltunen, J.K.; Zhu, Y.-X. Saturated Very-Long-Chain Fatty Acids Promote Cotton Fiber and Arabidopsis Cell Elongation by Activating Ethylene Biosynthesis. Plant Cell 2007, 19, 3692–3704. [Google Scholar] [CrossRef]
- Gou, J.Y.; Wang, L.J.; Chen, S.P.; Hu, W.L.; Chen, X.Y. Gene expression and metabolite profiles of cotton fiber during cell elongation and secondary cell wall synthesis. Cell Res. 2007, 17, 422–434. [Google Scholar] [CrossRef]
- Wanjie, S.W.; Welti, R.; Moreau, R.A.; Chapman, K.D. Identification and quantification of glycerolipids in cotton fibers: Reconciliation with metabolic pathway predictions from DNA databases. Lipids 2005, 40, 773–785. [Google Scholar] [CrossRef]
- Wang, Q.Q.; Liu, F.; Chen, X.S.; Ma, X.J.; Zeng, H.Q.; Yang, Z.M. Transcriptome profiling of early developing cotton fiber by deep-sequencing reveals significantly differential expression of genes in a fuzzless/lintless mutant. Genomics 2010, 96, 369–376. [Google Scholar] [CrossRef]
- Pang, C.-Y.; Wang, H.; Pang, Y.; Xu, C.; Jiao, Y.; Qin, Y.-M.; Western, T.L.; Yu, S.-X.; Zhu, Y.-X. Comparative Proteomics Indicates That Biosynthesis of Pectic Precursors Is Important for Cotton Fiber and Arabidopsis Root Hair Elongation. Mol. Cell. Proteomics 2010, 9, 2019–2033. [Google Scholar] [CrossRef]
- Trevisan, R.G.; Vilanova Júnior, N.S.; Eitelwein, M.T.; Molin, J.P. Management of plant growth regulators in cotton using active crop canopy sensors. Agriculture 2018, 8, 101. [Google Scholar] [CrossRef]
- Oosterhuis, D.M.; Kosmidou, K.K.; Cothren, J.T. Managing Cotton Growth and Development with Plant Growth Regulators. In Proceedings of the World Cotton R esearch Conference-2, Athens, Greece, 6–12 September 1998; pp. 46–68. [Google Scholar]
- Dodds, D.; Main, C.; Banks, C.; Barber, L.; Boman, R.; Brown, S.; Edmisten, K.; Faircloth, J.; Jones, M.; Lemon, R.; et al. Utility of Plant Growth Regulation in Cotton Production. Cotton 2010, 8–11. [Google Scholar]
- Sarwar, M.; Saleem, M.F.; Ullah, N.; Rizwan, M.; Ali, S.; Shahid, M.R.; Alamri, S.A.; Alyemeni, M.N.; Ahmad, P. Exogenously applied growth regulators protect the cotton crop from heat-induced injury by modulating plant defense mechanism. Sci. Rep. 2018, 8, 1–15. [Google Scholar] [CrossRef] [PubMed]
- da Costa, V.A.; Cothren, J.T. Cotton Flowering and Fruiting: Control and Modification With Plant Growth Regulators. Cott. Flower. Fruiting Control Modif. Plant Growth Regul. 2012, 79–108. [Google Scholar]
- Zhang, J.; Chen, H.; Wang, H.; Li, B.; Yi, Y.; Kong, F.; Liu, J.; Zhang, H. Constitutive Expression of a Tomato Small Heat Shock Protein Gene LeHSP21 Improves Tolerance to High-Temperature Stress by Enhancing Antioxidation Capacity in Tobacco. Plant Mol. Biol. Rep. 2015, 34, 399–409. [Google Scholar] [CrossRef]
- Sarwar, M.; Saleem, M.F.; Najeeb, U.; Shakeel, A.; Ali, S.; Bilal, M.F. Hydrogen peroxide reduces heat-induced yield losses in cotton (Gossypium hirsutum L.) by protecting cellular membrane damage. J. Agron. Crop Sci. 2017, 203, 429–441. [Google Scholar] [CrossRef]
- Tang, W.; Tu, L.; Yang, X.; Tan, J.; Deng, F.; Hao, J.; Guo, K.; Lindsey, K.; Zhang, X. The calcium sensor GhCaM7 promotes cotton fiber elongation by modulating reactive oxygen species (ROS) production. New Phytol. 2014, 202, 509–520. [Google Scholar] [CrossRef]
- Prasad, P.; Khatoon, U.; Verma, R.K.; Aalam, S.; Kumar, A.; Mohapatra, D.; Bhattacharya, P.; Bag, S.K.; Sawant, S.V. Transcriptional Landscape of Cotton Fiber Development and Its Alliance With Fiber-Associated Traits. Front. Plant Sci. 2022, 13, 1–18. [Google Scholar] [CrossRef]
- Achard, P.; Baghour, M.; Chapple, A.; Hedden, P.; Van Der Straeten, D.; Genschik, P.; Moritz, T.; Harberd, N.P. The plant stress hormone ethylene controls floral transition via DELLA-dependent regulation of floral meristem-identity genes. Proc. Natl. Acad. Sci. USA 2007, 104, 6484–6489. [Google Scholar] [CrossRef]
- Liao, W.; Ruan, M.; Cui, B.; Xu, N.; Lu, J.; Peng, M. Isolation and characterization of a GAI/RGA-like gene from Gossypium hirsutum. Plant Growth Regul. 2009, 58, 35–45. [Google Scholar] [CrossRef]
- Ahmed, M.; Iqbal, A.; Latif, A.; Din, S.U.; Sarwar, M.B.; Wang, X.; Rao, A.Q.; Husnain, T.; Ali Shahid, A. Overexpression of a Sucrose Synthase Gene Indirectly Improves Cotton Fiber Quality Through Sucrose Cleavage. Front. Plant Sci. 2020, 11, 476251. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Zhang, C.; Yang, X.; Liu, K.; Wu, Z.; Zhang, X.; Zheng, W.; Xun, Q.; Liu, C.; Lu, L.; et al. PAG1, a cotton brassinosteroid catabolism gene, modulates fiber elongation. New Phytol. 2014, 203, 437–448. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Dou, L.; Shang, H.; Li, H.; Yu, J.; Xiao, G. GhPIPLC2D promotes cotton fiber elongation by enhancing ethylene biosynthesis. iScience 2021, 24, 102199. [Google Scholar] [CrossRef] [PubMed]
- Samuel Yang, S.; Cheung, F.; Lee, J.J.; Ha, M.; Wei, N.E.; Sze, S.-H.; Stelly, D.M.; Thaxton, P.; Triplett, B.; Town, C.D.; et al. Accumulation of genome-specific transcripts, transcription factors and phytohormonal regulators during early stages of fiber cell development in allotetraploid cotton. Plant J. 2006, 47, 761–775. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Jogaiah, S.; Zhang, W.; Abdelrahman, M.; Fang, J.G. Spatio-temporal expression of miRNA159 family members and their GAMYB target gene during the modulation of gibberellin-induced grapevine parthenocarpy. J. Exp. Bot. 2018, 69, 3639–3650. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Triplett, B.A. Characterization of GhRac1 GTPase expressed in developing cotton (Gossypium hirsutum L.) fibers. Biochim. Biophys. Acta 2004, 1679, 214–221. [Google Scholar] [CrossRef]
- Ji, M.; Sun, K.; Fang, H.; Zhuang, Z.; Chen, H.; Chen, Q.; Cao, Z.; Wang, Y.; Ditta, A.; Khan, M.K.R.; et al. Genome-wide identification and characterization of the CLASP_N gene family in upland cotton (Gossypium hirsutum L.). PeerJ 2022, 9, 1–19. [Google Scholar] [CrossRef]
- Xiao, Y.-H.; Li, D.-M.; Yin, M.-H.; Li, X.-B.; Zhang, M.; Wang, Y.-J.; Dong, J.; Zhao, J.; Luo, M.; Luo, X.-Y.; et al. Gibberellin 20-oxidase promotes initiation and elongation of cotton fibers by regulating gibberellin synthesis. J. Plant Physiol. 2010, 167, 829–837. [Google Scholar] [CrossRef]
- Aleman, L.; Kitamura, J.; Abdel-Mageed, H.; Lee, J.; Sun, Y.; Nakajima, M.; Ueguchi-Tanaka, M.; Matsuoka, M.; Allen, R.D. Functional analysis of cotton orthologs of GA signal transduction factors GID1 and SLR1. Plant Mol. Biol. 2008, 68, 1–16. [Google Scholar] [CrossRef]
- Li, B.; Zhang, L.; Xi, J.; Hou, L.; Fu, X.; Pei, Y.; Zhang, M. An Unexpected Regulatory Sequence from Rho-Related GTPase6 Confers Fiber—Specific Expression in Upland Cotton. Int. J. Mol. Sci. 2022, 23, 1087. [Google Scholar] [CrossRef]
- Xu, S.-M.; Brill, E.; Llewellyn, D.J.; Furbank, R.T.; Ruan, Y.-L. Overexpression of a Potato Sucrose Synthase Gene in Cotton Accelerates Leaf Expansion, Reduces Seed Abortion, and Enhances Fiber Production. Mol. Plant 2012, 5, 430–441. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhu, Y.; Hu, W.; Zhang, X.; Cai, C.; Guo, W. Comparative Transcriptomics Reveals Jasmonic Acid-Associated Metabolism Related to Cotton Fiber Initiation. PLoS ONE 2015, 10, e0129854. [Google Scholar] [CrossRef] [PubMed]
- Demidchik, V.; Shabala, S.; Isayenkov, S.; Cuin, T.A.; Pottosin, I. Calcium transport across plant membranes: Mechanisms and functions. New Phytol. 2018, 220, 49–69. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Fokar, M.; Asami, T.; Yoshida, S.; Allen, R.D. Characterization of the brassinosteroid insensitive 1 genes of cotton. Plant Mol. Biol. 2004, 54, 221–232. [Google Scholar] [CrossRef]
- Sun, Y.; Veerabomma, S.; Abdel-Mageed, H.A.; Fokar, M.; Asami, T.; Yoshida, S.; Allen, R.D. Brassinosteroid Regulates Fiber Development on Cultured Cotton Ovules. Plant Cell Physiol. 2005, 46, 1384–1391. [Google Scholar] [CrossRef]
- Zhang, D.; Zhang, T.; Guo, W. Effect of H2O2 on fiber initiation using fiber retardation initiation mutants in cotton (Gossypium hirsutum). J. Plant Physiol. 2010, 167, 393–399. [Google Scholar] [CrossRef]
- Lu, R.; Zhang, J.; Liu, D.; Wei, Y.-L.; Wang, Y.; Li, X.-B. Characterization of bHLH/HLH genes that are involved in brassinosteroid (BR) signaling in fiber development of cotton (Gossypium hirsutum). BMC Plant Biol. 2018, 18, 304. [Google Scholar] [CrossRef]
- Luo, M.; Xiao, Y.; Li, X.; Lu, X.; Deng, W.; Li, D.; Hou, L.; Hu, M.; Li, Y.; Pei, Y. GhDET2, a steroid 5alpha-reductase, plays an important role in cotton fiber cell initiation and elongation. Plant J. 2007, 51, 419–430. [Google Scholar] [CrossRef]
- Walford, S.-A.; Wu, Y.; Llewellyn, D.J.; Dennis, E.S. Epidermal cell differentiation in cotton mediated by the homeodomain leucine zipper gene, GhHD-1. Plant J. 2012, 71, 464–478. [Google Scholar] [CrossRef]
- Chen, J.G.; Ullah, H.; Young, J.C.; Sussman, M.R.; Jones, A.M. ABP1 is required for organized cell elongation and division in Arabidopsis embryogenesis. Genes Dev. 2001, 15, 902–911. [Google Scholar] [CrossRef]
- Wu, H.; Hazak, O.; Cheung, A.Y.; Yalovsky, S. RAC/ROP GTPases and auxin signaling. Plant Cell 2011, 23, 1208–1218. [Google Scholar] [CrossRef] [PubMed]
- LiI, X.-B.; Xiao, Y.-H.; Luo, M.; Hou, L.; Li, D.-M.; Luo, X.-Y.; Pei, Y. Cloning and expression analysis of two Rac genes from cotton (Gossypium hirsutum L.). Acta Genet. Sin. 2005, 32, 72–78. [Google Scholar] [PubMed]
- Zhang, X.; Wang, L.; Xu, X.; Cai, C.; Guo, W. Genome-wide identification of mitogen-activated protein kinase gene family in Gossypium raimondii and the function of their corresponding orthologs in tetraploid cultivated cotton. BMC Plant Biol. 2014, 14, 345. [Google Scholar] [CrossRef]
- Zhao, J.; Peng, S.; Cui, H.; Li, P.; Li, T.; Liu, L.; Zhang, H.; Tian, Z.; Shang, H.; Xu, R. Dynamic Expression, Differential Regulation and Functional Diversity of the CNGC Family Genes in Cotton. Int. J. Mol. Sci. 2022, 23, 2041. [Google Scholar] [CrossRef]
- Xie, L.; Yang, C.; Wang, X. Brassinosteroids can regulate cellulose biosynthesis by controlling the expression of CESA genes in Arabidopsis. J. Exp. Bot. 2011, 62, 4495–4506. [Google Scholar] [CrossRef]
- Jutras, P.V.; Soldan, R.; Dodds, I.; Schuster, M.; Preston, G.M.; van der Hoorn, R.A.L. AgroLux: Bioluminescent Agrobacterium to improve molecular pharming and study plant immunity. Plant J. 2021, 108, 600–612. [Google Scholar] [CrossRef]
- Sati, D.; Pande, V.; Pandey, S.C.; Samant, M. Recent Advances in PGPR and Molecular Mechanisms Involved in Drought Stress Resistance. J. Soil Sci. Plant Nutr. 2022. [Google Scholar] [CrossRef]
- Zhang, H.; Shao, M.; Qiao, Z.; Yuan, S.; Wang, X.; Hua, S. Effect of phytohormones on fiber initiation of cotton ovule. Acta Physiol. Plant. 2009, 31, 979–986. [Google Scholar] [CrossRef]
- Gilbert, M.K.; Bland, J.M.; Shockey, J.M.; Cao, H.; Hinchliffe, D.J.; Fang, D.D.; Naoumkina, M. A Transcript Profiling Approach Reveals an Abscisic Acid-Specific Glycosyltransferase (UGT73C14) Induced in Developing Fiber of Ligon lintless-2 Mutant of Cotton (Gossypium hirsutum L.). PLoS ONE 2013, 8, e75268. [Google Scholar]
- Zhao, J.; Bai, W.; Zeng, Q.; Song, S.; Zhang, M.; Li, X.; Hou, L.; Xiao, Y.; Luo, M.; Li, D.; et al. Moderately enhancing cytokinin level by down-regulation of GhCKX expression in cotton concurrently increases fiber and seed yield. Mol. Breed. 2015, 35, 60. [Google Scholar] [CrossRef]
- Bose, J.; Pottosin, I.; Shabala, S.; Palmgren, M.; Shabala, S. Calcium Efflux Systems in Stress Signaling and Adaptation in Plants. Front. Plant Sci. 2011, 2, 85. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Han, M.; Zhang, C.; Yao, L.; Sun, J.; Zhang, T. Comparative proteomic analysis reveals the mechanisms governing cotton fiber differentiation and initiation. J. Proteomics 2012, 75, 845–856. [Google Scholar] [CrossRef] [PubMed]
- Mei, W.; Qin, Y.; Song, W.; Li, J.; Zhu, Y. Cotton GhPOX1 encoding plant class III peroxidase may be responsible for the high level of reactive oxygen species production that is related to cotton fiber elongation. J. Genet. Genomics 2009, 36, 141–150. [Google Scholar] [CrossRef]
- Zhao, P.-M.; Wang, L.-L.; Han, L.-B.; Wang, J.; Yao, Y.; Wang, H.-Y.; Du, X.-M.; Luo, Y.-M.; Xia, G.-X. Proteomic Identification of Differentially Expressed Proteins in the Ligon lintless Mutant of Upland Cotton (Gossypium hirsutum L.). J. Proteome Res. 2010, 9, 1076–1087. [Google Scholar] [CrossRef] [PubMed]
- Li, H.-B.; Qin, Y.-M.; Pang, Y.; Song, W.-Q.; Mei, W.-Q.; Zhu, Y.-X. A cotton ascorbate peroxidase is involved in hydrogen peroxide homeostasis during fibre cell development. New Phytol. 2007, 175, 462–471. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Lu, L.; Yang, Z.; Wu, Z.; Qin, W.; Yu, D.; Ren, Z.; Li, Y.; Wang, L.; Li, F.; et al. GhCaM7-like, a calcium sensor gene, influences cotton fiber elongation and biomass production. Plant Physiol. Biochem. PPB 2016, 109, 128–136. [Google Scholar] [CrossRef] [PubMed]
- Huang, Q.-S.; Wang, H.-Y.; Gao, P.; Wang, G.-Y.; Xia, G.-X. Cloning and characterization of a calcium-dependent protein kinase gene associated with cotton fiber development. Plant Cell Rep. 2008, 27, 1869–1875. [Google Scholar] [CrossRef]
- Wang, H.-Y.; Wang, J.; Gao, P.; Jiao, G.-L.; Zhao, P.-M.; Li, Y.; Wang, G.-L.; Xia, G.-X. Down-regulation of GhADF1 gene expression affects cotton fibre properties. Plant Biotechnol. J. 2008, 7, 13–23. [Google Scholar] [CrossRef]
- Prakash, P.; Srivastava, R.; Prasad, P.; Kumar Tiwari, V.; Kumar, A.; Pandey, S.; Sawant, S.V. Trajectories of cotton fiber initiation: A regulatory perspective. Preprints 2020, 1–41. [Google Scholar] [CrossRef]
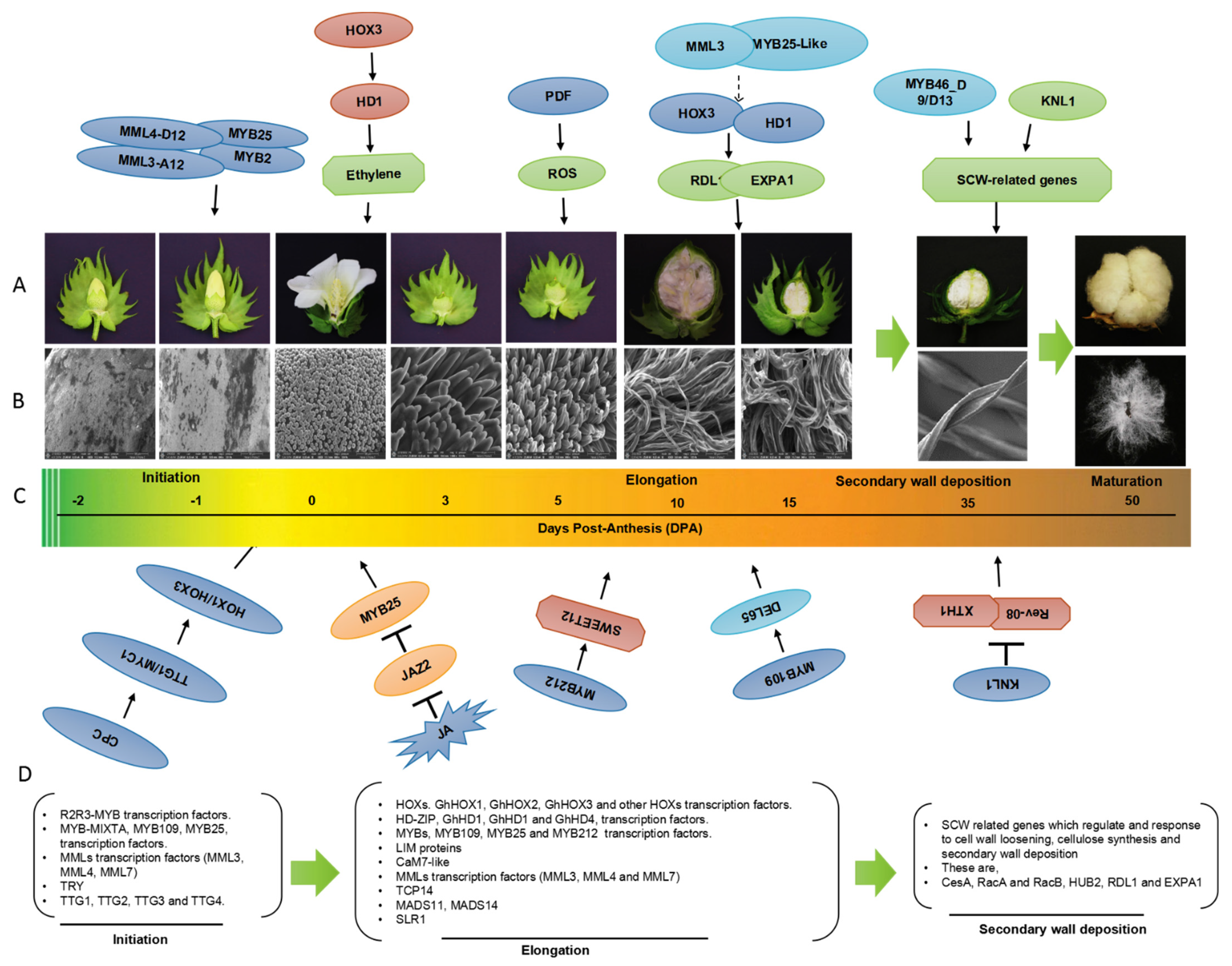


| Gene Name | Gene Family | Function Stage of Fiber Development |
|---|---|---|
| CIPK1 | CBL-interacting protein kinase | Expressed during fiber elongation |
| Exp1 | a-Expansion 1 | Positive role in secondary cell wall deposition |
| ACT1 | Actin1 | Expressed during fiber elongation |
| BG | b-1,4-Glucanase | Positive role in secondary cell wall deposition |
| ManA2 | b-Mannosidase, | Expressed during fiber development in elongation |
| Pel | Pectate lyase | Degradation of de-esterified pectin and has a role in fiber elongation |
| POD2 | Bacterial-induced peroxidase | Expressed during fiber initiation and elongation |
| RacA | Small GTPase | Expressed during fiber elongation |
| RacB | Small GTPase | Expressed during secondary cell wall thickening |
| Sus1 | Sucrose synthase | Expressed in fiber initiation and elongation |
| LTP3 | Lipid transfer protein gene | Cutin synthesis during fiber primary cell wall synthesis |
| 14-3-3L | 14-3-3 | Expressed at early stages of fiber elongation |
| CelA1 | Cellulose synthase | Expressed in secondary cell wall synthesis, involved in the synthesis of cellulose |
| CelA3 | Cellulose synthase | Involved in cellulose biosynthesis stage in developing cotton fibers |
| MYB109 | MYB | Positive in initiation and elongation |
| MYB25 | MYB | Positive role fiber initiation |
| MML3-A12 | MYB | Positive in fuzz fiber initiation |
| MML4-D12 | MYB | Positive in lint fiber initiation |
| MYB212 | MYB | Positive in elongation |
| MYB46-D13 | MYB | Positive role in secondary cell wall deposition |
| MYB46-D9 | MYB | Positive role in secondary cell wall deposition |
| CPC | MYB | Expressed during fiber initiation and negative role in the initiation |
| TRY | MYB | Expressed during fiber initiation and negative role in the initiation |
| HD1 | HD-ZIP IV | Positive in initiation |
| HOX3 | HD-ZIP IV | Positive in initiation |
| TTG1 | WD repeat | Positive in initiation |
| TTG2 | WD repeat | Positive in initiation |
| TTG3 | WD repeat | Positive in initiation |
| TTG4 | WD repeat | Positive in initiation |
| SLR1 | DELLA | Expressed during fiber elongation and negative role in elongation |
| BZR1 | BES1_N | Positive in initiation |
| MADS11 | MADS-box | Positive in elongation |
| MADS14 | MADS-box | Expressed during fiber elongation and negative role in elongation |
| TCP14 | TCP | Positive in initiation and elongation |
| JAZ2 | JASMONATE-ZIN-DOMAIN | Negative in lint and fuzz fiber initiation |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jan, M.; Liu, Z.; Guo, C.; Sun, X. Molecular Regulation of Cotton Fiber Development: A Review. Int. J. Mol. Sci. 2022, 23, 5004. https://doi.org/10.3390/ijms23095004
Jan M, Liu Z, Guo C, Sun X. Molecular Regulation of Cotton Fiber Development: A Review. International Journal of Molecular Sciences. 2022; 23(9):5004. https://doi.org/10.3390/ijms23095004
Chicago/Turabian StyleJan, Masood, Zhixin Liu, Chenxi Guo, and Xuwu Sun. 2022. "Molecular Regulation of Cotton Fiber Development: A Review" International Journal of Molecular Sciences 23, no. 9: 5004. https://doi.org/10.3390/ijms23095004
APA StyleJan, M., Liu, Z., Guo, C., & Sun, X. (2022). Molecular Regulation of Cotton Fiber Development: A Review. International Journal of Molecular Sciences, 23(9), 5004. https://doi.org/10.3390/ijms23095004






